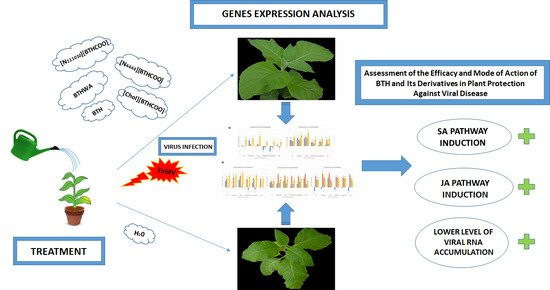
Assessment of the Efficacy and Mode of Action of Benzo(1,2,3)-Thiadiazole-7-Carbothioic Acid S-Methyl Ester (BTH) and Its Derivatives in Plant Protection Against Viral Disease
Abstract
1. Introduction
2. Results
2.1. Phytotoxicity of BTH and Its Derivatives after Soil Application
2.2. Symptoms of Viral Infection in the Absence and Presence of Resistance Inducers
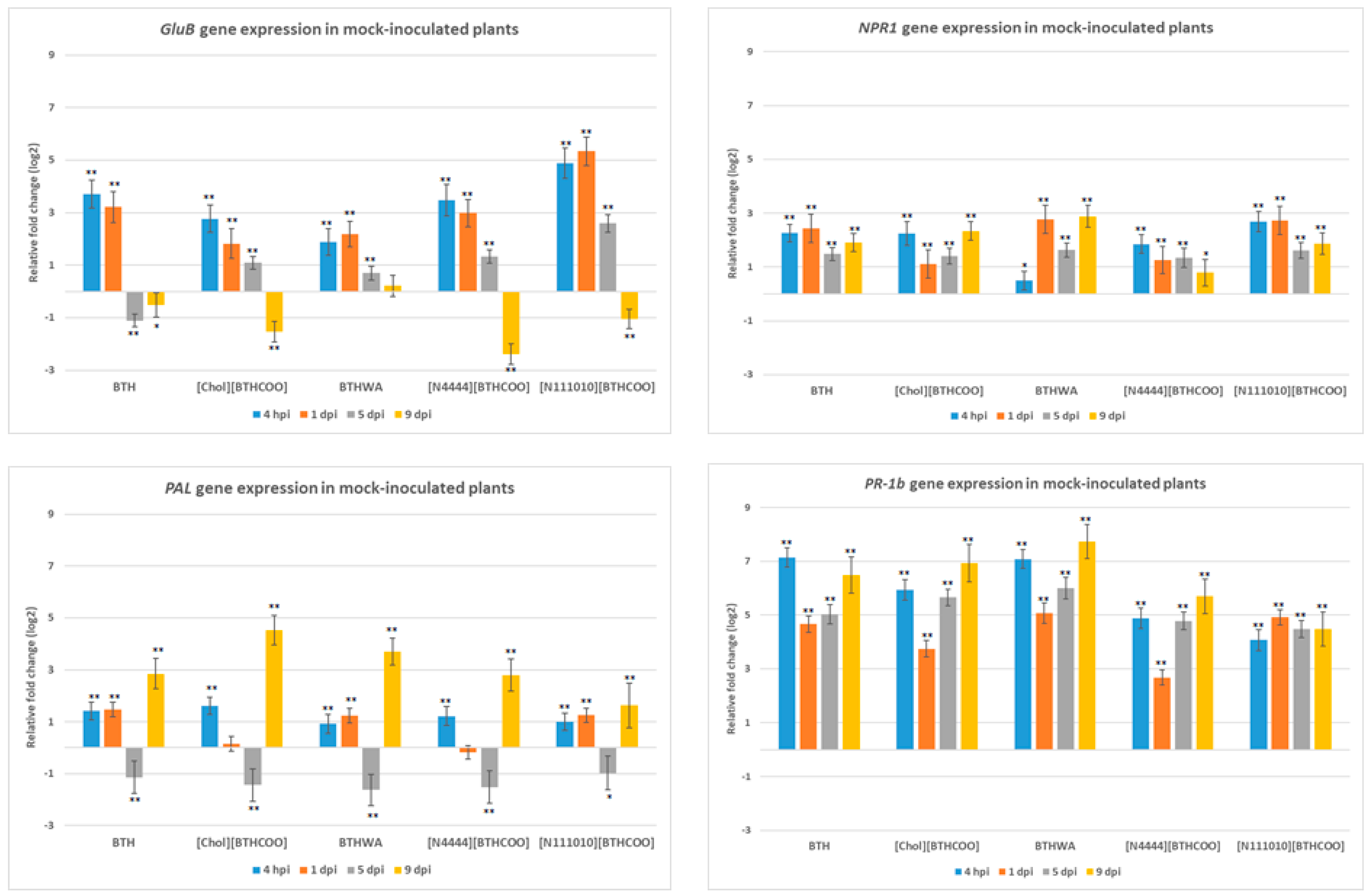
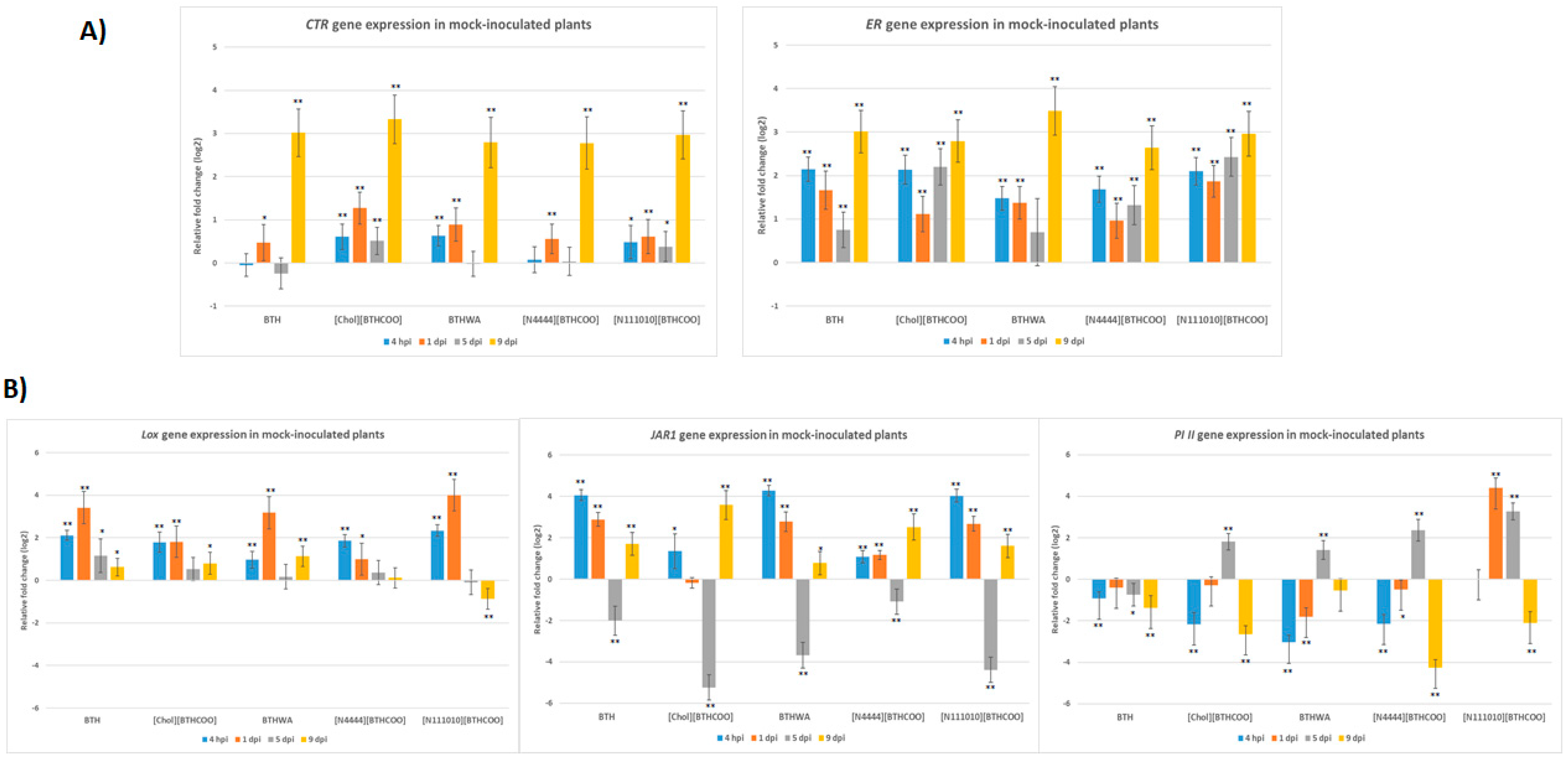
2.3. Analysis of the Expression of Defence-Related Marker Genes after BTH and Its Derivatives Treatments in the Absence of Pathogens
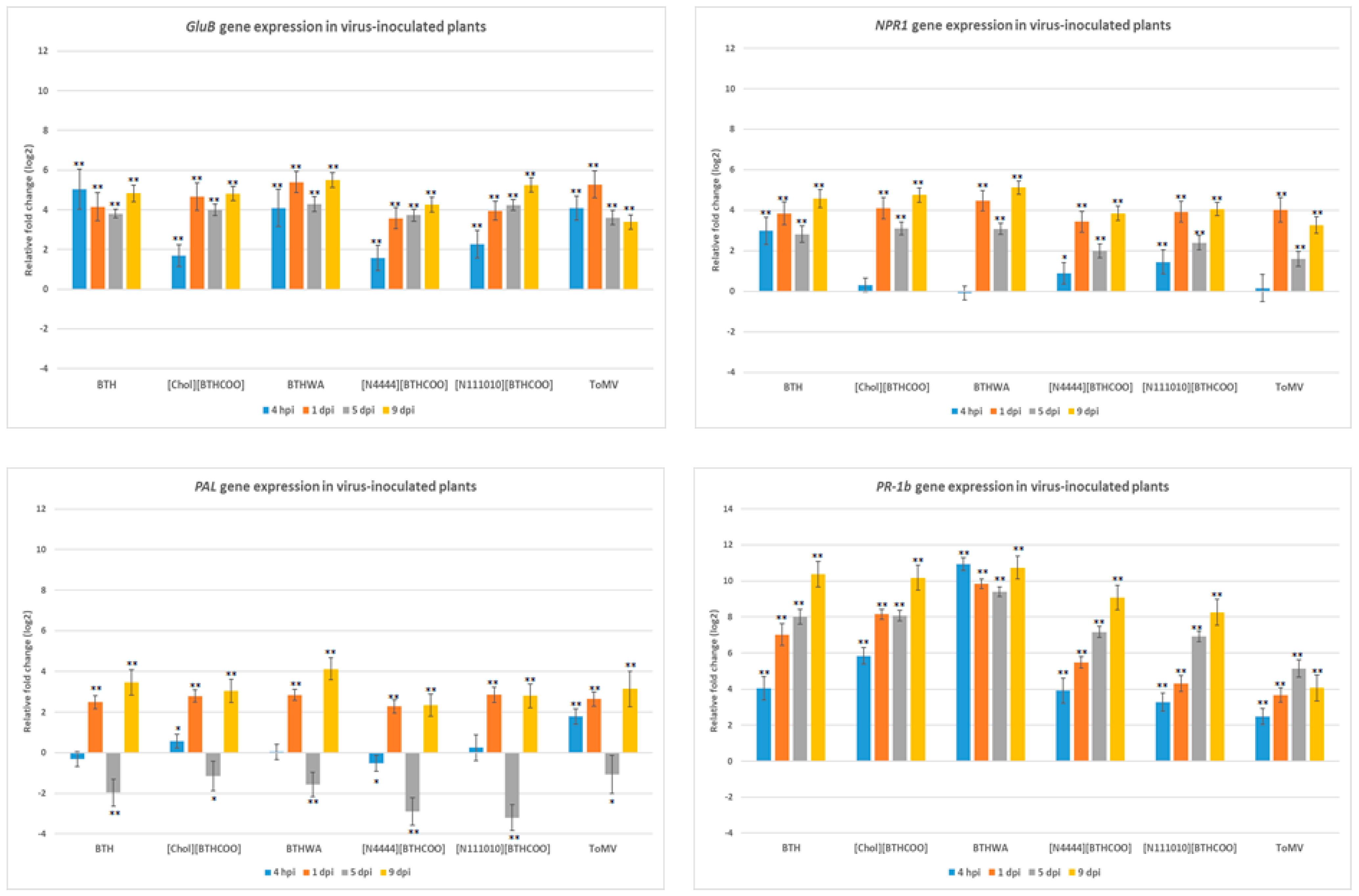
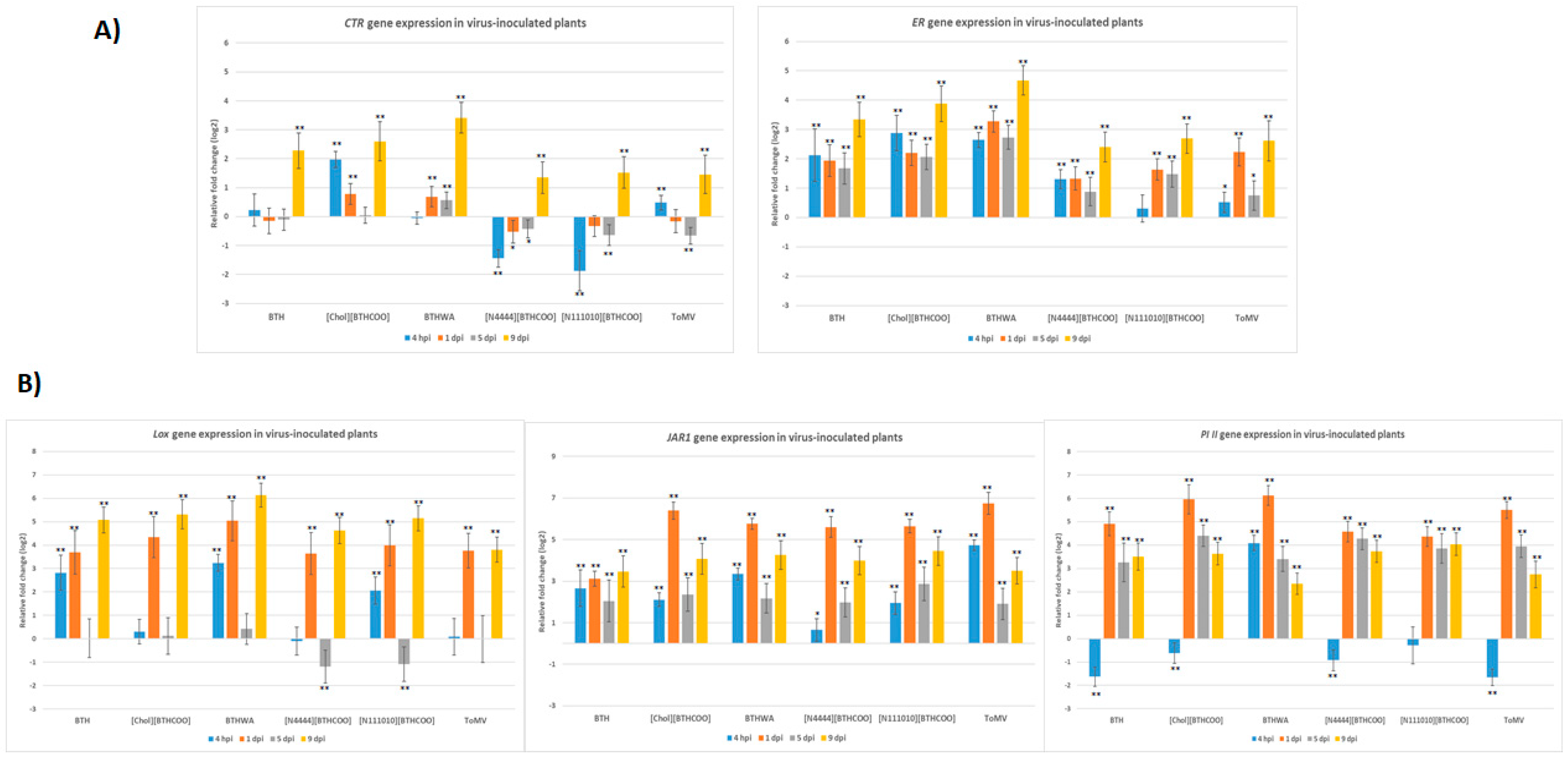
2.4. Analysis of the Expression of Defence-Related Marker Genes after BTH and Its Derivatives Treatments in Virus-Infected Plants
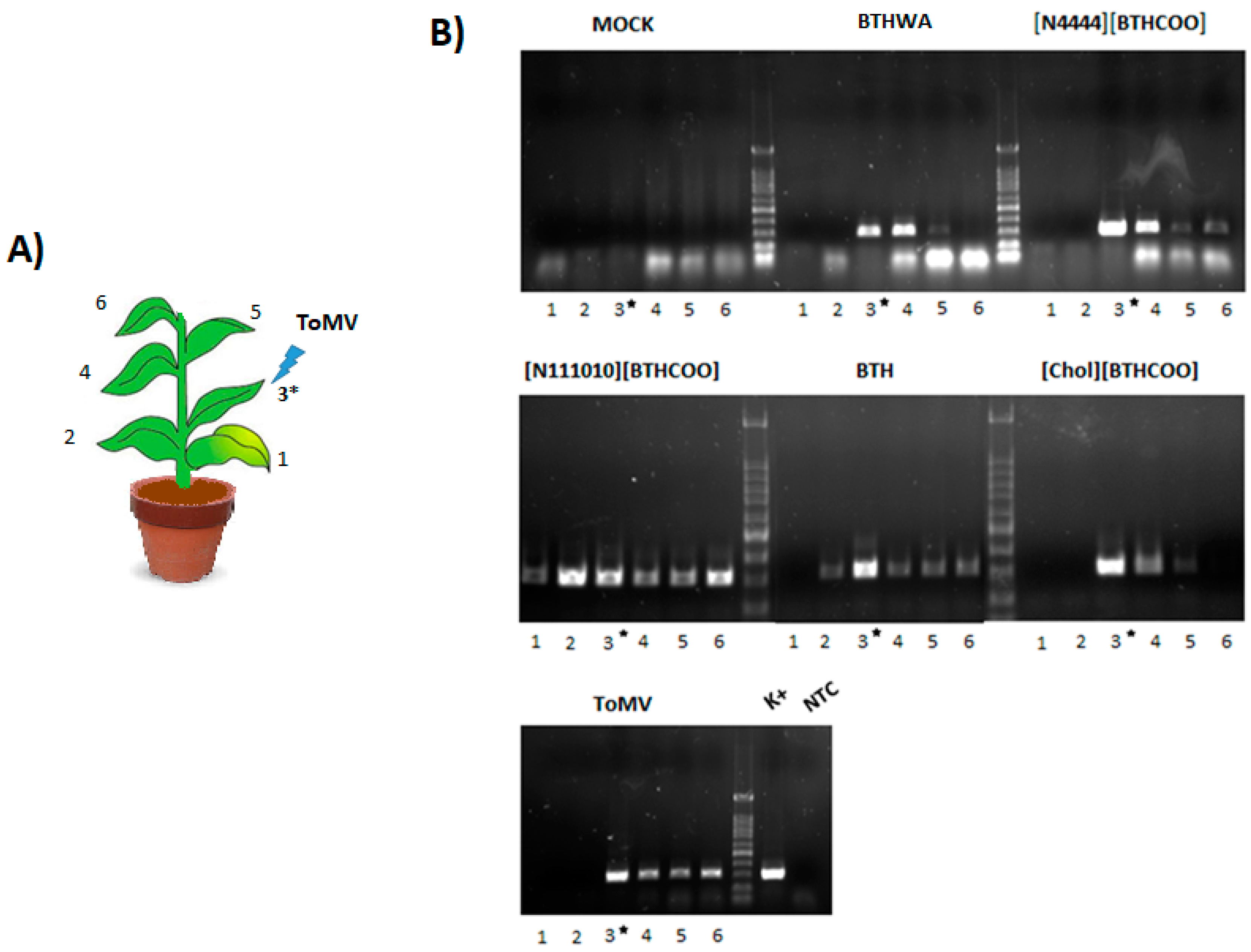
2.5. Viral RNA Accumulation in Plants after Treatment with BTH and Its Derivatives
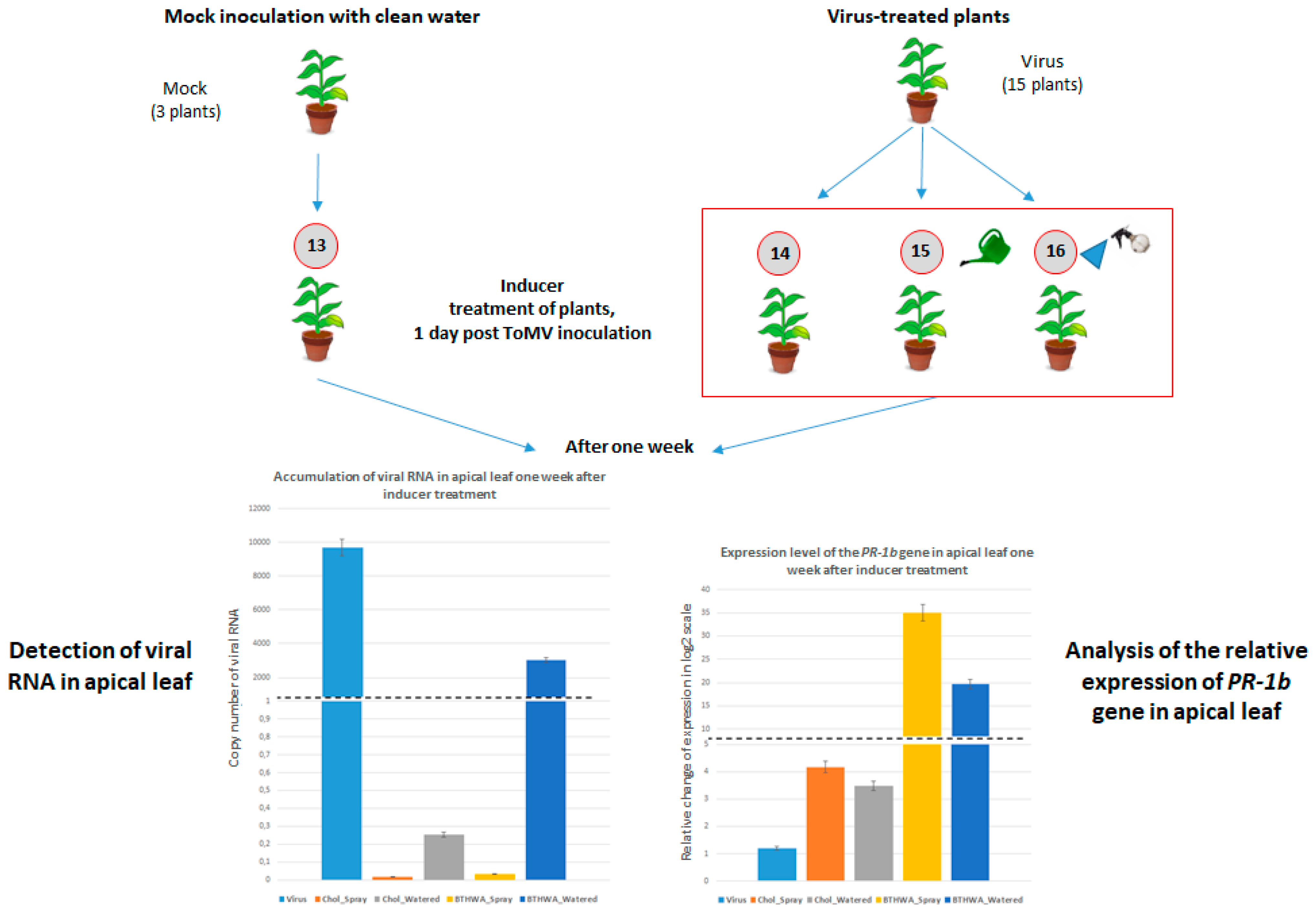
2.6. Effectiveness of Resistance Inducer Application (BTHWA or[Chol][BTHCOO]) to Plants Previously Infected with Virus
3. Discussion
4.1. BTH and Its Derivatives
4.2. Plants and Virus Materials
4.3. BTH and Its Derivatives Treatments of Plants Followed by Subsequent Virus Inoculation
4.4. BTH and Its Derivatives Treatments of Plants Following Prior Virus Inoculation
4.5. RNA Isolation and cDNA Synthesis
4.6. Analysis of SA, JA, and ET Pathway-Related Gene Expression
4.7. Analysis of Viral RNA Accumulation
4.8. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| BTH | benzo(1,2,3)-thiadiazole-7-carbothioic acid S-methyl ester |
| SAR | systemic acquired resistance |
| SA | salicylic acid |
| PAL | phenylalanine ammonia-lyase |
| NPR1 | non-expressor of PR genes |
| PR | pathogenesis-related |
| JA | jasmonic acid |
| ET | ethylene |
| LAR | localized acquired resistance |
| ISR | induced systemic resistance |
| TMV | Tobacco mosaic virus |
| PGPR | plant growth-promoting rhizobacteria |
| MeSA | methyl salicylic acid |
| AzA | azelaic acid |
| DA | dihydroabetinal |
| G3P | glycerol-3-phosphate |
| PIP | pipecolic acid |
| GluB | glucan endo-1,3-beta-glucanase |
| TGA | basic leucine zipper transcription factor |
| ER | ethylene receptor |
| CTR | serine/threonine protein kinase |
| JAR1 | jasmonic acid-amido synthetase 1 |
| LOX | lipoxygenase |
| PI II | proteinase inhibitor II |
| HR | hypersensitive response |
| PCD | programmed cell death |
| OPDA | oxophytodienoic acid |
| ToMV | Tomato mosaic virus |
| MP | movement protein |
| CP | coat protein |
| JAZ | jasmonate ZIM domain |
| [Chol][BTHCOO] | liquid cholinium benzo(1,2,3)-thiadiazole-7-carboxylate |
| [N4444][BTHCOO] | tetrabutylammonium benzo(1,2,3)-thiadiazole-7-carboxylate |
| [N111010][BTHCOO] | didecyldimethylammonium benzo(1,2,3)-thiadiazole-7-carboxylate |
| BTHWA | amide derivative of benzo(1,2,3)-thiadiazole-7-carbothioic acid |
| α-MMC | alpha-momorcharin |
| ASM | acibenzolar-S-methyl |
| SABP2 | salicylic acid binding protein 2 |
| PBZ | probenazole |
| ICS | isochorismate synthase |
References
- Pelham, J. Resistance in tomato to Tobaco mosaic virus. Euphytica 1966, 15, 258–267. [Google Scholar] [CrossRef]
- Obrępalska-Stęplowska, A.; Renaut, J.; Planchon, S.; Przybylska, A.; Wieczorek, P.; Barylski, J.; Palukaitis, P. Effect of temperature on the pathogenesis, accumulation of viral and satellite RNAs and on plant proteome in peanut stunt virus and satellite RNA-infected plants. Front. Plant Sci. 2015, 6, 903. [Google Scholar] [CrossRef] [PubMed]
- Erb, M. Volatiles as inducers and suppressors of plant defense and immunity—Origins, specificity, perception and signaling. Curr. Opin. Plant Biol. 2018, 44, 117–121. [Google Scholar] [CrossRef] [PubMed]
- Beauverie, J. Essaisd’immunisation des vegetauxcontre les maladies cryptogamiques. CR Acad. Sci. Ser. III 1901, 133, 107–110. [Google Scholar]
- Jones, J.D.; Dangl, J.L. The plant immune system. Nature 2006, 444, 323–329. [Google Scholar] [CrossRef]
- Doods, P.N.; Rathjen, J.P. Plant immunity: Towards an integrated view of plant-pathogen interactions. Nat. Rev. Genet. 2010, 11, 539–548. [Google Scholar] [CrossRef]
- Gómez-Gómez, L.; Boller, T. Flagellin perception: A paradigm for innate immunity. Trends Plant Sci. 2002, 7, 251–256. [Google Scholar] [CrossRef]
- Flor, H.H. Current status of the gene for gene concept. Annu. Rev. Phytopathol. 1971, 9, 275–296. [Google Scholar] [CrossRef]
- Bektas, Y.; Eulgem, T. Synthetic plant defense elicitors. Front. Plant Sci. 2015, 5, 804. [Google Scholar] [CrossRef] [PubMed]
- Ross, A.F. Systemic acquired resistance induced by localized virus infections in plants. Virology 1961, 14, 340–358. [Google Scholar] [CrossRef]
- Choudhary, D.K.; Johri, B.N. Interactions of Bacillus spp.; plants—With special reference to induced systemic resistance (ISR). Microbiol. Res. 2009, 5, 493–513. [Google Scholar] [CrossRef] [PubMed]
- Choudhary, D.K.; Prakash, A.; Johri, B.N. Induced systemic resistance (ISR) in plants: Mechanism of action. Indian J. Microbiol. 2007, 4, 289–297. [Google Scholar] [CrossRef]
- van Wees, S.C.M.; Luijendijk, M.; Smoorenburg, I.; van Loon, L.C.; Pieterse, C.M.J. Rhizobacteria-mediated induced systemic resistance (ISR) in Arabidopsis is not associated with a direct effect on expression of known defense-related genes but stimulates the expression of the jasmonate-inducible gene Atvsp upon challenge. Plant Mol. Biol. 1999, 4, 537–549. [Google Scholar] [CrossRef]
- Heil, M.; Bostock, R.M. Induced Systemic Resistance (ISR) Against Pathogens in the Context of Induced Plant Defences. Ann. Bot. 2002, 5, 503–512. [Google Scholar] [CrossRef]
- Kuć, J. Concepts and Direction of Induced Systemic Resistance in Plants and its Application. Eur. J. Plant Pathol. 2001, 1, 7–12. [Google Scholar] [CrossRef]
- Tuzun, S. The Relationship Between Pathogen-induced Systemic Resistance (ISR) and Multigenic (horizontal) Resistance in Plants. Eur. J. Plant Pathol. 2001, 1, 85–93. [Google Scholar] [CrossRef]
- Felton, G.W.; Korth, K.L.; Bi, J.L.; Wesley, S.V.; Huhman, D.V.; Mathews, M.C.; Murphy, J.B.; Lamb, C.; Dixon, R.A. Inverse relationship between systemic resistance of plants to microorganisms and to insect herbivory. Curr. Biol. 1999, 6, 317–320. [Google Scholar] [CrossRef]
- Oostendorp, M.; Kunz, W.; Dietrich, B.; Staub, T. Induced Disease Resistance in Plants by Chemicals. Eur. J. Plant Pathol. 2001, 1, 19–28. [Google Scholar] [CrossRef]
- Gaffney, T.; Friedrich, L.; Vernooij, B.; Negrotto, D.; Nye, G.; Uknes, S.; Ward, E.; Kessmann, H.; Ryals, J. Requirement of Salicylic Acid for the Induction of Systemic Acquired Resistance. Science 1993, 5122, 754–756. [Google Scholar] [CrossRef] [PubMed]
- Min, L.; Huan, C.; Jian, C.; Ming, C.; Palmer, I.A.; Walter, G.; Fengquan, L.; Qing, F.Z. TCP Transcription Factors Interact with NPR1 and Contribute Redundantly to Systemic Acquired Resistance. Front. Plant Sci. 2018, 9, 1153. [Google Scholar]
- Klessig, D.F.; Choi, H.W.; Dempsey, D.M. Systemic Acquired Resistance and Salicylic Acid: Past, Present, and Future. Mol. Plant-Microbe Interact. 2018, 9, 871–888. [Google Scholar] [CrossRef] [PubMed]
- Wani, M.Y.; Mehraj, S.; Rather, R.A.; Rani, S.; Hajam, O.A.; Ganie, N.A.; Mir, M.R.; Baqual, M.F.; Kamili, A.S. Systemic acquired resistance (SAR): A novel strategy for plant protection with reference to mulberry. Int. J. Chem. Stud. 2018, 2, 1184–1192. [Google Scholar]
- Pospieszny, H. Systemic Acquired Resistance (SAR) in integrated plant protection. Prog. Plant Prot. 2016, 4, 436–442. [Google Scholar]
- Ross, A.F. Localized acquired resistance to plant virus infection in hypersensitive hosts. Virology 1961, 14, 329–339. [Google Scholar] [CrossRef]
- Ryals, J.A.; Neuenschwander, U.H.; Willits, M.G.; Molina, A.; Steiner, H.Y.; Hunt, M.D. Systemic acquired resistance. Plant Cell. 1996, 8, 1809–1819. [Google Scholar] [CrossRef]
- Conrath, U. Systemic Acquired Resistance. Plant Signalinbehav. 2006, 4, 179–184. [Google Scholar] [CrossRef]
- Kachroo, A.; Robin, G.P. Systemic signaling during plant defense. Curr. Opin. Plant Biol. 2013, 16, 527–533. [Google Scholar] [CrossRef] [PubMed]
- Bernsdorff, F.; Döring, A.C.; Gruner, K.; Schuck, S.; Bräutigam, A.; Zeier, J. Pipecolic Acid Orchestrates Plant Systemic Acquired Resistance and Defense Priming via Salicylic Acid-Dependent and -Independent Pathways. Plant Cell 2016, 28, 102–129. [Google Scholar] [CrossRef] [PubMed]
- Friedrich, L.; Lawton, K.; Ruess, W.; Masner, P.; Specker, N.; Gut Rella, M.; Meier, B.; Dincher, S.; Staub, T.; Uknes, S.; et al. A benzothiadiazole derivative induces systemic acquired resistance in tobacco. Plant J. 1996, 1, 61–70. [Google Scholar] [CrossRef]
- Lawton, K.; Friedrich, L.; Hunt, M.; Weymann, K.; Delaney, T.; Kessmann, H.; Staub, T.; Ryals, J. Benzothiadiazole induces disease resistance in Arabidopsis by activation of the systemic acquired resistance signal transduction pathway. Plant J. 1996, 10, 71–82. [Google Scholar] [CrossRef]
- Nischwitz, C.; Csinos, A.S.; Mullis, S.W.; Hickman, L.L.; Stevenson, K.L.; Gitaitis, R.D. Effect transplant age, tobacco cultivar Acibenzolar S-methyl, and imidacloprid on tomato spotted wilt infection in flue-cured tobacco. Plant Dis. 2008, 92, 1524–1528. [Google Scholar] [CrossRef]
- Anfoka, G.H. Benzo-(1,2,3)-thiadiazole-7-carbothioic acid S-methyl ester induces systemic resistance in tomato (Lycopersicum esculentum mill cv. Vollendung) to cucumber mosaic virus. Crop Protect. 2000, 19, 401–405. [Google Scholar] [CrossRef]
- Smith-Becker, J.; Keena, N.T.; Becker, J.O. Acibenzolar-S-methyl induces resistance to Colletotrichum lagenarium and Cucumber mosaic virus in cantaloupe. Crop Protect. 2003, 22, 769–774. [Google Scholar] [CrossRef]
- Görlach, J.; Volrath, S.; Knauf-Beiter, G.; Hengy, G.; Beckhove, U.; Kogel, K.H.; Oostendorp, M.; Staub, T.; Ward, E.; Kessmann, H.; et al. Benzothiadiazole, a novel class of inducers of systemic acquired resistance, activates gene expression and disease resistance in wheat. Plant Cell 1996, 8, 629–643. [Google Scholar] [CrossRef] [PubMed]
- Kunz, W.; Schurter, R.; Maetzke, T. The Chemistry of Benzothiadiazole Plant Activators. Pestic. Sci. 1997, 50, 275–282. [Google Scholar] [CrossRef]
- Heil, M.; Hilpert, A.; Kaiser, W.; Linesnmair, K.E. Reduced growth and seed set following chemical induction of pathogen defence: Does systemic acquired resistance (SAR) incur allocation costs? J. Ecol. 2000, 88, 645–654. [Google Scholar] [CrossRef]
- Smiglak, M.; Lewandowski, P.; Kukawka, R.; Budziszewska, M.; Krawczyk, K.; Obrepalska-Steplowska, A.; Pospieszny, H. Dual functional salts of benzo[1.2.3]thiadiazole-7-carboxylates as a highly efficient weapon against viral plant diseases. ACS Sustain. Chem. Eng. 2017, 5, 4197–4204. [Google Scholar] [CrossRef]
- Kukawka, R.; Czerwoniec, P.; Lewandkowski, P.; Pospieszny, H.; Smiglak, M. New ionic liquids based on systemic acquired resistance inducers combined with the phytotoxicity reducing cholinium cation. New J. Chem. 2018, 14, 11984–11990. [Google Scholar] [CrossRef]
- Kumar, D.; Haq, I.; Chapagai, D.; Tripathi, D.; Donald, D.; Hossain, M.; Devaiah, S. Hormone Signaling: Current Perspectives on the Roles of Salicylic Acid and Its Derivatives in Plants. Form. Struct. Act. Phytochem. Recent Adv. Phytochem. 2015, 45. [Google Scholar] [CrossRef]
- Glazebrook, J. Contrasting Mechanisms of Defense Against Biotrophic and Necrotrophic Pathogens. Annu. Rev. Phytopathol. 2005, 43, 205–227. [Google Scholar] [CrossRef]
- Nahar, K.; Kyndt, T.; De Vleesschauwer, D.; Höfte, M.; Gheysen, G. The Jasmonate Pathway Is a Key Player in Systemically Induced Defense against Root Knot Nematodes in Rice. Plant Physiol. 2011, 1, 305–316. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Chang, P.L.C.; Liu, D.; Narasimhan, M.L.; Kashchandra, G.R.; Hasegawa, P.M.; Bressan, R.A. Plant defense genes are synergistically induced by ethylene and methyl jasmonate. Plant Cell. 1994, 6, 1077–1085. [Google Scholar] [CrossRef]
- Schenk, P.M.; Kazan, K.; Wilson, I.; Anderson, J.P.; Richmond, T.; Somerville, S.C.; Manners, J.M. Coordinated plant defense responses in Arabidopsis revealed by microarray analysis. Proc. Natl. Acad. Sci. USA 2000, 21, 11655–11660. [Google Scholar] [CrossRef] [PubMed]
- Kunkel, B.N.; Brooks, D.M. Cross talk between signaling pathways in pathogen defense. Curr. Opin. Plant Biol. 2002, 5, 325–331. [Google Scholar] [CrossRef]
- Yu, X.; Armstrong, C.M.; Zhou, M.; Duan, Y. Bismerthiazol Inhibits Xanthomonas citri subsp. Citri Growth and Induces Differential Expression of Citrus Defense-Related Genes. Phytopatology 2016, 106, 693–701. [Google Scholar] [CrossRef] [PubMed]
- Das, S.; Chakraborty, P.; Mandal, P.; Saha, D.; Saha, A. Phenylalanine ammonia-lyase gene induction with benzothiadiazole elevates defence against Lasiodiplodia theobromae in tea in India. J. Phytopathol. 2017, 165, 755–761. [Google Scholar] [CrossRef]
- Liu, S.-I.; Wu, J.; Zhang, P.; His, G.; Huang, Y.; Lu, J.; Zhang, Y.-I. Response of phytohormones and correlation of SAR signal pathway genes to the different resistance levels of grapevine against Plasmopara viticola infection. Plant Physiol. Biochem. 2016, 107, 56–66. [Google Scholar] [CrossRef] [PubMed]
- Loake, G.; Grant, M. Salicylic acid in plant defense—The players and protagonists. J. Curr. Opin. Plant Biol. 2007, 10, 466–472. [Google Scholar] [CrossRef] [PubMed]
- Carr, J.P.; Lewsey, M.G.; Palukaitis, P. Signalling in Induced Resistance. Adv. Virus Res. 2010, 76, 57–121. [Google Scholar] [PubMed]
- Maleck, K.; Dietrich, R.A. Defense on multiple fronts: How do plants cope with diverse enemies? Trends Plant Sci. 1999, 6, 215–219. [Google Scholar] [CrossRef]
- An, G.; Mitra, A.; Choi, H.K.; Costa, M.A.; An, K.; Thornburg, R.W.; Ryan, C.A. Functional analysis of the 3’ control region of the potato wound-inducible proteinase inhibitor II gene. Plant Cell 1989, 1, 115–122. [Google Scholar] [PubMed]
- Stone, J.M.; Walker, J.C. Plant Protein Kinase Families and Signal Transduction. Plant Physiol. 1995, 108, 451–457. [Google Scholar] [CrossRef] [PubMed]
- Kubota, K.; Tsuda, S.; Tamai, A.; Meshi, T. Tomato Mosaic Virus Replication Protein Suppresses Virus-Targeted Posttranscriptional Gene Silencing. J. Virol. 2003, 20, 11016–11026. [Google Scholar] [CrossRef]
- He, M.; He, C.-Q.; Ding, N.-Z. Natural recombination between tobacco and tomato mosaic viruses. Virus Res. 2012, 163, 374–379. [Google Scholar] [CrossRef] [PubMed]
- Hussein, W.; Awad, H.; Fahim, S. Systemic Resistance Induction of Tomato Plants against ToMV Virus by Surfactin Produced from Bacillus subtilis BMG02. Am. J. Microbiol. Res. 2016, 4, 153–158. [Google Scholar]
- Roossinck, M.J.; García-Arenal, F. Ecosystem simplification, biodiversity loss and plant virus emergence. Curr. Opin. Virol. 2015, 10, 56–62. [Google Scholar] [CrossRef] [PubMed]
- Wren, J.D.; Roossinck, M.J.; Nelson, R.S.; Scheets, K.; Palmer, M.W.; Melcher, U. Plant Virus Biodiversity and Ecology. Plosbiology 2006, 3, e80. [Google Scholar] [CrossRef] [PubMed]
- Moriones, E.; Navas-Castillo, J. Tomato yellow leaf curl virus, an emerging virus complex causing epidemics worldwide. Virus Res. 2000, 1–2, 123–134. [Google Scholar] [CrossRef]
- Hanssen, I.M.; Paeleman, A.; Vandewoestijne, E.; Van Bergen, L.; Bragard, C.; Lievens, B.; Vanachter, A.C.R.C.; Thomma, B.P.H.J. Pepino mosaic virus isolates and differential symptomatology in tomato. Plant Pathol. 2009, 58, 450–460. [Google Scholar] [CrossRef]
- Whitham, S.; McCormick, S.; Baker, B. The N gene of tobacco confers resistance to tobacco mosaic virus in transgenic tomato. Plant Biol. 1996, 93, 8776–8781. [Google Scholar] [CrossRef]
- Broadbent, L. Epidemiology and control of Tomato Mosaic Virus. Annu. Rev. 1976, 1. [Google Scholar] [CrossRef]
- Hammerschmidt, R.; Kuc, J. Induced Resistance to Disease in Plants; Kluwer Academic Publishers: Dordrecht, The Netherlands, 1995. [Google Scholar]
- Kuc, J. Induced immunity to plant disease. Bioscience 1982, 32, 854–860. [Google Scholar]
- Shine, M.B.; Xiao, X.; Kachroo, P.; Kachroo, A. Signaling mechanisms underlying systemic acquired resistance to microbal pathogens. Plant Sci. 2018. [Google Scholar] [CrossRef]
- Mauch-Mani, B.; Baccelli, I.; Luna, E.; Flors, V. Defense Priming: An Adaptive Part of Induce Resistance. Annu. Rev. Plant. Biol. 2017, 68, 16.1–16.28. [Google Scholar] [CrossRef]
- Thakur, M.; Sohal, B.S. Role of Elicitors in Inducing Resistance in Plants against Pathogen Infection: A Review. ISRN Biochem. 2013. [Google Scholar] [CrossRef] [PubMed]
- Smiglak, M.; Pospieszny, H.; Maciejewski, H.; Kukawka, R.; Lewandowski, P. Środek do ochrony roślin. PL Patent 230659, 30 September 2013. [Google Scholar]
- Smiglak, M.; Pospieszny, H.; Maciejewski, H.; Kukawka, R.; Lewandowski, P. Środek do ochrony roślin. PL Patent 231217, 1 October 2013. [Google Scholar]
- Smiglak, M.; Pospieszny, H.; Kukawka, R.; Lewandowski, P.; Stolarska, O.; Maciejewski, H. Application of 7-Carboxybenzo[1,2,3]thiadiazole Amides as Plant Stimulants. PCT/IB2016/054491, 26 July 2016. [Google Scholar]
- Kouzai, Y.; Noutoshi, Y.; Inoue, K.; Shimizu, M.; Onda, Y.; Mochida, K. Benzothiadiazole, a plant defense inducer, negatively regulates sheath blight resistance in Brachypodium distachyon. Sci. Rep. 2018, 26, 17358. [Google Scholar] [CrossRef]
- Durner, J.; Wendehenne, D.; Klessig, D.F. Defense gene induction in tobacco by nitric oxide, cyclic GMP, and cyclic ADP-ribose. Plant Biol. 1998, 95, 10328–10333. [Google Scholar] [CrossRef]
- Pallas, J.A.; Paiva, N.L.; Lamb, C.; Dixon, R.A. Tobacco plants epigenetically suppressed in phenylalanine ammonia-lyase expression do not develop systemic acquired resistance in response to infection by tobacco mosaic virus. Plant J. 1996, 2, 281–293. [Google Scholar] [CrossRef]
- Chen, Z.; Zheng, Z.; Huang, J.; Lai, Z.; Fan, B. Biosynthesis of salicylic acid in plants. Plant Signal Behav. 2009, 4, 493–496. [Google Scholar] [CrossRef]
- Yu, X.Z.; Fan, W.J.; Lin, Y.J.; Zhang, F.-F.; Gupta, D.K. Differential expression of the PAL gene family in rice seedlings exposed to chromium by microarray analysis. Ecotoxicology 2018, 27, 325. [Google Scholar] [CrossRef]
- Yu, J.; Gao, J.; Wang, X.Y.; Wei, Q.; Yang, L.F.; Qiu, K.; Kuai, B.K. The Pathway and Regulation of Salicylic Acid Biosynthesis in Probenazole-Treated Arabidopsis. J. Plant Biol. 2010, 53, 417–424. [Google Scholar] [CrossRef]
- Wong, C.E.; Carson, R.A.J.; Carr, J.P. Chemically-induced virus resistance in Arabidopsis thaliana is independent of pathogenesis-related protein expression and the NPR1 gene. Mol. Plant-Microbe Interact. 2002, 15, 75–81. [Google Scholar] [CrossRef] [PubMed]
- Mou, Z.; Fan, W.; Dong, X. Inducers of Plant Systemic Acquired Resistance Regulate NPR1 Function through Redox Changes. Cell 2003, 113, 935–944. [Google Scholar] [CrossRef]
- Wiśniewska, A.; Kamińska, K.; Nawrocka, K. The role of salicylic and jasmonic acids in plant response. Postępy Biol. Komórki 2015, 42, 743–764. [Google Scholar]
- Ng, G.; Seabolt, S.; Zhang, C.; Salimian, S.; Watkins, T.A.; Lu, H. Genetic Dissection of Salicylic Acid-Mediated Defense Signaling Networks in Arabidopsis. Genetics 2011, 189, 851–859. [Google Scholar] [CrossRef] [PubMed]
- Nakayama, A.; Fukushima, S.; Goto, S.; Matsushita, A.; Shimono, M.; Sugano, S.; Jiang, C.-J.; Akagi, A.; Yamazaki, M.; Inoue, H.; et al. Genome-wide identification of WRKY45-regulated genes that mediate benzothiadiazole-induced defense responses in rice. BMC Plant Biol. 2013, 13, 150. [Google Scholar] [CrossRef] [PubMed]
- Kauffmann, S.; Legrand, M.; Geoffroy, P.; Fritig, B. Biological function of ‘pathogenesis-related’ proteins: Four PR proteins of tobacco have 1,3-f-glucanase activity. EMBO J. 1987, 11, 3209–3212. [Google Scholar] [CrossRef]
- Livne, B.; Faktor, O.; Zeitoune, S.; Edelbauma, O.; Sela, I. TMV-induced expression of tobacco β-glucanase promoter activity is mediated by a single, inverted, GCC motif. Plant Sci. 1997, 2, 159–169. [Google Scholar] [CrossRef]
- Bucher, G.L.; Tarina, C.; Heinlein, M.; Di Serio, F.; Meins, F., Jr.; Iglesias, V.A. Local expression of enzymatically active class I β-1, 3-glucanase enhances symptoms of TMV infection in tobacco. Plant J. 2001, 28, 361–369. [Google Scholar] [CrossRef] [PubMed]
- Zavaliev, R.; Levy, A.; Gera, A.; Epel, B.L. Subcellular Dynamics and Role of Arabidopsis β-1,3-Glucanases in Cell-to-Cell Movement of Tobamoviruses. Mol Plant-Microbe Interact. 2013, 26, 1016–1030. [Google Scholar] [CrossRef]
- Levy, A.; Guenoune-Gelbart, D.; Epel, B.L. β-1,3-Glucanases. Plasmodesmal Gate Keepers for Intercellular Communication. Plant Signal Behav. 2007, 5, 404–407. [Google Scholar] [CrossRef]
- Palukaitis, P.; Carr, J.P. Plant resistance responses to viruses. J. Plant Pathol. 2008, 2, 153–171. [Google Scholar]
- Chini, A.; Fonseca, S.; Fernández, G.; Adie, B.; Chico, J.M.; Lorenzo, O.; García-Casado, G.; López-Vidriero, I.; Lozano, F.M.; Ponce, M.R.; Micol, J.L.; et al. The JAZ family of repressors is the missing link in jasmonate signaling. Nature 2007, 448, 666–671. [Google Scholar] [CrossRef] [PubMed]
- Chung, H.S.; Koo, A.J.K.; Gao, X.; Jayanty, S.; Thines, B.; Jones, A.D.; Howe, G.A. Regulation and function of Arabidopsis JASMONATE ZIM-domain genes in response to wounding and herbivory. Plant Physiol. 2008, 146, 952–964. [Google Scholar] [CrossRef] [PubMed]
- Park, K.-S.; Cheong, J.-J.; Lee, S.-J.; Suh, M.-C.; Choi, D. A novel proteinase inhibitor gene transiently induced by tobacco mosaic virus infection. Biochim. Biophys. Acta 2000, 1492, 509–512. [Google Scholar] [CrossRef]
- Merchante, C.; Alonso, J.M.; Stepanova, A.N. Ethylene signaling: Simple ligand, complex regulation. Curr. Opin. Plant Biol. 2013, 16, 554–560. [Google Scholar] [CrossRef] [PubMed]
- Yanga, T.; Zhua, L.; Mengb, Y.; Lva, R.; Zhoua, Z.; Zhua, L.; Lina, H.; Xia, D. Alpha-momorcharin enhances Tobacco mosaic virus resistance in tobacco NN by manipulating jasmonic acid-salicylic acid crosstalk. J. Plant Physiol. 2018, 223, 116–126. [Google Scholar] [CrossRef]
- Lenk, M.; Wenig, M.; Mengel, F.; Häußler, F.; Vlot, C. Arabidopsis thaliana Immunity-Related Compounds Modulate Disease Susceptibility in Barley. Agronomy 2018, 142. [Google Scholar] [CrossRef]
- Pieterse, C.M.J.; van Wees, S.C.M.; van Pelt, J.A.; Knoester, M.; Laan, R.; Gerrits, H.; Weisbeek, P.J.; van Loon, L.C. A Novel Signaling Pathway Controlling Induced Systemic Resistance in Arabidopsis. Plant Cell 1998, 10, 1571–1580. [Google Scholar] [CrossRef]
- Schweiger, R.; Heise, A.-M.; Persicke, M.; Müller, C. Interactions between the jasmonic and salicylic acid pathway modulate the plant metabolome and affect herbivores of different feeding types. Plant Cell Environ. 2014, 37, 1574–1585. [Google Scholar] [CrossRef]
- Johnson, R.; Narvez, J.; An, G.; Ryan, C. Expression of proteinase inhibitors I and II in transgenic tobacco plants: Effects on natural defense against Manduca sexta larvae. Botany 1989, 86, 9871–9875. [Google Scholar] [CrossRef]
- Inbar, M.; Doostdar, H.; Gerling, D.; Mayer, R.T. Induction of systemic acquired resistance in cotton by BTH has a negligible effect on phytophagous insects. Entomol. Exp. Appl. 2001, 99, 65–70. [Google Scholar] [CrossRef]
- Van Loon, L.C. Induced resistance in plants and the role of pathogenesis-related proteins. Eur. J. Plant Pathol. 1997, 103, 753–765. [Google Scholar] [CrossRef]
- Oliveira, M.D.M.; Varanda, C.M.R.; Félix, M.R.F. Induced resistance during the interaction pathogen x plant and the use of resistance inducers. Phytochem. Lett. 2016, 15, 152–158. [Google Scholar] [CrossRef]
- Mandadi, K.K.; Scholthof, K.-B.G. Plant Immune Responses Against Viruses: How Does a Virus Cause Disease? Plant Cell 2013, 5, 1489–1505. [Google Scholar] [CrossRef] [PubMed]
- Trejo-Saavedra, D.L.; García-Neria, M.A.; Rivera-Bustamante, R.F. Benzothiadiazole (BTH) induces resistance to Pepper golden mosaic virus (PepGMV) in pepper (Capsicum annuum L.). Biol. Res. 2013, 4, 333–340. [Google Scholar] [CrossRef] [PubMed]
- Kessmann, H.; Staub, T.; Hofmann, C.; Maetzke, T.; Herzog, J.; Ward, E.; Uknes, S.; Ryals, J. Induction of systemic acquired resistance in plants by chemicals. Ann. Rev. Phytopathol. 1994, 32, 439–459. [Google Scholar] [CrossRef]
- Smith, K.M. Plant Viruses, 6th ed.; John Wiley & Sons: Hoboken, NJ, USA, 1977; pp. 43–53. [Google Scholar]
- Wieczorek, P.; Wrzesińska, B.; Obrępalska-Stęplowska, A. Assessment of reference gene stability influenced by extremely divergent disease symptoms in Solannumlycopersicum L. J. Virol. Methods 2013, 194, 161–168. [Google Scholar] [CrossRef]
- Untergasser, A.; Cutcutache, I.; Koressaar, T.; Ye, J.; Faircloth, B.C.; Remm, M.; Rozen, S.G. Primer3—New capabilities and interfaces. Nucleic Acids Res. 2012, 15, e115. [Google Scholar] [CrossRef]

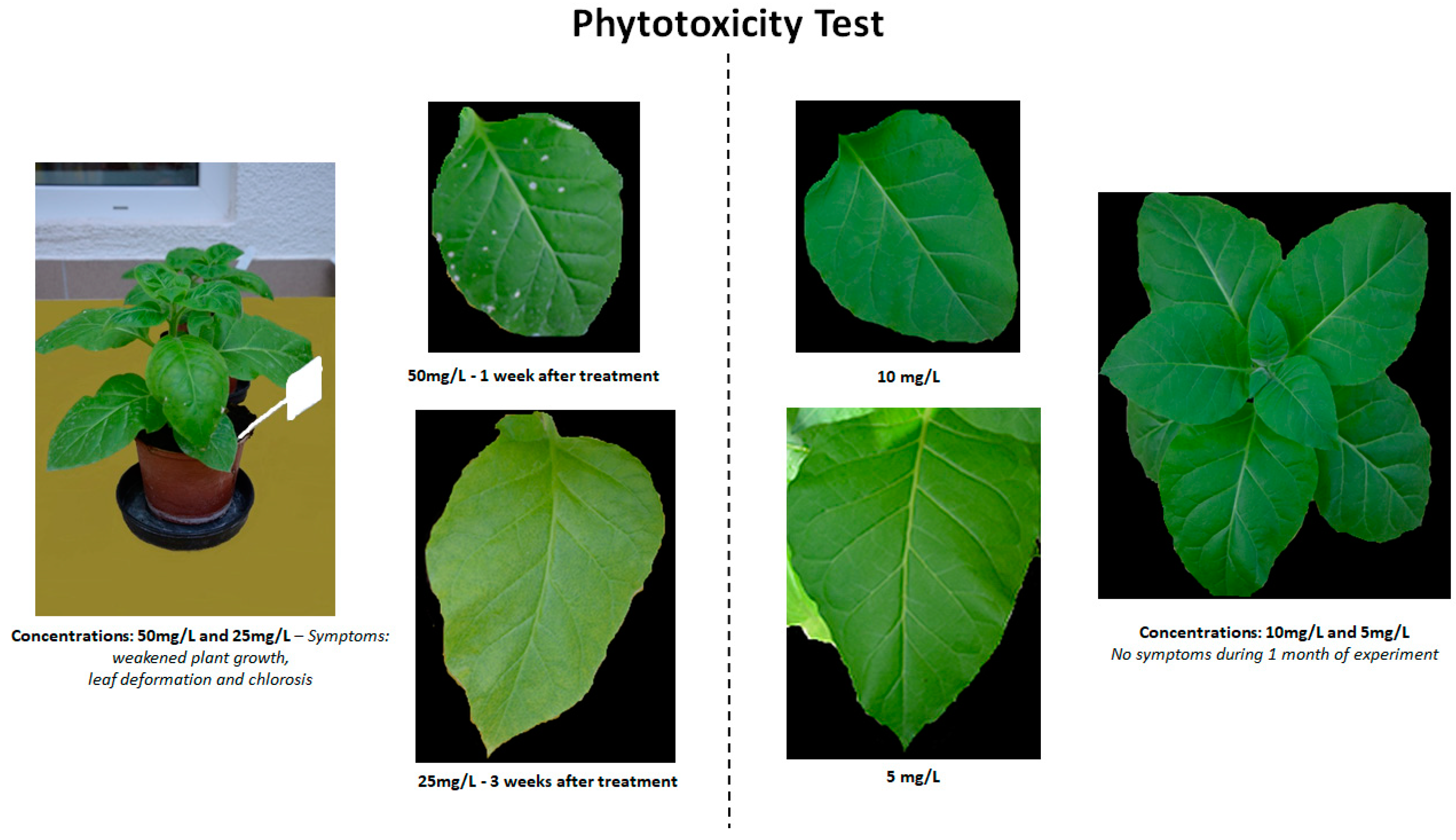


| Compound/Concentration | 5 mg/L | 10 mg/L | 25 mg/L | 50 mg/L |
|---|---|---|---|---|
| BTH | - | - | * | + |
| [Chol][BTHCOO] | - | - | * | + |
| BTHWA | - | - | * | + |
| [N4444][BTHCOO] | - | - | * | + |
| [N111010][BTHCOO] | - | - | * | + |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Frąckowiak, P.; Pospieszny, H.; Smiglak, M.; Obrępalska-Stęplowska, A. Assessment of the Efficacy and Mode of Action of Benzo(1,2,3)-Thiadiazole-7-Carbothioic Acid S-Methyl Ester (BTH) and Its Derivatives in Plant Protection Against Viral Disease. Int. J. Mol. Sci. 2019, 20, 1598. https://doi.org/10.3390/ijms20071598
Frąckowiak P, Pospieszny H, Smiglak M, Obrępalska-Stęplowska A. Assessment of the Efficacy and Mode of Action of Benzo(1,2,3)-Thiadiazole-7-Carbothioic Acid S-Methyl Ester (BTH) and Its Derivatives in Plant Protection Against Viral Disease. International Journal of Molecular Sciences. 2019; 20(7):1598. https://doi.org/10.3390/ijms20071598
Chicago/Turabian StyleFrąckowiak, Patryk, Henryk Pospieszny, Marcin Smiglak, and Aleksandra Obrępalska-Stęplowska. 2019. "Assessment of the Efficacy and Mode of Action of Benzo(1,2,3)-Thiadiazole-7-Carbothioic Acid S-Methyl Ester (BTH) and Its Derivatives in Plant Protection Against Viral Disease" International Journal of Molecular Sciences 20, no. 7: 1598. https://doi.org/10.3390/ijms20071598
APA StyleFrąckowiak, P., Pospieszny, H., Smiglak, M., & Obrępalska-Stęplowska, A. (2019). Assessment of the Efficacy and Mode of Action of Benzo(1,2,3)-Thiadiazole-7-Carbothioic Acid S-Methyl Ester (BTH) and Its Derivatives in Plant Protection Against Viral Disease. International Journal of Molecular Sciences, 20(7), 1598. https://doi.org/10.3390/ijms20071598