Comparative Transcriptomic Studies on a Cadmium Hyperaccumulator Viola baoshanensis and Its Non-Tolerant Counterpart V. inconspicua
Abstract
1. Introduction
2. Results
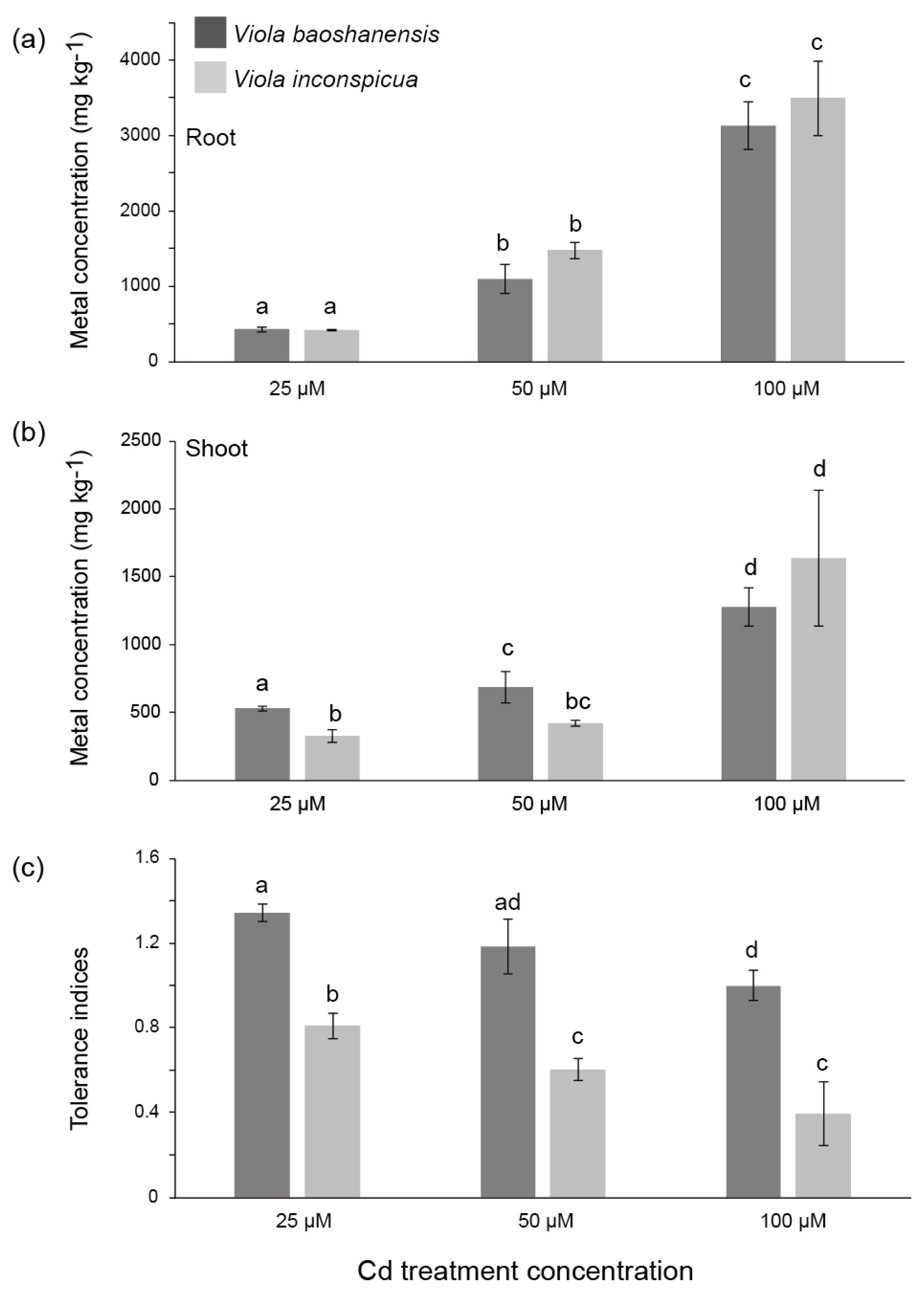
2.1. Metal Accumulation and Tolerance in the Two Viola Species from Hydroponic Experiments
2.2. Summary of the Data for Transcriptome Sequencing, De Novo Assembling, and Annotation
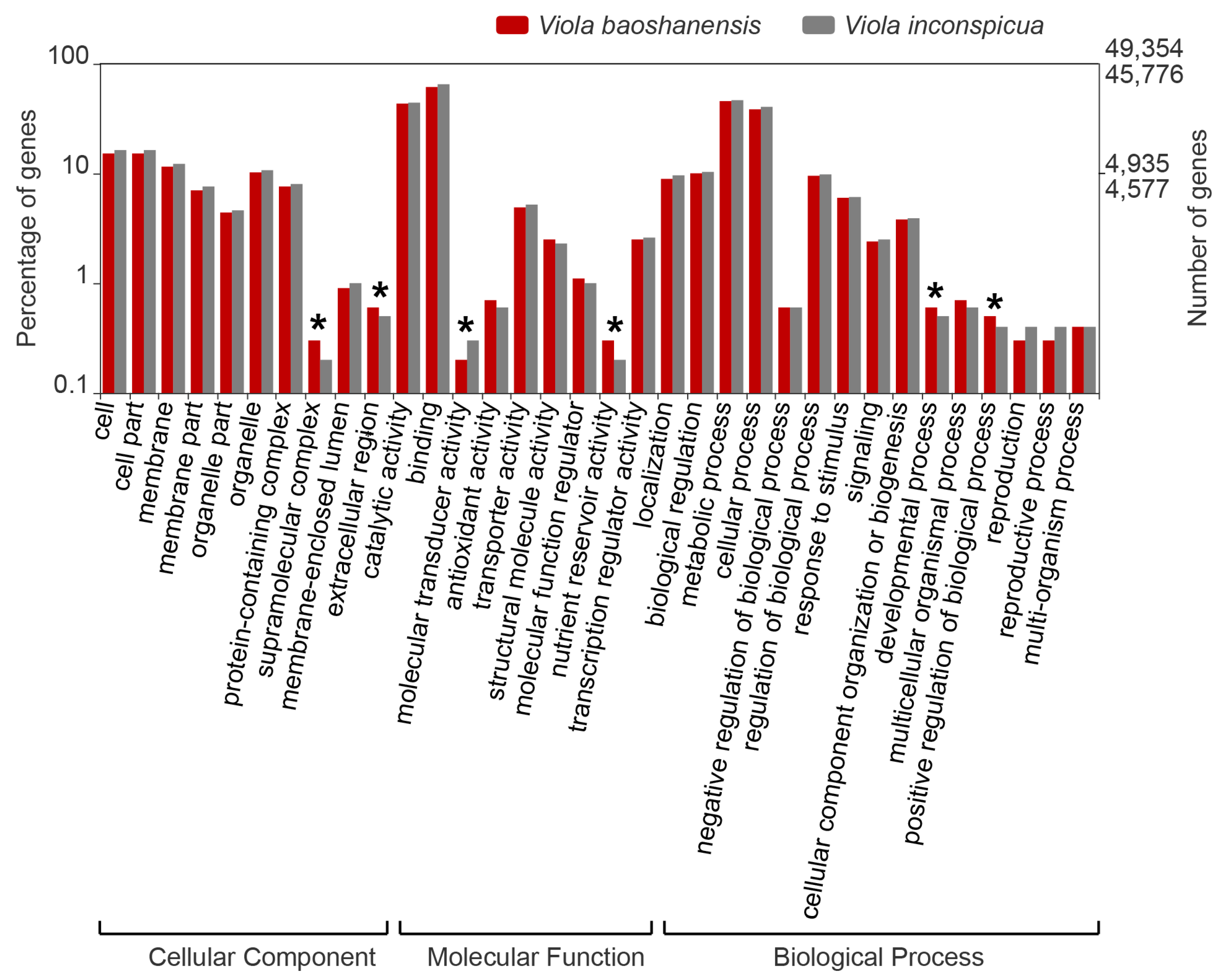
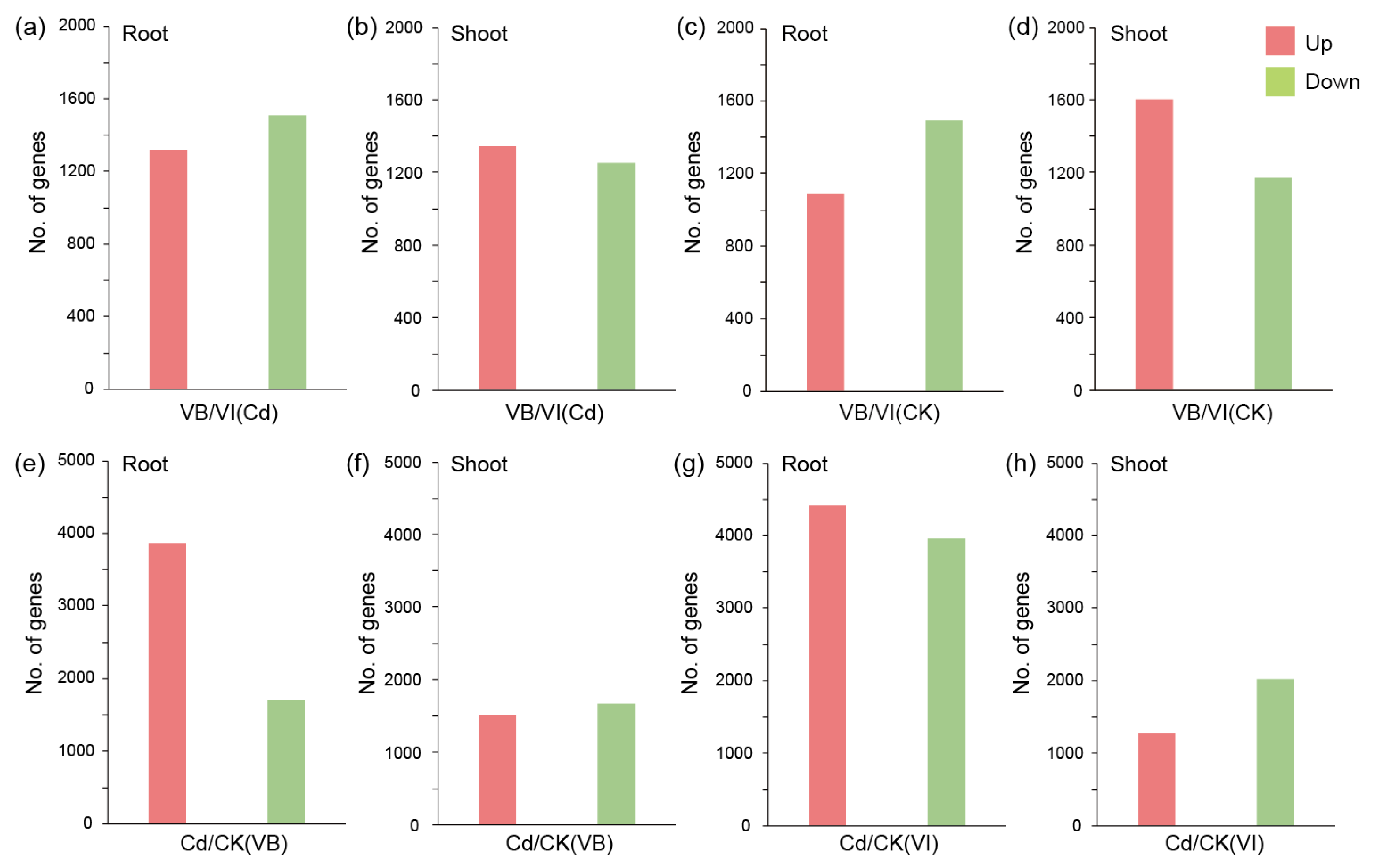
2.3. Analyses of Differential Expression (DE) and Functional Enrichment
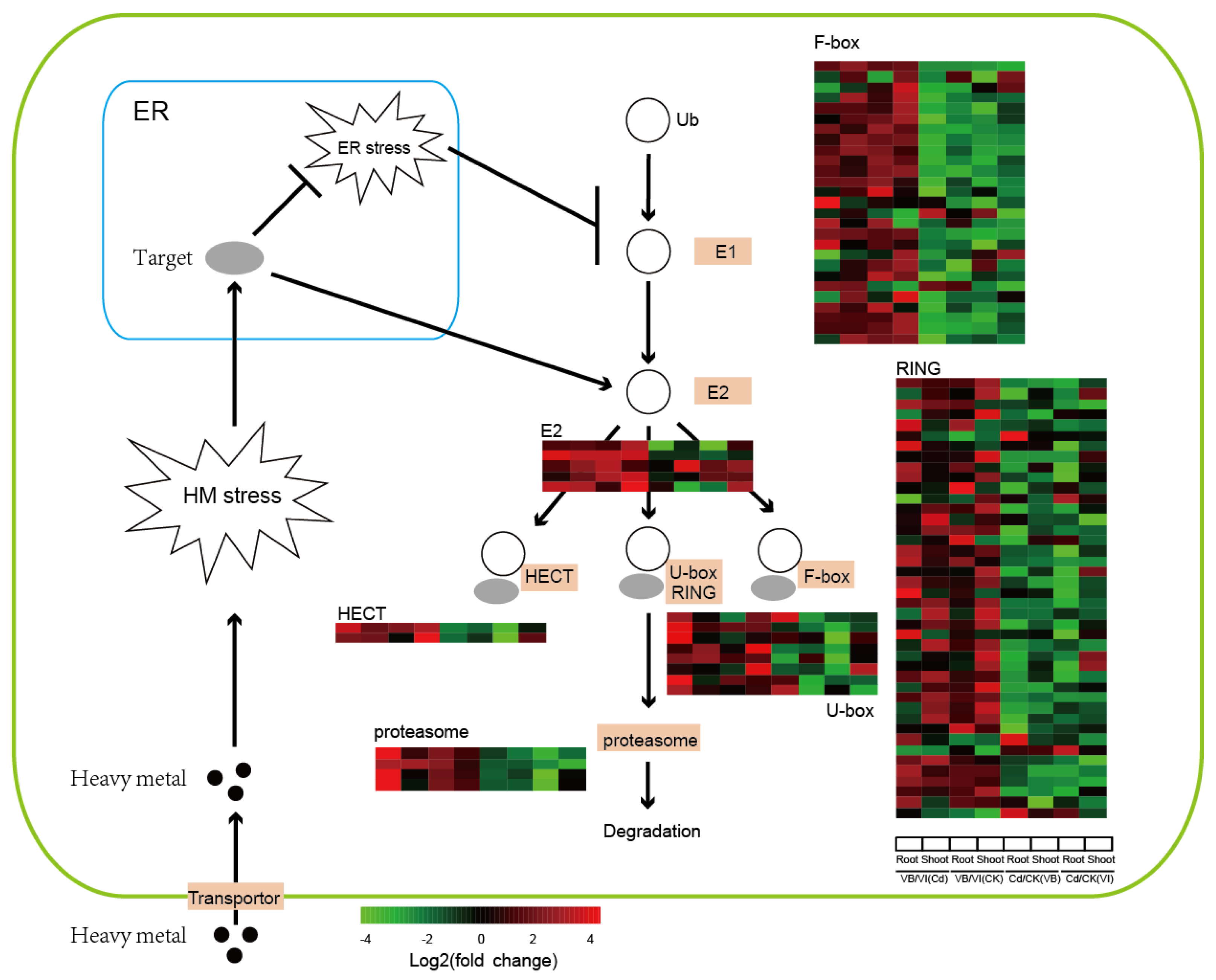
2.4. Ubiquitin Proteosome System (UPS) Pathway-Related Genes: Response to Cd Stress
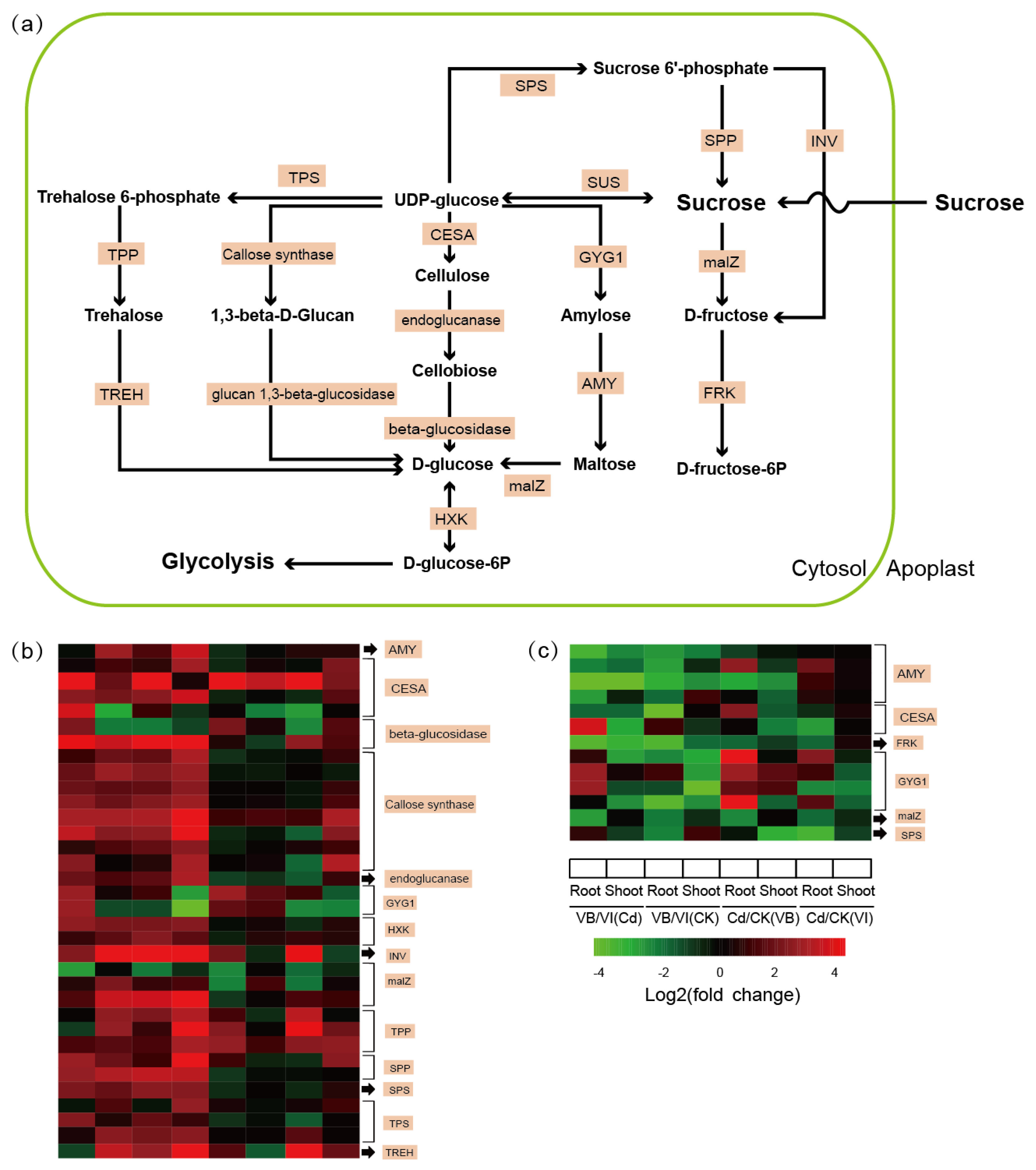
2.5. DEGs Involved in Sucrose Metabolism
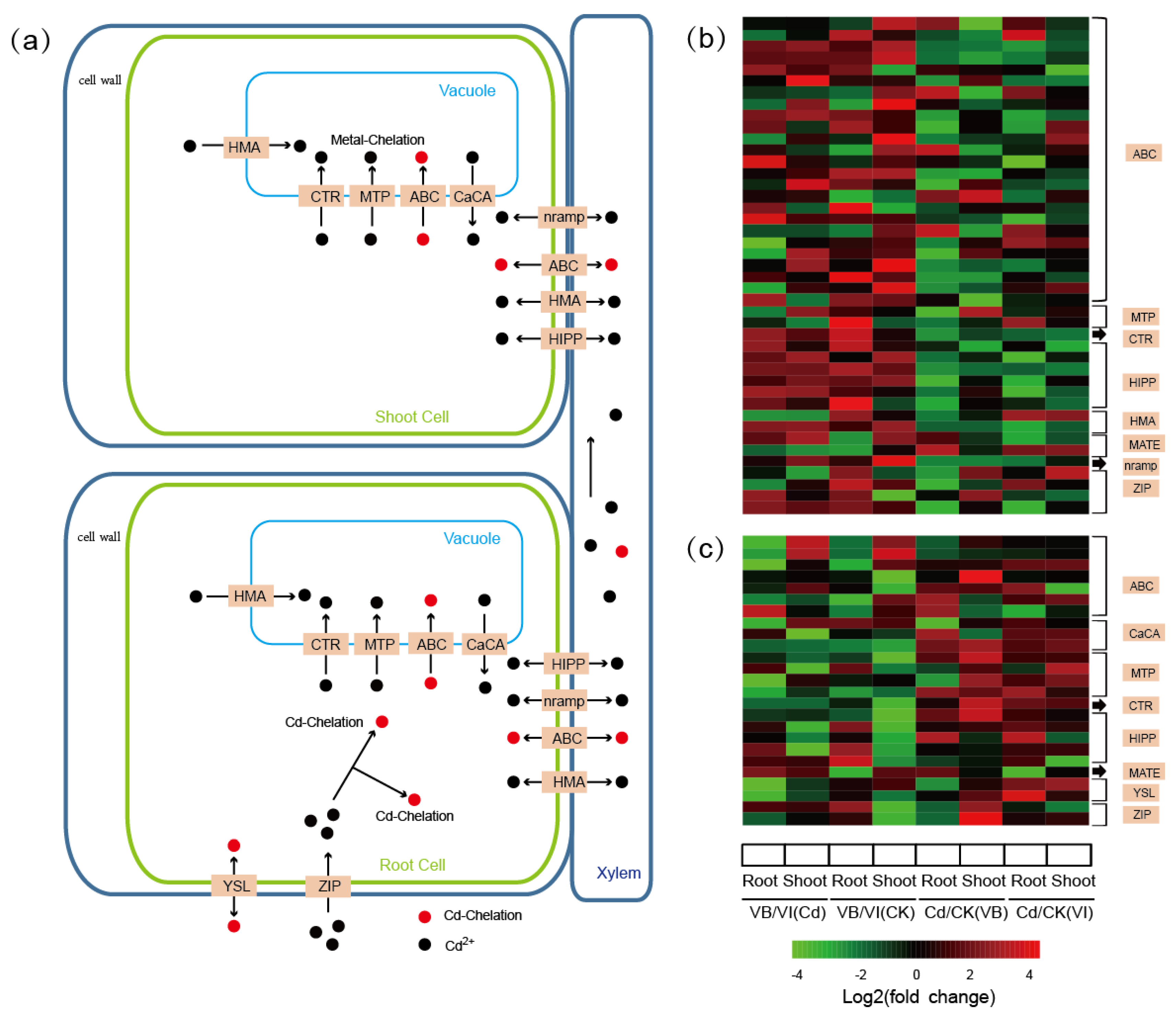
2.6. DEGs Encoding Metal Transporter Proteins
3. Discussion
3.1. How Does the UPS Pathway Enhance Heavy-metal Resistance in Plants?
3.2. Relations of Sucrose Metabolism with Heavy-Metal Stress in Plants
3.3. Contributions of Transporter Proteins to Heavy Metal Tolerance in Plants
4. Materials and Methods
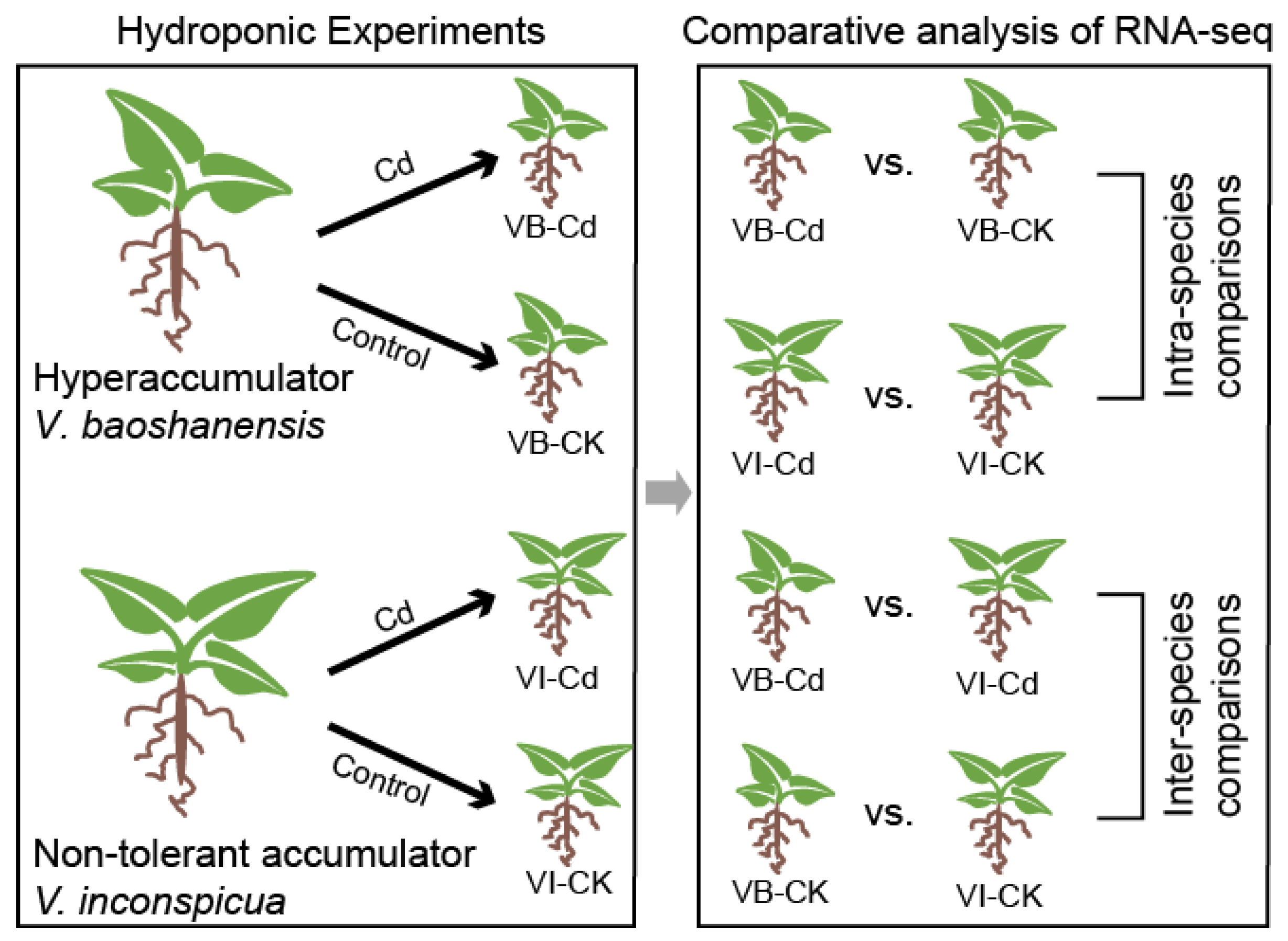
4.1. Collection of Plant Samples
4.2. Hydroponic Experiments
4.3. Elemental Analysis of the Hydroponic Samples
4.4. RNA Extraction and cDNA Library Preparation
4.5. Transcriptome Sequencing, Assembling, and Annotation
4.6. Differential Expression and Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| ABC | ATP-binding cassette transporters |
| AMY | alpha-amylase |
| bglX | beta-glucosidase |
| BUSCO | Benchmarking Universal Single-Copy Orthologs |
| Cd | cadmium |
| CaCA | the Ca2+: cation antiporter Family |
| CESA | cellulose synthase |
| CTR | the Copper transporter family |
| DEG | differential expression gene |
| FRK | fructokinase |
| GO | Gene Ontology |
| GYG1 | glycogenin |
| HMA | heavy metal ATPase |
| HIPP | heavy metal-associated isoprenylated plant protein |
| HXK | hexokinase |
| INV | beta-fructofuranosidase |
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
| malZ | alpha-glucosidase |
| MATE | multi-antimicrobial extrusion protein |
| MTP | metal tolerance protein |
| Ni | nickel |
| Nramp | natural resistance-associated macrophage protein |
| OM | organic matter |
| Pb | lead |
| SPP | sucrose-6-phosphatase |
| SPS | sucrose-phosphate synthase |
| TPP | trehalose 6-phosphate phosphatase |
| TPS | trehalose 6-phosphate synthase |
| TREH | alpha, alpha-trehalase |
| UPS | ubiquitin proteosome system |
| YSL | yellow stripe-like family |
| ZIP | the Zinc/Iron permease family |
| Zn | zinc |
References
- Buha, A.; Matovic, V.; Antonijevic, B.; Bulat, Z.; Curcic, M.; Renieri, E.; Tsatsakis, A.; Schweitzer, A.; Wallace, D. Overview of cadmium thyroid disrupting effects and mechanisms. Int. J. Mol. Sci. 2018, 19, 1501. [Google Scholar] [CrossRef] [PubMed]
- Clemens, S.; Aarts, M.G.; Thomine, S.; Verbruggen, N. Plant science: The key to preventing slow cadmium poisoning. Trends Plant Sci. 2013, 18, 92–99. [Google Scholar] [CrossRef] [PubMed]
- Ali, H.; Khan, E.; Sajad, M.A. Phytoremediation of heavy metals-Concepts and applications. Chemosphere 2013, 91, 869–881. [Google Scholar] [CrossRef] [PubMed]
- Rascio, N.; Navari-Izzo, F. Heavy metal hyperaccumulating plants: How and why do they do it? And what makes them so interesting? Plant Sci. 2011, 180, 169–181. [Google Scholar] [CrossRef] [PubMed]
- Nathalie, V.; Christian, H.; Henk, S. Molecular mechanisms of metal hyperaccumulation in plants. New Phytol. 2009, 181, 759–776. [Google Scholar] [CrossRef]
- Krämer, U. Metal Hyperaccumulation in Plants. Annu. Rev. Plant Biol. 2010, 61, 517–534. [Google Scholar] [CrossRef] [PubMed]
- Li, J.T.; Gurajala, H.K.; Wu, L.; Antony, V.D.E.; Qiu, R.L.; Baker, A.J.M.; Tang, Y.T.; Yang, X.; Shu, W. Hyperaccumulator plants from China: A synthesis of the current state of knowledge. Environ. Sci. Technol. 2018, 52, 11980–11994. [Google Scholar] [CrossRef]
- Kusznierewicz, B.; Bączek-Kwinta, R.; Bartoszek, A.; Piekarska, A.; Huk, A.; Manikowska, A.; Antonkiewicz, J.; Namieśnik, J.; Konieczka, P. The dose-dependent influence of zinc and cadmium contamination of soil on their uptake and glucosinolate content in white cabbage (Brassica oleracea var. capitata f. alba). Environ. Toxicol. Chem. 2012, 31, 2482–2489. [Google Scholar] [CrossRef]
- Shahid, M.; Dumat, C.; Khalid, S.; Schreck, E.; Xiong, T.; Niazi, N.K. Foliar heavy metal uptake, toxicity and detoxification in plants: A comparison of foliar and root metal uptake. J. Hazard Mater. 2016, 325, 36–58. [Google Scholar] [CrossRef]
- Cobbett, C.S. Phytochelatins and their roles in heavy metal detoxification. Plant Physiol. 2000, 123, 825–832. [Google Scholar] [CrossRef] [PubMed]
- Luigi, P.; Gea, G.; Kjell, S.; Giampiero, C.; Jean-Francois, H. Target or barrier? The cell wall of early- and later-diverging plants vs cadmium toxicity: Differences in the response mechanisms. Front. Plant Sci. 2015, 6, 133. [Google Scholar] [CrossRef]
- Hossain, M.A.; Piyatida, P.; da Silva, J.A.T.; Fujita, M. Molecular mechanism of heavy metal toxicity and tolerance in plants: Central role of glutathione in detoxification of reactive oxygen species and methylglyoxal and in heavy metal chelation. J. Bot. 2012, 2012, 872875. [Google Scholar] [CrossRef]
- Verbruggen, N.; Hanikenne, M.; Clemens, S. A more complete picture of metal hyperaccumulation through next-generation sequencing technologies. Front. Plant Sci. 2013, 4, 388. [Google Scholar] [CrossRef] [PubMed]
- Halimaa, P.; Blande, D.; Aarts, M.G.; Tuomainen, M.; Tervahauta, A.; Karenlampi, S. Comparative transcriptome analysis of the metal hyperaccumulator Noccaea caerulescens. Front. Plant Sci. 2014, 5, 213. [Google Scholar] [CrossRef][Green Version]
- Cappa, J.J.; Pilon-Smits, E.A.H. Evolutionary aspects of elemental hyperaccumulation. Planta 2014, 239, 267–275. [Google Scholar] [CrossRef]
- Goolsby, E.W.; Mason, C.M. Toward a more physiologically and evolutionarily relevant definition of metal hyperaccumulation in plants. Front. Plant Sci. 2015, 6, 33. [Google Scholar] [CrossRef]
- Weber, M.; Trampczynska, A.; Clemens, S. Comparative transcriptome analysis of toxic metal responses in Arabidopsis thaliana and the Cd(2+)-hypertolerant facultative metallophyte Arabidopsis halleri. Plant Cell Environ. 2010, 29, 950–963. [Google Scholar] [CrossRef]
- Peng, J.-S.; Wang, Y.-J.; Ding, G.; Ma, H.-L.; Zhang, Y.-J.; Gong, J.-M. A pivotal role of cell wall in cadmium accumulation in the Crassulaceae hyperaccumulator Sedum plumbizincicola. Mol. Plant 2017, 10, 771–774. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Shu, W.; Lan, C. Viola baoshanensis, a plant that hyperaccumulates cadmium. Chin. Sci. Bull. 2004, 49, 29–32. [Google Scholar] [CrossRef]
- Tonin, C.; Vandenkoornhuyse, P.; Joner, E.J.; Straczek, J.; Leyval, C. Assessment of arbuscular mycorrhizal fungi diversity in the rhizosphere of Viola calaminaria and effect of these fungi on heavy metal uptake by clover. Mycorrhiza 2001, 10, 161–168. [Google Scholar] [CrossRef]
- Sychta, K.; Słomka, A.; Suski, S.; Fiedor, E.; Gregoraszczuk, E.; Kuta, E. Suspended cells of metallicolous and nonmetallicolous Viola species tolerate, accumulate and detoxify zinc and lead. Plant Physiol. Biochem. 2018, 132, 666–674. [Google Scholar] [CrossRef] [PubMed]
- Fernando, E.S.; Quimado, M.O.; Doronila, A.I. Rinorea niccolifera (Violaceae), a new, nickel-hyperaccumulating species from Luzon Island, Philippines. PhytoKeys 2014, 37, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.A.; Liao, B.; Wang, S.L.; Zhang, J.; Li, J.T. Pb and Zn accumulation in a Cd-hyperaccumulator (Viola baoshanensis). Int. J. Phytoremediat. 2010, 12, 574–585. [Google Scholar] [CrossRef]
- Simão, F.A.; Waterhouse, R.M.; Ioannidis, P.; Kriventseva, E.V.; Zdobnov, E.M. BUSCO: Assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 2015, 31, 3210–3212. [Google Scholar] [CrossRef]
- Parekh, S.; Vieth, B.; Ziegenhain, C.; Enard, W.; Hellmann, I. Strategies for quantitative RNA-seq analyses among closely related species. bioRxiv 2018, 297408. [Google Scholar] [CrossRef]
- Song, Y.; Jin, L.; Wang, X. Cadmium absorption and transportation pathways in plants. Int. J. Phytoremediat. 2017, 19, 133–141. [Google Scholar] [CrossRef] [PubMed]
- Ari, S.; Mark, B.; Richard, E.; Jack, L.; Stuart, N. The ubiquitin-proteasome system: Central modifier of plant signalling. New Phytol. 2012, 196, 13–28. [Google Scholar] [CrossRef]
- Jungmann, J.; Reins, H.A.; Schobert, C.; Jentsch, S. Resistance to cadmium mediated by ubiquitin-dependent proteolysis. Nature 1993, 361, 369. [Google Scholar] [CrossRef]
- Chai, T.Y.; Zhang, Y.X. Expression analysis of polyubiquitin genes from bean in response to heavy metals. Acta Bot. Sin. 1999, 41, 1052–1057. [Google Scholar]
- Oono, Y.; Yazawa, T.; Kanamori, H.; Sasaki, H.; Mori, S.; Handa, H.; Matsumoto, T. Genome-Wide Transcriptome Analysis of Cadmium Stress in Rice. BioMed Res. Int. 2016, 2016, 1–9. [Google Scholar] [CrossRef]
- Flick, K.; Kaiser, P. Protein degradation and the stress response. Cell Dev. Biol. 2012, 23, 515–522. [Google Scholar] [CrossRef] [PubMed]
- Amm, I.; Sommer, T.; Wolf, D.H. Protein quality control and elimination of protein waste: The role of the ubiquitin-proteasome system. BBA-Mol. Cell. Res. 2014, 1843, 182–196. [Google Scholar] [CrossRef]
- Hasan, M.K.; Cheng, Y.; Kanwar, M.K.; Chu, X.Y.; Ahammed, G.J.; Qi, Z.Y. Responses of Plant Proteins to Heavy Metal Stress—A Review. Front. Plant Sci. 2017, 8, 1492. [Google Scholar] [CrossRef]
- Sharma, S.K.; Goloubinoff, P.; Christen, P. Cellular Effects of Heavy Metals; Springer: New York, NY, USA, 2011; pp. 263–274. [Google Scholar]
- Chen, C.C.; Chen, Y.Y.; Tang, I.C.; Liang, H.M.; Lai, C.C.; Chiou, J.M.; Yeh, K.C. Arabidopsis SUMO E3 Ligase SIZ1 Is Involved in Excess Copper Tolerance. Plant Physiol. 2011, 156, 2225–2234. [Google Scholar] [CrossRef]
- Lim, S.D.; Jin, G.H.; Han, A.R.; Yong, C.P.; Lee, C.; Yong, S.O.; Jang, C.S. Positive regulation of rice RING E3 ligase OsHIR1 in arsenic and cadmium uptakes. Plant Mol. Biol. 2014, 85, 365. [Google Scholar] [CrossRef] [PubMed]
- Dametto, A.; Buffon, G.; dos Reis Blasi, Ã.A.; Sperotto, R.A. Ubiquitination pathway as a target to develop abiotic stress tolerance in rice. Plant Signal. Behav. 2015, 10, e1057369. [Google Scholar] [CrossRef] [PubMed]
- Yongling, R. Sucrose metabolism: Gateway to diverse carbon use and sugar signaling. Annu. Rev. Plant Biol. 2014, 65, 33–67. [Google Scholar] [CrossRef]
- Antonkiewicz, J.; Para, A. The use of dialdehyde starch derivatives in the phytoremediation of soils contaminated with heavy metals. Int. J. Phytoremediat. 2016, 18, 245–250. [Google Scholar] [CrossRef]
- Baccio, D.D.; Galla, G.; Bracci, T.; Andreucci, A.; Barcaccia, G.; Tognetti, R.; Sebastiani, L. Transcriptome analyses of Populus × euramericana clone I-214 leaves exposed to excess zinc. Tree Physiol. 2011, 31, 1293–1308. [Google Scholar] [CrossRef]
- Feng, J.; Jia, W.; Lv, S.; Bao, H.; Miao, F.; Zhang, X.; Wang, J.; Li, J.; Li, D.; Zhu, C. Comparative transcriptome combined with morpho-physiological analyses revealed key factors for differential cadmium accumulation in two contrasting sweet sorghum genotypes. Plant Biotechnol. J. 2017, 16, 558–571. [Google Scholar] [CrossRef]
- Huang, Y.Y.; Shen, C.; Chen, J.X.; He, C.T.; Zhou, Q.; Tan, X.; Yuan, J.; Yang, Z. Comparative transcriptome analysis of two Ipomoea aquatica Forsk. cultivars targeted to explore possible mechanism of genotype dependent accumulation of cadmium. J. Agric. Food Chem. 2016, 64, 5241–5250. [Google Scholar] [CrossRef] [PubMed]
- Granot, D. Role of tomato hexose kinases. Funct. Plant Biol. 2007, 34, 564–570. [Google Scholar] [CrossRef]
- Melissa, R.; Lorenz, G.; David, S.; András, G.; Mattias, H.M.; Manoj, K.; Marie Caroline, S.; Regina, F.; Geoffrey, D.; Mark, S. Fructokinase is required for carbon partitioning to cellulose in aspen wood. Plant J. 2012, 70, 967–977. [Google Scholar] [CrossRef]
- Laurence, L.; Judith, W.; Marjorie, P.; Joanna Marie-France, C.; Pascal, T.; Alain, G. Oxidative pentose phosphate pathway-dependent sugar sensing as a mechanism for regulation of root ion transporters by photosynthesis. Plant Physiol. 2008, 146, 2036–2053. [Google Scholar] [CrossRef]
- Chen, X.-Y.; Kim, J.-Y. Callose synthesis in higher plants. Plant Signal. Behav. 2009, 4, 489–492. [Google Scholar] [CrossRef] [PubMed]
- Bo, X.; Xiaomin, W.; Maosheng, Z.; Zhongming, Z.; Zonglie, H. CalS7 encodes a callose synthase responsible for callose deposition in the phloem. Plant J. 2011, 65, 1–14. [Google Scholar] [CrossRef]
- Vatén, A.; Dettmer, J.; Shuang, W.; Stierhof, Y.D.; Miyashima, S.; Yadav, S.R.; Roberts, C.; Campilho, A.; Bulone, V.; Lichtenberger, R. Callose Biosynthesis Regulates Symplastic Trafficking during Root Development. Dev. Cell 2011, 21, 1144–1155. [Google Scholar] [CrossRef]
- Levy, A.; Erlanger, M.; Rosenthal, M.; Epel, B.L. A plasmodesmata-associated β-1,3-glucanase in Arabidopsis. Plant J. 2010, 49, 669–682. [Google Scholar] [CrossRef] [PubMed]
- Jens, M.; Theresa, T.; Marcus, H.; Janine, T.; Moore, K.L.; Gerd, H.; Dhurvas Chandrasekaran, D.; Katharina, B.; Steffen, A. Iron-dependent callose deposition adjusts root meristem maintenance to phosphate availability. Dev. Cell 2015, 33, 216–230. [Google Scholar] [CrossRef]
- Mira, H.; Martinez-Garcia, F.; Penarrubia, L. Evidence for the plant-specific intercellular transport of the Arabidopsiscopper chaperone CCH. Plant J. 2010, 25, 521–528. [Google Scholar] [CrossRef]
- Zhang, H.; Shi, W.L.; You, J.F.; Bian, M.D.; Qin, X.M.; Hui, Y.U.; Liu, Q.; Ryan, P.R.; Yang, Z.M. Transgenic Arabidopsis thaliana plants expressing a β-1,3-glucanase from sweet sorghum (Sorghum bicolor L.) show reduced callose deposition and increased tolerance to aluminium toxicity. Plant Cell Environ. 2015, 38, 1178–1188. [Google Scholar] [CrossRef] [PubMed]
- Keunen, E.; Peshev, D.; Vangronsveld, J.; Van, D.E.W.; Cuypers, A. Plant sugars are crucial players in the oxidative challenge during abiotic stress: Extending the traditional concept. Plant Cell Environ. 2013, 36, 1242–1255. [Google Scholar] [CrossRef] [PubMed]
- Sami, F.; Yusuf, M.; Faizan, M.; Faraz, A.; Hayat, S. Role of sugars under abiotic stress. Plant Physiol. Biochem. 2016, 109, 54–61. [Google Scholar] [CrossRef]
- Mostofa, M.G.; Hossain, M.A.; Fujita, M.; Tran, L.S. Physiological and biochemical mechanisms associated with trehalose-induced copper-stress tolerance in rice. Sci. Rep. 2015, 5, 11433. [Google Scholar] [CrossRef]
- Martins, L.L.; Mourato, M.P.; Baptista, S.; Reis, R.; Carvalheiro, F.; Almeida, A.M.; Fevereiro, P.; Cuypers, A. Response to oxidative stress induced by cadmium and copper in tobacco plants (Nicotiana tabacum) engineered with the trehalose-6-phosphate synthase gene (AtTPS1). Acta Physiol. Plant. 2014, 36, 755–765. [Google Scholar] [CrossRef]
- Ali, Q.; Ashraf, M. Induction of Drought Tolerance in Maize (Zea mays L.) due to Exogenous Application of Trehalose: Growth, Photosynthesis, Water Relations and Oxidative Defence Mechanism. J. Agron. Crop Sci. 2011, 197, 258–271. [Google Scholar] [CrossRef]
- Benaroudj, N.; Lee, D.; Goldberg, A. Trehalose accumulation during cellular stress protects cells and cellular proteins from damage by oxygen radicals. J. Biol. Chem. 2001, 276, 24261–24267. [Google Scholar] [CrossRef]
- Lux, A.; Martinka, M.; Vaculík, M.; White, P.J. Root responses to cadmium in the rhizosphere: A review. J. Exp. Bot. 2011, 62, 21–37. [Google Scholar] [CrossRef] [PubMed]
- Pence, N.S.; Larsen, P.B.; Ebbs, S.D.; Letham, D.L.; Lasat, M.M.; Garvin, D.F.; Eide, D.; Kochian, L.V. The molecular physiology of heavy metal transport in the Zn/Cd hyperaccumulator Thlaspi caerulescens. Proc. Natl. Acad. Sci. USA 2000, 97, 4956–4960. [Google Scholar] [CrossRef]
- Hanikenne, M.; Talke, I.N.; Haydon, M.J.; Lanz, C.; Nolte, A.; Motte, P.; Kroymann, J.; Weigel, D.; Kramer, U. Evolution of metal hyperaccumulation required cis-regulatory changes and triplication of HMA4. Nature 2008, 453, 391–395. [Google Scholar] [CrossRef]
- Li, J.; Wang, L.; Zheng, L.; Wang, Y.; Chen, X.; Zhang, W. A Functional Study Identifying Critical Residues Involving Metal Transport Activity and Selectivity in Natural Resistance-Associated Macrophage Protein 3 in Arabidopsis thaliana. Int. J. Mol. Sci. 2018, 19, 1430. [Google Scholar] [CrossRef]
- Sharma, S.S.; Dietz, K.J.; Mimura, T. Vacuolar compartmentalization as indispensable component of heavy metal detoxification in plants. Plant Cell Environ. 2016, 39, 1112–1126. [Google Scholar] [CrossRef] [PubMed]
- Gendre, D.; Czernic, P.; Conéjéro, G.; Pianelli, K.; Briat, J.F.; Lebrun, M.; Mari, S. TcYSL3, a member of the YSL gene family from the hyper-accumulator Thlaspi caerulescens, encodes a nicotianamine-Ni/Fe transporter. Plant J. 2010, 49, 1–15. [Google Scholar] [CrossRef]
- Dai, J.; Wang, N.; Xiong, H.; Qiu, W.; Nakanishi, H.; Kobayashi, T.; Nishizawa, N.K.; Zuo, Y. The Yellow Stripe-Like (YSL) Gene Functions in Internal Copper Transport in Peanut. Genes 2018, 9, 635. [Google Scholar] [CrossRef]
- Li, J.T.; Deng, D.M.; Peng, G.T.; Deng, J.C.; Zhang, J.; Liao, B. Successful micropropagation of the cadmium hyperaccumulator Viola baoshanensis (Violaceae). Int. J. Phytoremediat. 2010, 12, 761–771. [Google Scholar] [CrossRef]
- Hoagland, D.R.; Arnon, D.I. The water-culture method for growing plants without soil. Circ. Calif. Agric. Exp. Stn. 1950, 347, 357–359. [Google Scholar]
- Allen, S.E.; Grimshaw, H.M.; Parkinson, J.A.; Quarmby, C. Chemical analysis of ecological materials. J. Appl. Ecol. 1974, 13, 650. [Google Scholar]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Xian, A.; Lin, F.; Raychowdhury, R.; Zeng, Q. Trinity: Reconstructing a full-length transcriptome without a genome from RNA-Seq data. Nat. Biotechnol. 2011, 29, 644–652. [Google Scholar] [CrossRef]
- Pertea, G.; Huang, X.; Liang, F.; Antonescu, V.; Sultana, R.; Karamycheva, S.; Lee, Y.; White, J.; Cheung, F.; Parvizi, B. TIGR Gene Indices clustering tools (TGICL): A software system for fast clustering of large EST datasets. Bioinformatics 2003, 19, 651–652. [Google Scholar] [CrossRef] [PubMed]
- Consortium, U.P. The Universal Protein Resource (UniProt) 2009. Nucleic Acids Res. 2009, 37, 169–174. [Google Scholar] [CrossRef]
- Zdobnov, E.M.; Apweiler, R. InterProScan—An integration platform for the signature-recognition methods in InterPro. Bioinformatics 2001, 17, 847–848. [Google Scholar] [CrossRef] [PubMed]
- Ye, J.; Zhang, Y.; Cui, H.; Liu, J.; Wu, Y.; Cheng, Y.; Xu, H.; Huang, X.; Li, S.; Zhou, A. WEGO 2.0: A web tool for analyzing and plotting GO annotations, 2018 update. Nucleic Acids Res. 2018, 46, W71–W75. [Google Scholar] [CrossRef]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef] [PubMed]
- Bo, L.; Dewey, C.N. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 2011, 12, 323. [Google Scholar] [CrossRef]
- Moreno-Hagelsieb, G.; Latimer, K. Choosing BLAST options for better detection of orthologs as reciprocal best hits. Bioinformatics 2008, 24, 319–324. [Google Scholar] [CrossRef] [PubMed]
- Robinson, M.D.; Mccarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Q.; Wang, X.-J. GOEAST: A web-based software toolkit for Gene Ontology enrichment analysis. Nucleic Acids Res. 2008, 36, 358–363. [Google Scholar] [CrossRef] [PubMed]
- Yu, G.; Wang, L.G.; Han, Y.; He, Q.Y. clusterProfiler: An R package for comparing biological themes among gene clusters. Omics 2012, 16, 284–287. [Google Scholar] [CrossRef]
| Parameter | Viola baoshanensis | Viola inconspicua |
|---|---|---|
| No. of contigs (before filtering) | 105,280 | 101,616 |
| No. of contigs | 82,854 | 80,059 |
| Maximum length of contigs (bp) | 67,368 | 42,149 |
| Average length of contigs (bp) | 1984 | 1348 |
| Contig N50 (bp) | 2778 | 1709 |
| GC content (%) | 43.8 | 42.5 |
| Annotated in KEGG | 31,772 | 31,141 |
| Annotated in GO | 49,354 | 45,646 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shu, H.; Zhang, J.; Liu, F.; Bian, C.; Liang, J.; Liang, J.; Liang, W.; Lin, Z.; Shu, W.; Li, J.; et al. Comparative Transcriptomic Studies on a Cadmium Hyperaccumulator Viola baoshanensis and Its Non-Tolerant Counterpart V. inconspicua. Int. J. Mol. Sci. 2019, 20, 1906. https://doi.org/10.3390/ijms20081906
Shu H, Zhang J, Liu F, Bian C, Liang J, Liang J, Liang W, Lin Z, Shu W, Li J, et al. Comparative Transcriptomic Studies on a Cadmium Hyperaccumulator Viola baoshanensis and Its Non-Tolerant Counterpart V. inconspicua. International Journal of Molecular Sciences. 2019; 20(8):1906. https://doi.org/10.3390/ijms20081906
Chicago/Turabian StyleShu, Haoyue, Jun Zhang, Fuye Liu, Chao Bian, Jieliang Liang, Jiaqi Liang, Weihe Liang, Zhiliang Lin, Wensheng Shu, Jintian Li, and et al. 2019. "Comparative Transcriptomic Studies on a Cadmium Hyperaccumulator Viola baoshanensis and Its Non-Tolerant Counterpart V. inconspicua" International Journal of Molecular Sciences 20, no. 8: 1906. https://doi.org/10.3390/ijms20081906
APA StyleShu, H., Zhang, J., Liu, F., Bian, C., Liang, J., Liang, J., Liang, W., Lin, Z., Shu, W., Li, J., Shi, Q., & Liao, B. (2019). Comparative Transcriptomic Studies on a Cadmium Hyperaccumulator Viola baoshanensis and Its Non-Tolerant Counterpart V. inconspicua. International Journal of Molecular Sciences, 20(8), 1906. https://doi.org/10.3390/ijms20081906





