Dynamics of Dual Specificity Phosphatases and Their Interplay with Protein Kinases in Immune Signaling
Abstract
1. Introduction
2. Results
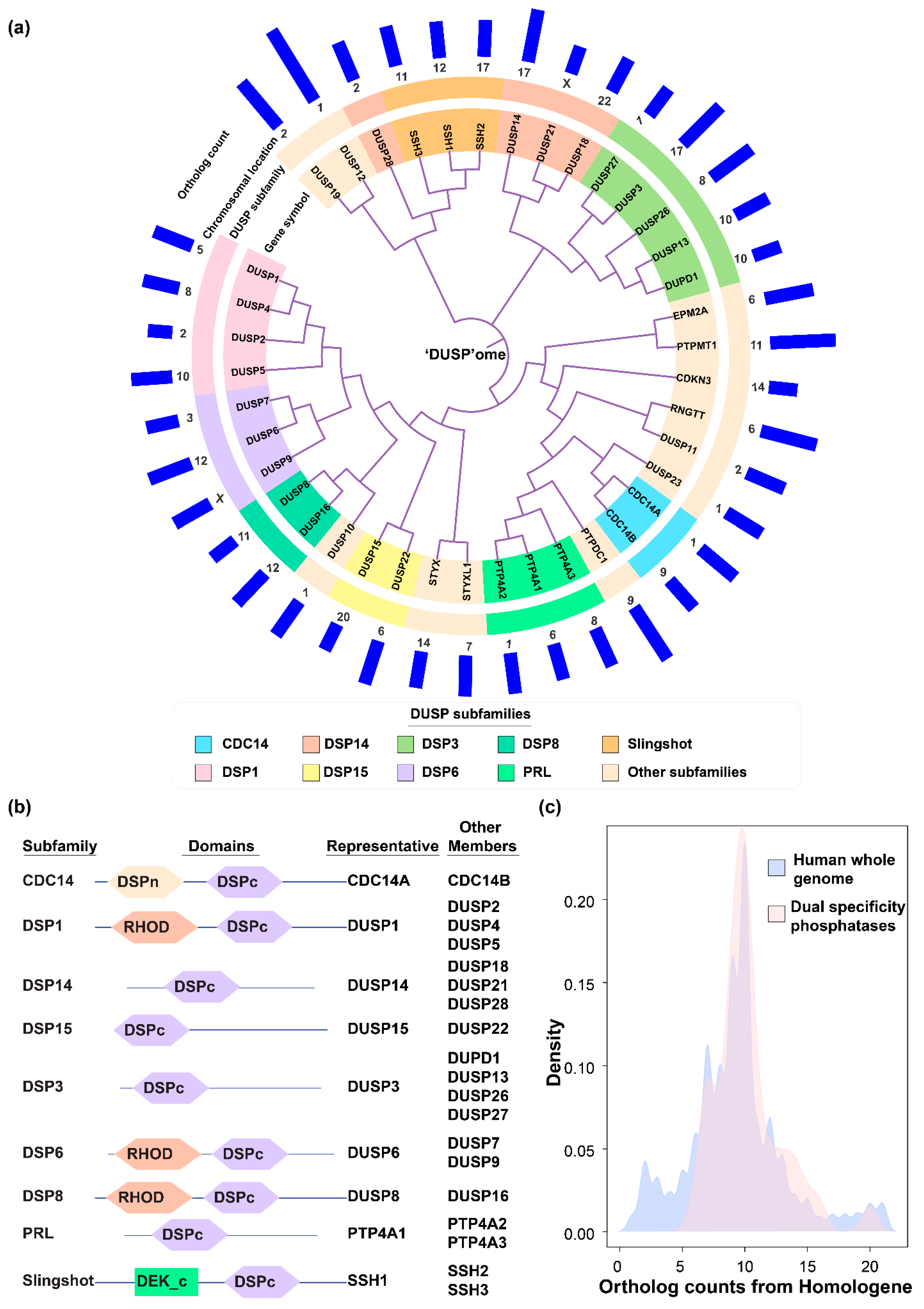
2.1. DUSP Classification into Subfamilies and Evolutionary Conservation
2.2. Expression of Dual Specificity Phosphatases and Protein Kinases in Hematopoietic Cells, Primary and Secondary Lymphoid Organs
2.3. Correlation of Dual Specificity Phosphatase Activity with Protein Kinase Activity in Immune Cells
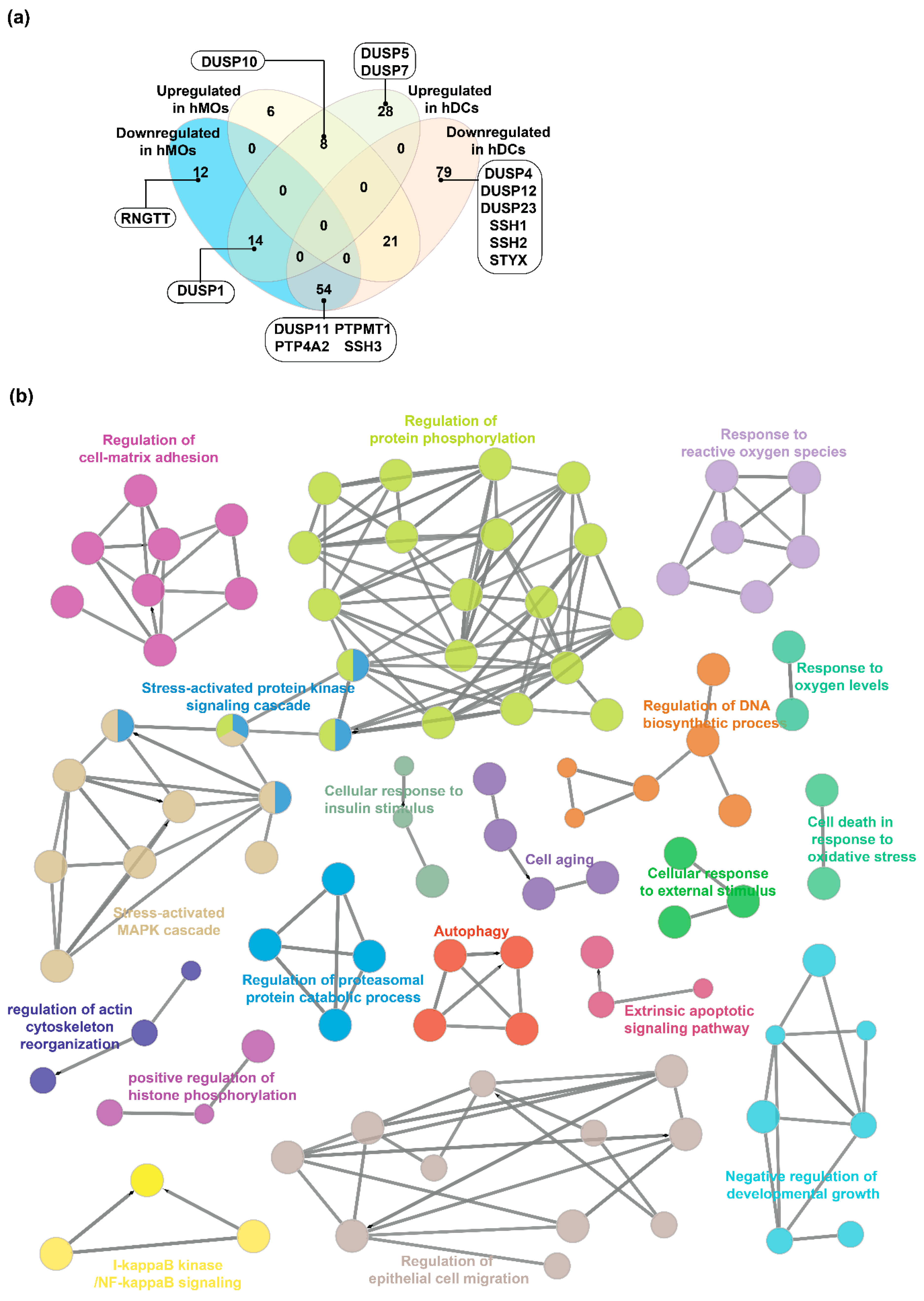
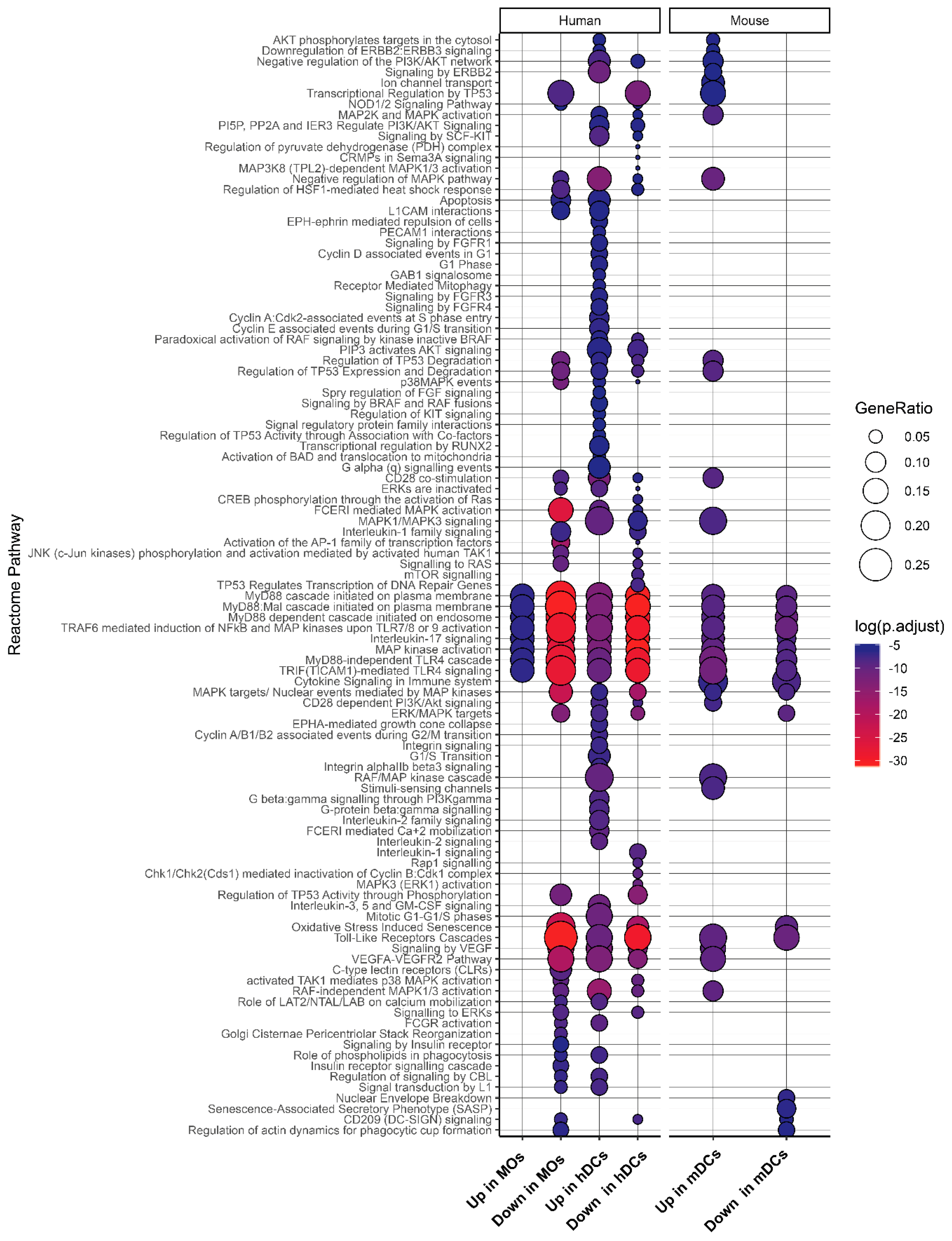
2.4. Expression Landscape and Signaling Dynamics of DUSPs and Kinases in Activated Immune Cells
3. Discussion
4. Materials and Methods
4.1. Datasets
4.2. DUSP and Kinase Lists for Analysis
4.3. Classification of DUSP Family Members, Domain Analysis and Species Conservation Analysis of DUSP Sequences
4.4. Landscape of DUSPs and Kinases in Immune Cells
4.5. Landscape of DUSPs and Kinases in Primary and Secondary Lymphoid Organs
4.6. Baseline DUSP Interactome
4.7. Landscape of DUSPs and Kinases in Activated Immune Cells
4.8. Functional, Pathway and Network Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| DC | Dendritic cells |
| MO | Monocytes |
| LPS | Lipopolysaccharides |
| DUSP | Dual Specificity Phosphatase |
| PK | Protein Kinase |
| GO | Gene Ontology |
References
- Humphrey, S.J.; James, D.E.; Mann, M. Protein Phosphorylation: A Major Switch Mechanism for Metabolic Regulation. Trends Endocrinol. Metab. 2015, 26, 676–687. [Google Scholar] [CrossRef] [PubMed]
- Cohen, P. The origins of protein phosphorylation. Nat. Cell Biol. 2002, 4, E127–E130. [Google Scholar] [CrossRef]
- Ardito, F.; Giuliani, M.; Perrone, D.; Troiano, G.; Lo Muzio, L. The crucial role of protein phosphorylation in cell signaling and its use as targeted therapy (Review). Int. J. Mol. Med. 2017, 40, 271–280. [Google Scholar] [CrossRef] [PubMed]
- Manning, G.; Whyte, D.B.; Martinez, R.; Hunter, T.; Sudarsanam, S. The protein kinase complement of the human genome. Science 2002, 298, 1912–1934. [Google Scholar] [CrossRef]
- Griffith, M.; Griffith, O.L.; Coffman, A.C.; Weible, J.V.; McMichael, J.F.; Spies, N.C.; Koval, J.; Das, I.; Callaway, M.B.; Eldred, J.M.; et al. DGIdb: Mining the druggable genome. Nat. Methods 2013, 10, 1209–1210. [Google Scholar] [CrossRef]
- Alonso, A.; Sasin, J.; Bottini, N.; Friedberg, I.; Friedberg, I.; Osterman, A.; Godzik, A.; Hunter, T.; Dixon, J.; Mustelin, T. Protein tyrosine phosphatases in the human genome. Cell 2004, 117, 699–711. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.J.; Dixon, J.E.; Manning, G. Genomics and evolution of protein phosphatases. Sci. Signal. 2017, 10, 474. [Google Scholar] [CrossRef]
- Lang, R.; Hammer, M.; Mages, J. DUSP meet immunology: dual specificity MAPK phosphatases in control of the inflammatory response. J. Immunol. 2006, 177, 7497–7504. [Google Scholar] [CrossRef]
- Liu, Y.; Shepherd, E.G.; Nelin, L.D. MAPK phosphatases--regulating the immune response. Nat. Rev. Immunol. 2007, 7, 202–212. [Google Scholar] [CrossRef] [PubMed]
- Jeffrey, K.L.; Camps, M.; Rommel, C.; Mackay, C.R. Targeting dual-specificity phosphatases: manipulating MAP kinase signalling and immune responses. Nat. Rev. Drug Discov. 2007, 6, 391–403. [Google Scholar] [CrossRef] [PubMed]
- Abraham, S.M.; Clark, A.R. Dual-specificity phosphatase 1: A critical regulator of innate immune responses. Biochem. Soc. Trans. 2006, 34, 1018–1023. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Reynolds, J.M.; Chang, S.H.; Martin-Orozco, N.; Chung, Y.; Nurieva, R.I.; Dong, C. MKP-1 is necessary for T cell activation and function. J. Biol. Chem. 2009, 284, 30815–30824. [Google Scholar] [CrossRef]
- Korhonen, R.; Huotari, N.; Hommo, T.; Leppanen, T.; Moilanen, E. The expression of interleukin-12 is increased by MAP kinase phosphatase-1 through a mechanism related to interferon regulatory factor 1. Mol. Immunol. 2012, 51, 219–226. [Google Scholar] [CrossRef] [PubMed]
- Hammer, M.; Mages, J.; Dietrich, H.; Servatius, A.; Howells, N.; Cato, A.C.; Lang, R. Dual specificity phosphatase 1 (DUSP1) regulates a subset of LPS-induced genes and protects mice from lethal endotoxin shock. J. Exp. Med. 2006, 203, 15–20. [Google Scholar] [CrossRef] [PubMed]
- Qian, F.; Deng, J.; Cheng, N.; Welch, E.J.; Zhang, Y.; Malik, A.B.; Flavell, R.A.; Dong, C.; Ye, R.D. A non-redundant role for MKP5 in limiting ROS production and preventing LPS-induced vascular injury. EMBO J. 2009, 28, 2896–2907. [Google Scholar] [CrossRef] [PubMed]
- Jeffrey, K.L.; Brummer, T.; Rolph, M.S.; Liu, S.M.; Callejas, N.A.; Grumont, R.J.; Gillieron, C.; Mackay, F.; Grey, S.; Camps, M.; Rommel, C.; et al. Positive regulation of immune cell function and inflammatory responses by phosphatase PAC-1. Nat. Immunol. 2006, 7, 274–283. [Google Scholar] [CrossRef] [PubMed]
- Kovanen, P.E.; Bernard, J.; Al-Shami, A.; Liu, C.; Bollenbacher-Reilley, J.; Young, L.; Pise-Masison, C.; Spolski, R.; Leonard, W.J. T-cell development and function are modulated by dual specificity phosphatase DUSP5. J. Biol. Chem. 2008, 283, 17362–17369. [Google Scholar] [CrossRef]
- Boulding, T.; Wu, F.; McCuaig, R.; Dunn, J.; Sutton, C.R.; Hardy, K.; Tu, W.; Bullman, A.; Yip, D.; Dahlstrom, J.E.; Rao, S. Differential Roles for DUSP Family Members in Epithelial-to-Mesenchymal Transition and Cancer Stem Cell Regulation in Breast Cancer. PLoS ONE 2016, 11, e0148065. [Google Scholar] [CrossRef] [PubMed]
- Kaltenmeier, C.T.; Vollmer, L.L.; Vernetti, L.A.; Caprio, L.; Davis, K.; Korotchenko, V.N.; Day, B.W.; Tsang, M.; Hulkower, K.I.; Lotze, M.T.; Vogt, A. A Tumor Cell-Selective Inhibitor of Mitogen-Activated Protein Kinase Phosphatases Sensitizes Breast Cancer Cells to Lymphokine-Activated Killer Cell Activity. J. Pharmacol. Exp. Ther. 2017, 361, 39–50. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Hyer, J.M.; Yu, H.; D’Silva, N.J.; Kirkwood, K.L. DUSP1 phosphatase regulates the proinflammatory milieu in head and neck squamous cell carcinoma. Cancer Res. 2014, 74, 7191–7197. [Google Scholar] [CrossRef] [PubMed]
- Guo, K.; Li, J.; Tang, J.P.; Tan, C.P.; Hong, C.W.; Al-Aidaroos, A.Q.; Varghese, L.; Huang, C.; Zeng, Q. Targeting intracellular oncoproteins with antibody therapy or vaccination. Sci. Transl. Med. 2011, 3, 99ra85. [Google Scholar] [CrossRef]
- Guo, K.; Tang, J.P.; Tan, C.P.; Wang, H.; Zeng, Q. Monoclonal antibodies target intracellular PRL phosphatases to inhibit cancer metastases in mice. Cancer Biol. Ther. 2008, 7, 750–757. [Google Scholar] [CrossRef] [PubMed]
- Guo, K.; Li, J.; Wang, H.; Osato, M.; Tang, J.P.; Quah, S.Y.; Gan, B.Q.; Zeng, Q. PRL-3 initiates tumor angiogenesis by recruiting endothelial cells in vitro and in vivo. Cancer Res. 2006, 66, 9625–9635. [Google Scholar] [CrossRef]
- Rios, P.; Nunes-Xavier, C.E.; Tabernero, L.; Kohn, M.; Pulido, R. Dual-specificity phosphatases as molecular targets for inhibition in human disease. Antioxid. Redox Signal. 2014, 20, 2251–2273. [Google Scholar] [CrossRef]
- Subbannayya, Y.; Pinto, S.M.; Gowda, H.; Prasad, T.S. Proteogenomics for understanding oncology: recent advances and future prospects. Expert. Rev. Proteomics 2016, 13, 297–308. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.E.; Song, J.; Bosl, K.; Muller, A.C.; Vitko, D.; Bennett, K.L.; Superti-Furga, G.; Pandey, A.; Kandasamy, R.K.; Kim, M.S. Proteogenomic Analysis to Identify Missing Proteins from Haploid Cell Lines. Proteomics 2018, 18, e1700386. [Google Scholar] [CrossRef] [PubMed]
- Pinto, S.M.; Verma, R.; Advani, J.; Chatterjee, O.; Patil, A.H.; Kapoor, S.; Subbannayya, Y.; Raja, R.; Gandotra, S.; Prasad, T.S.K. Integrated Multi-Omic Analysis of Mycobacterium tuberculosis H37Ra Redefines Virulence Attributes. Front. Microbiol. 2018, 9, 1314. [Google Scholar] [CrossRef]
- Kandasamy, R.K.; Vladimer, G.I.; Snijder, B.; Muller, A.C.; Rebsamen, M.; Bigenzahn, J.W.; Moskovskich, A.; Sabler, M.; Stefanovic, A.; Scorzoni, S.; et al. A time-resolved molecular map of the macrophage response to VSV infection. NPJ Syst. Biol. Appl. 2016, 2, 16027. [Google Scholar] [CrossRef] [PubMed]
- Rieckmann, J.C.; Geiger, R.; Hornburg, D.; Wolf, T.; Kveler, K.; Jarrossay, D.; Sallusto, F.; Shen-Orr, S.S.; Lanzavecchia, A.; Mann, M.; et al. Social network architecture of human immune cells unveiled by quantitative proteomics. Nat. Immunol. 2017, 18, 583–593. [Google Scholar] [CrossRef] [PubMed]
- Chevrier, N.; Mertins, P.; Artyomov, M.N.; Shalek, A.K.; Iannacone, M.; Ciaccio, M.F.; Gat-Viks, I.; Tonti, E.; DeGrace, M.M.; Clauser, K.R.; et al. Systematic discovery of TLR signaling components delineates viral-sensing circuits. Cell 2011, 147, 853–867. [Google Scholar] [CrossRef]
- Elpek, K.G.; Cremasco, V.; Shen, H.; Harvey, C.J.; Wucherpfennig, K.W.; Goldstein, D.R.; Monach, P.A.; Turley, S.J. The tumor microenvironment shapes lineage, transcriptional, and functional diversity of infiltrating myeloid cells. Cancer Immunol. Res. 2014, 2, 655–667. [Google Scholar] [CrossRef]
- Gat-Viks, I.; Chevrier, N.; Wilentzik, R.; Eisenhaure, T.; Raychowdhury, R.; Steuerman, Y.; Shalek, A.K.; Hacohen, N.; Amit, I.; Regev, A. Deciphering molecular circuits from genetic variation underlying transcriptional responsiveness to stimuli. Nat. Biotechnol. 2013, 31, 342–349. [Google Scholar] [CrossRef][Green Version]
- Jovanovic, M.; Rooney, M.S.; Mertins, P.; Przybylski, D.; Chevrier, N.; Satija, R.; Rodriguez, E.H.; Fields, A.P.; Schwartz, S.; Raychowdhury, R.; et al. Immunogenetics. Dynamic profiling of the protein life cycle in response to pathogens. Science 2015, 347, 1259038. [Google Scholar] [CrossRef] [PubMed]
- Kamal, A.H.M.; Fessler, M.B.; Chowdhury, S.M. Comparative and network-based proteomic analysis of low dose ethanol- and lipopolysaccharide-induced macrophages. PLoS ONE 2018, 13, e0193104. [Google Scholar] [CrossRef] [PubMed]
- Mertins, P.; Przybylski, D.; Yosef, N.; Qiao, J.; Clauser, K.; Raychowdhury, R.; Eisenhaure, T.M.; Maritzen, T.; Haucke, V.; Satoh, T.; et al. An Integrative Framework Reveals Signaling-to-Transcription Events in Toll-like Receptor Signaling. Cell Rep. 2017, 19, 2853–2866. [Google Scholar] [CrossRef] [PubMed]
- Mingueneau, M.; Kreslavsky, T.; Gray, D.; Heng, T.; Cruse, R.; Ericson, J.; Bendall, S.; Spitzer, M.H.; Nolan, G.P.; Kobayashi, K.; et al. The transcriptional landscape of alphabeta T cell differentiation. Nat. Immunol. 2013, 14, 619–632. [Google Scholar] [CrossRef] [PubMed]
- Muller, M.M.; Lehmann, R.; Klassert, T.E.; Reifenstein, S.; Conrad, T.; Moore, C.; Kuhn, A.; Behnert, A.; Guthke, R.; Driesch, D.; et al. Global analysis of glycoproteins identifies markers of endotoxin tolerant monocytes and GPR84 as a modulator of TNFalpha expression. Sci. Rep. 2017, 7, 838. [Google Scholar] [CrossRef] [PubMed]
- Shalek, A.K.; Satija, R.; Adiconis, X.; Gertner, R.S.; Gaublomme, J.T.; Raychowdhury, R.; Schwartz, S.; Yosef, N.; Malboeuf, C.; Lu, D.; et al. Single-cell transcriptomics reveals bimodality in expression and splicing in immune cells. Nature 2013, 498, 236–240. [Google Scholar] [CrossRef] [PubMed]
- Coordinators, N.R. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2016, 44, D7–D19. [Google Scholar]
- Arthur, J.S.; Ley, S.C. Mitogen-activated protein kinases in innate immunity. Nat. Rev. Immunol. 2013, 13, 679–692. [Google Scholar] [CrossRef]
- Yuan, X.; Wu, H.; Bu, H.; Zhou, J.; Zhang, H. Targeting the immunity protein kinases for immuno-oncology. Eur. J. Med. Chem. 2019, 163, 413–427. [Google Scholar] [CrossRef]
- Michel, T.; Poli, A.; Cuapio, A.; Briquemont, B.; Iserentant, G.; Ollert, M.; Zimmer, J. Human CD56bright NK Cells: An Update. J. Immunol. 2016, 196, 2923–2931. [Google Scholar] [CrossRef] [PubMed]
- Tucker, K.L.; Sage, T.; Stevens, J.M.; Jordan, P.A.; Jones, S.; Barrett, N.E.; St-Arnaud, R.; Frampton, J.; Dedhar, S.; Gibbins, J.M. A dual role for integrin-linked kinase in platelets: regulating integrin function and alpha-granule secretion. Blood 2008, 112, 4523–4531. [Google Scholar] [CrossRef]
- Bhattacharjee, A.; Pal, S.; Feldman, G.M.; Cathcart, M.K. Hck is a key regulator of gene expression in alternatively activated human monocytes. J. Biol. Chem. 2011, 286, 36709–36723. [Google Scholar] [CrossRef] [PubMed]
- Boehm, T.; Hess, I.; Swann, J.B. Evolution of lymphoid tissues. Trends Immunol. 2012, 33, 315–321. [Google Scholar] [CrossRef] [PubMed]
- Pabst, R. Plasticity and heterogeneity of lymphoid organs. What are the criteria to call a lymphoid organ primary, secondary or tertiary? Immunol. Lett. 2007, 112, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Girard-Madoux, M.J.H.; Gomez de Aguero, M.; Ganal-Vonarburg, S.C.; Mooser, C.; Belz, G.T.; Macpherson, A.J.; Vivier, E. The immunological functions of the Appendix: An example of redundancy? Semin Immunol. 2018, 36, 31–44. [Google Scholar] [CrossRef]
- Kooij, I.A.; Sahami, S.; Meijer, S.L.; Buskens, C.J.; Te Velde, A.A. The immunology of the vermiform appendix: A review of the literature. Clin. Exp. Immunol. 2016, 186, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Kawaji, H.; Kasukawa, T.; Forrest, A.; Carninci, P.; Hayashizaki, Y. The FANTOM5 collection, a data series underpinning mammalian transcriptome atlases in diverse cell types. Sci. Data 2017, 4, 170113. [Google Scholar] [CrossRef] [PubMed]
- Consortium, G.T. Human genomics. The Genotype-Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science 2015, 348, 648–660. [Google Scholar] [CrossRef] [PubMed]
- Uhlen, M.; Zhang, C.; Lee, S.; Sjostedt, E.; Fagerberg, L.; Bidkhori, G.; Benfeitas, R.; Arif, M.; Liu, Z.; Edfors, F.; et al. A pathology atlas of the human cancer transcriptome. Science 2017, 357, 6352. [Google Scholar] [CrossRef] [PubMed]
- Uhlen, M.; Fagerberg, L.; Hallstrom, B.M.; Lindskog, C.; Oksvold, P.; Mardinoglu, A.; Sivertsson, A.; Kampf, C.; Sjostedt, E.; Asplund, A.; et al. Proteomics. Tissue-based map of the human proteome. Science 2015, 347, 1260419. [Google Scholar] [CrossRef]
- Barbosa-Morais, N.L.; Irimia, M.; Pan, Q.; Xiong, H.Y.; Gueroussov, S.; Lee, L.J.; Slobodeniuc, V.; Kutter, C.; Watt, S.; Colak, R.; et al. The evolutionary landscape of alternative splicing in vertebrate species. Science 2012, 338, 1587–1593. [Google Scholar] [CrossRef] [PubMed]
- Roadmap Epigenomics Consortium; Kundaje, A.; Meuleman, W.; Ernst, J.; Bilenky, M.; Yen, A.; Heravi-Moussavi, A.; Kheradpour, P.; Zhang, Z.; Wang, J.; et al. Integrative analysis of 111 reference human epigenomes. Nature 2015, 518, 317–330. [Google Scholar] [CrossRef] [PubMed]
- Lin, S.; Lin, Y.; Nery, J.R.; Urich, M.A.; Breschi, A.; Davis, C.A.; Dobin, A.; Zaleski, C.; Beer, M.A.; Chapman, W.C.; et al. Comparison of the transcriptional landscapes between human and mouse tissues. Proc. Natl. Acad. Sci. USA 2014, 111, 17224–17229. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.H.; Xiang, J.; Yan, M.; Zhang, Y.; Zhao, Y.; Yue, C.F.; Xu, J.; Zheng, F.M.; Chen, J.N.; Kang, Z.; et al. The mitotic kinase Aurora-A induces mammary cell migration and breast cancer metastasis by activating the Cofilin-F-actin pathway. Cancer Res. 2010, 70, 9118–9128. [Google Scholar] [CrossRef] [PubMed]
- Lopes, L.J.S.; Tesser-Gamba, F.; Petrilli, A.S.; de Seixas Alves, M.T.; Garcia-Filho, R.J.; Toledo, S.R.C. MAPK pathways regulation by DUSP1 in the development of osteosarcoma: Potential markers and therapeutic targets. Mol. Carcinog. 2017, 56, 1630–1641. [Google Scholar] [CrossRef] [PubMed]
- Theodosiou, A.; Smith, A.; Gillieron, C.; Arkinstall, S.; Ashworth, A. MKP5, a new member of the MAP kinase phosphatase family, which selectively dephosphorylates stress-activated kinases. Oncogene 1999, 18, 6981–6988. [Google Scholar] [CrossRef]
- Matsuguchi, T.; Musikacharoen, T.; Johnson, T.R.; Kraft, A.S.; Yoshikai, Y. A novel mitogen-activated protein kinase phosphatase is an important negative regulator of lipopolysaccharide-mediated c-Jun N-terminal kinase activation in mouse macrophage cell lines. Mol. Cell. Biol. 2001, 21, 6999–7009. [Google Scholar] [CrossRef] [PubMed]
- Bosl, K.; Giambelluca, M.; Haug, M.; Bugge, M.; Espevik, T.; Kandasamy, R.K.; Bergstrom, B. Coactivation of TLR2 and TLR8 in Primary Human Monocytes Triggers a Distinct Inflammatory Signaling Response. Front. Physiol. 2018, 9, 618. [Google Scholar] [CrossRef] [PubMed]
- Amit, I.; Garber, M.; Chevrier, N.; Leite, A.P.; Donner, Y.; Eisenhaure, T.; Guttman, M.; Grenier, J.K.; Li, W.; Zuk, O.; et al. Unbiased reconstruction of a mammalian transcriptional network mediating pathogen responses. Science 2009, 326, 257–263. [Google Scholar] [CrossRef] [PubMed]
- Vijayan, V.; Pradhan, P.; Braud, L.; Fuchs, H.R.; Gueler, F.; Motterlini, R.; Foresti, R.; Immenschuh, S. Human and murine macrophages exhibit differential metabolic responses to lipopolysaccharide—A divergent role for glycolysis. Redox Biol. 2019, 22, 101147. [Google Scholar] [CrossRef] [PubMed]
- Sakuta, T.; Matsushita, K.; Yamaguchi, N.; Oyama, T.; Motani, R.; Koga, T.; Nagaoka, S.; Abeyama, K.; Maruyama, I.; Takada, H.; et al. Enhanced production of vascular endothelial growth factor by human monocytic cells stimulated with endotoxin through transcription factor SP-1. J. Med. Microbiol. 2001, 50, 233–237. [Google Scholar] [CrossRef][Green Version]
- Huang, C.Y.; Tan, T.H. DUSPs, to MAP kinases and beyond. Cell Biosci. 2012, 2, 24. [Google Scholar] [CrossRef] [PubMed]
- Bhore, N.; Wang, B.J.; Chen, Y.W.; Liao, Y.F. Critical Roles of Dual-Specificity Phosphatases in Neuronal Proteostasis and Neurological Diseases. Int. J. Mol. Sci. 2017, 18, 1963. [Google Scholar] [CrossRef] [PubMed]
- Alonso, A.; Pulido, R. The extended human PTPome: A growing tyrosine phosphatase family. FEBS J. 2016, 283, 1404–1429. [Google Scholar] [CrossRef] [PubMed]
- Hatzihristidis, T.; Liu, S.; Pryszcz, L.; Hutchins, A.P.; Gabaldon, T.; Tremblay, M.L.; Miranda-Saavedra, D. PTP-central: A comprehensive resource of protein tyrosine phosphatases in eukaryotic genomes. Methods 2014, 65, 156–164. [Google Scholar] [CrossRef] [PubMed]
- Rios, P.; Li, X.; Kohn, M. Molecular mechanisms of the PRL phosphatases. FEBS J. 2013, 280, 505–524. [Google Scholar] [CrossRef]
- Huang, L.C.; Ross, K.E.; Baffi, T.R.; Drabkin, H.; Kochut, K.J.; Ruan, Z.; D’Eustachio, P.; McSkimming, D.; Arighi, C.; Chen, C.; et al. Integrative annotation and knowledge discovery of kinase post-translational modifications and cancer-associated mutations through federated protein ontologies and resources. Sci. Rep. 2018, 8, 6518. [Google Scholar] [CrossRef]
- Pula, G.; Schuh, K.; Nakayama, K.; Nakayama, K.I.; Walter, U.; Poole, A.W. PKCdelta regulates collagen-induced platelet aggregation through inhibition of VASP-mediated filopodia formation. Blood 2006, 108, 4035–4044. [Google Scholar] [CrossRef]
- Meinders, M.; Kulu, D.I.; van de Werken, H.J.; Hoogenboezem, M.; Janssen, H.; Brouwer, R.W.; van Ijcken, W.F.; Rijkers, E.J.; Demmers, J.A.; Kruger, I.; et al. Sp1/Sp3 transcription factors regulate hallmarks of megakaryocyte maturation and platelet formation and function. Blood 2015, 125, 1957–1967. [Google Scholar] [CrossRef]
- Hitchcock, I.S.; Fox, N.E.; Prevost, N.; Sear, K.; Shattil, S.J.; Kaushansky, K. Roles of focal adhesion kinase (FAK) in megakaryopoiesis and platelet function: Studies using a megakaryocyte lineage specific FAK knockout. Blood 2008, 111, 596–604. [Google Scholar] [CrossRef][Green Version]
- Wu, J.; Mei, C.; Vlassara, H.; Striker, G.E.; Zheng, F. Oxidative stress-induced JNK activation contributes to proinflammatory phenotype of aging diabetic mesangial cells. Am. J. Physiol. Renal. Physiol. 2009, 297, F1622–F1631. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Blattman, J.N.; Kennedy, N.J.; Duong, J.; Nguyen, T.; Wang, Y.; Davis, R.J.; Greenberg, P.D.; Flavell, R.A.; Dong, C. Regulation of innate and adaptive immune responses by MAP kinase phosphatase 5. Nature 2004, 430, 793–797. [Google Scholar] [CrossRef]
- Hammer, M.; Mages, J.; Dietrich, H.; Schmitz, F.; Striebel, F.; Murray, P.J.; Wagner, H.; Lang, R. Control of dual-specificity phosphatase-1 expression in activated macrophages by IL-10. Eur. J. Immunol. 2005, 35, 2991–3001. [Google Scholar] [CrossRef] [PubMed]
- Nizamutdinova, I.T.; Kim, Y.M.; Lee, J.H.; Chang, K.C.; Kim, H.J. MKP-7, a negative regulator of JNK, regulates VCAM-1 expression through IRF-1. Cell Signal. 2012, 24, 866–872. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Nallaparaju, K.C.; Liu, X.; Jiao, H.; Reynolds, J.M.; Wang, Z.X.; Dong, C. MAPK phosphatase 7 regulates T cell differentiation via inhibiting ERK-mediated IL-2 expression. J. Immunol. 2015, 194, 3088–3095. [Google Scholar] [CrossRef] [PubMed]
- Niedzielska, M.; Bodendorfer, B.; Munch, S.; Eichner, A.; Derigs, M.; da Costa, O.; Schweizer, A.; Neff, F.; Nitschke, L.; Sparwasser, T.; et al. Gene trap mice reveal an essential function of dual specificity phosphatase Dusp16/MKP-7 in perinatal survival and regulation of Toll-like receptor (TLR)-induced cytokine production. J. Biol. Chem. 2014, 289, 2112–2126. [Google Scholar] [CrossRef]
- Peng, H.Z.; Yun, Z.; Wang, W.; Ma, B.A. Dual specificity phosphatase 1 has a protective role in osteoarthritis fibroblastlike synoviocytes via inhibition of the MAPK signaling pathway. Mol. Med. Rep. 2017, 16, 8441–8447. [Google Scholar] [CrossRef] [PubMed]
- Aggelou, H.; Chadla, P.; Nikou, S.; Karteri, S.; Maroulis, I.; Kalofonos, H.P.; Papadaki, H.; Bravou, V. LIMK/cofilin pathway and Slingshot are implicated in human colorectal cancer progression and chemoresistance. Virchows Arch. 2018, 472, 727–737. [Google Scholar] [CrossRef] [PubMed]
- Maimaiti, Y.; Maimaitiming, M.; Li, Y.; Aibibula, S.; Ainiwaer, A.; Aili, A.; Sun, Z.; Abudureyimu, K. SSH1 expression is associated with gastric cancer progression and predicts a poor prognosis. BMC Gastroenterol. 2018, 18, 12. [Google Scholar] [CrossRef]
- Caution, K.; Gavrilin, M.A.; Tazi, M.; Kanneganti, A.; Layman, D.; Hoque, S.; Krause, K.; Amer, A.O. Caspase-11 and caspase-1 differentially modulate actin polymerization via RhoA and Slingshot proteins to promote bacterial clearance. Sci. Rep. 2015, 5, 18479. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.T.; Chang, M.C.; Chen, Y.L.; Chen, T.C.; Chen, C.A.; Cheng, W.F. Insulin-like growth factors inhibit dendritic cell-mediated anti-tumor immunity through regulating ERK1/2 phosphorylation and p38 dephosphorylation. Cancer Lett. 2015, 359, 117–126. [Google Scholar] [CrossRef] [PubMed]
- Spadaro, O.; Camell, C.D.; Bosurgi, L.; Nguyen, K.Y.; Youm, Y.H.; Rothlin, C.V.; Dixit, V.D. IGF1 Shapes Macrophage Activation in Response to Immunometabolic Challenge. Cell Rep. 2017, 19, 225–234. [Google Scholar] [CrossRef] [PubMed]
- Cook, D.A.; Kannarkat, G.T.; Cintron, A.F.; Butkovich, L.M.; Fraser, K.B.; Chang, J.; Grigoryan, N.; Factor, S.A.; West, A.B.; Boss, J.M.; et al. LRRK2 levels in immune cells are increased in Parkinson’s disease. NPJ Parkinsons Dis. 2017, 3, 11. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.; James, W.S.; Cowley, S.A. LRRK2 in peripheral and central nervous system innate immunity: its link to Parkinson’s disease. Biochem. Soc. Trans. 2017, 45, 131–139. [Google Scholar] [CrossRef] [PubMed]
- Moehle, M.S.; Webber, P.J.; Tse, T.; Sukar, N.; Standaert, D.G.; DeSilva, T.M.; Cowell, R.M.; West, A.B. LRRK2 inhibition attenuates microglial inflammatory responses. J. Neurosci. 2012, 32, 1602–1611. [Google Scholar] [CrossRef] [PubMed]
- Miranda-Saavedra, D.; Barton, G.J. Classification and functional annotation of eukaryotic protein kinases. Proteins 2007, 68, 893–914. [Google Scholar] [CrossRef]
- Hunter, T. Signaling—2000 and beyond. Cell 2000, 100, 113–127. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. 20 years of the SMART protein domain annotation resource. Nucleic Acids Res. 2018, 46, D493–D496. [Google Scholar] [CrossRef] [PubMed]
- Mudunuri, U.; Che, A.; Yi, M.; Stephens, R.M. bioDBnet: the biological database network. Bioinformatics 2009, 25, 555–556. [Google Scholar] [CrossRef] [PubMed]
- Reimand, J.; Arak, T.; Adler, P.; Kolberg, L.; Reisberg, S.; Peterson, H.; Vilo, J. g:Profiler-a web server for functional interpretation of gene lists (2016 update). Nucleic Acids Res. 2016, 44, W83–W89. [Google Scholar] [CrossRef] [PubMed]
- Veres, D.V.; Gyurko, D.M.; Thaler, B.; Szalay, K.Z.; Fazekas, D.; Korcsmaros, T.; Csermely, P. ComPPI: A cellular compartment-specific database for protein-protein interaction network analysis. Nucleic Acids Res. 2015, 43, D485–D493. [Google Scholar] [CrossRef] [PubMed]
- Assenov, Y.; Ramirez, F.; Schelhorn, S.E.; Lengauer, T.; Albrecht, M. Computing topological parameters of biological networks. Bioinformatics 2008, 24, 282–284. [Google Scholar] [CrossRef] [PubMed]
- Pavlopoulos, G.A.; Secrier, M.; Moschopoulos, C.N.; Soldatos, T.G.; Kossida, S.; Aerts, J.; Schneider, R.; Bagos, P.G. Using graph theory to analyze biological networks. BioData Min. 2011, 4, 10. [Google Scholar] [CrossRef]
- Bindea, G.; Mlecnik, B.; Hackl, H.; Charoentong, P.; Tosolini, M.; Kirilovsky, A.; Fridman, W.H.; Pages, F.; Trajanoski, Z.; Galon, J. ClueGO: A Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics 2009, 25, 1091–1093. [Google Scholar] [CrossRef]
- Fabregat, A.; Jupe, S.; Matthews, L.; Sidiropoulos, K.; Gillespie, M.; Garapati, P.; Haw, R.; Jassal, B.; Korninger, F.; May, B.; et al. The Reactome Pathway Knowledgebase. Nucleic Acids Res. 2018, 46, D649–D655. [Google Scholar] [CrossRef] [PubMed]
- Huber, W.; Carey, V.J.; Gentleman, R.; Anders, S.; Carlson, M.; Carvalho, B.S.; Bravo, H.C.; Davis, S.; Gatto, L.; Girke, T.; et al. Orchestrating high-throughput genomic analysis with Bioconductor. Nat. Methods 2015, 12, 115–121. [Google Scholar] [CrossRef] [PubMed]
- Yu, G.; Wang, L.G.; Han, Y.; He, Q.Y. clusterProfiler: An R package for comparing biological themes among gene clusters. OMICS 2012, 16, 284–287. [Google Scholar] [CrossRef] [PubMed]


| DUSP-Protein Kinase Pair (Spearman’s Rank Correlation Coefficient ρ) | Relationship | Known/Novel |
|---|---|---|
| SSH1-AURKA (0.94) | Aurora Kinase A (AURKA) overexpression increased slingshot kinase 1 (SSH1) expression in breast cancer cells [56] | Known |
| DUSP1-MAPK7 (0.79) | DUSP1 gene silencing increased MAPK7 expression in osteosarcoma cells [57] | Known |
| DUSP1-MAPK8 (0.82) | DUSP1 gene silencing increased MAPK8 expression in osteosarcoma cells [57] | Known |
| DUSP10-MAPK8 (0.81) | DUSP10 (MKP-5) dephosphorylates MAPK8 (JNK) [58] | Known |
| Dusp16-Mapk8 (0.83) | DUSP-16 (MKP-7) regulates MAPK8 (JNK) in LPS-activated macrophages [59] | Known |
| SSH1-CDK17(0.94) | Novel | |
| DUSP12-CLK3(0.97) | Novel | |
| SSH1-LMTK2 (0.97) | Novel | |
| DUSP12-AURKA (0.96) | Novel | |
| Ptpmt1-Mast3 (0.97) | Novel | |
| Dusp1-Egfr (0.73) | Novel | |
| Dusp16-Lrrk2 (0.86) | Novel | |
| Dusp10-Igf1r (0.99) | Novel |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Subbannayya, Y.; Pinto, S.M.; Bösl, K.; Prasad, T.S.K.; Kandasamy, R.K. Dynamics of Dual Specificity Phosphatases and Their Interplay with Protein Kinases in Immune Signaling. Int. J. Mol. Sci. 2019, 20, 2086. https://doi.org/10.3390/ijms20092086
Subbannayya Y, Pinto SM, Bösl K, Prasad TSK, Kandasamy RK. Dynamics of Dual Specificity Phosphatases and Their Interplay with Protein Kinases in Immune Signaling. International Journal of Molecular Sciences. 2019; 20(9):2086. https://doi.org/10.3390/ijms20092086
Chicago/Turabian StyleSubbannayya, Yashwanth, Sneha M. Pinto, Korbinian Bösl, T. S. Keshava Prasad, and Richard K. Kandasamy. 2019. "Dynamics of Dual Specificity Phosphatases and Their Interplay with Protein Kinases in Immune Signaling" International Journal of Molecular Sciences 20, no. 9: 2086. https://doi.org/10.3390/ijms20092086
APA StyleSubbannayya, Y., Pinto, S. M., Bösl, K., Prasad, T. S. K., & Kandasamy, R. K. (2019). Dynamics of Dual Specificity Phosphatases and Their Interplay with Protein Kinases in Immune Signaling. International Journal of Molecular Sciences, 20(9), 2086. https://doi.org/10.3390/ijms20092086






