A Smart Imaging Workflow for Organ-Specific Screening in a Cystic Kidney Zebrafish Disease Model
Abstract
1. Introduction
2. Results
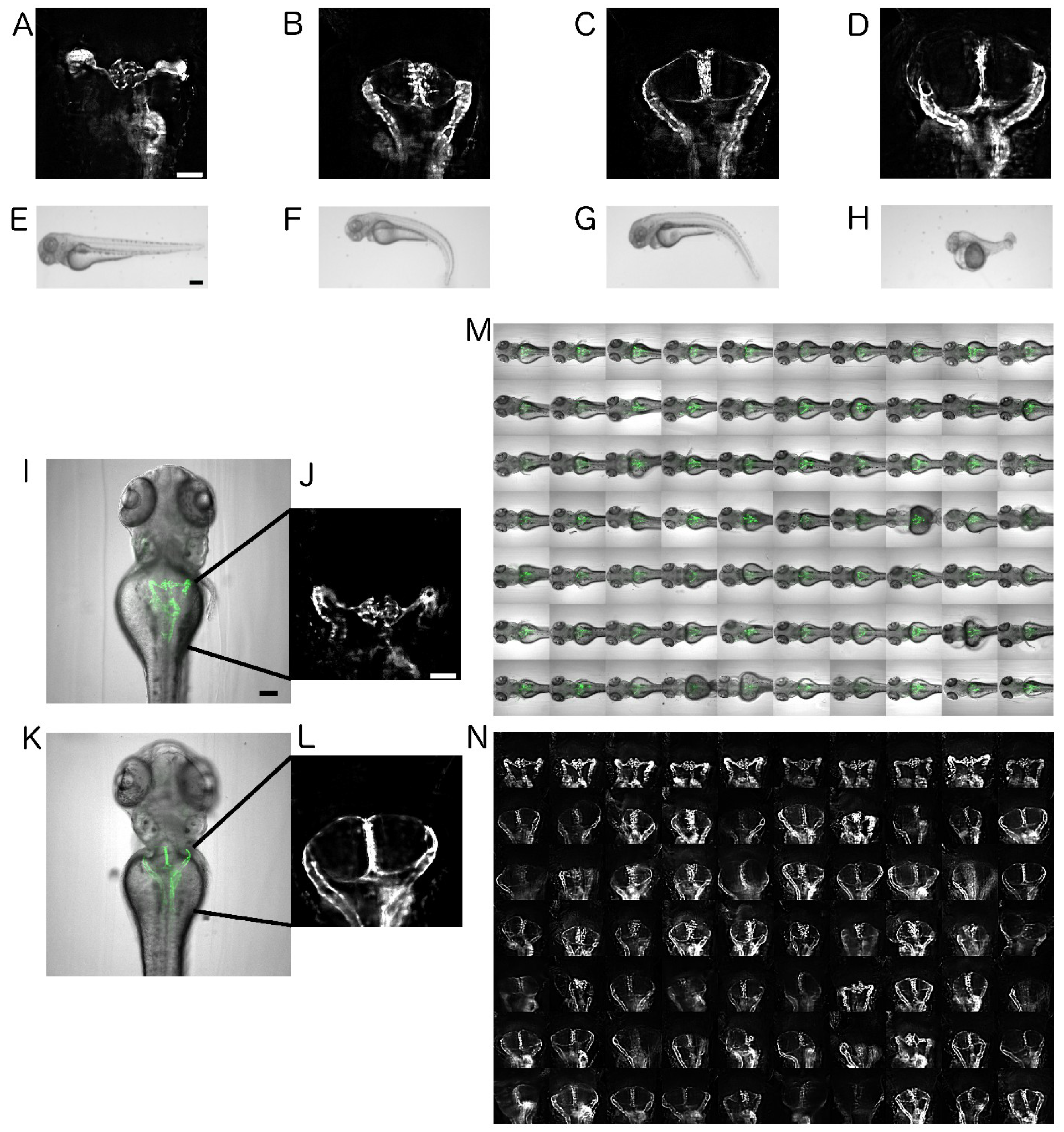
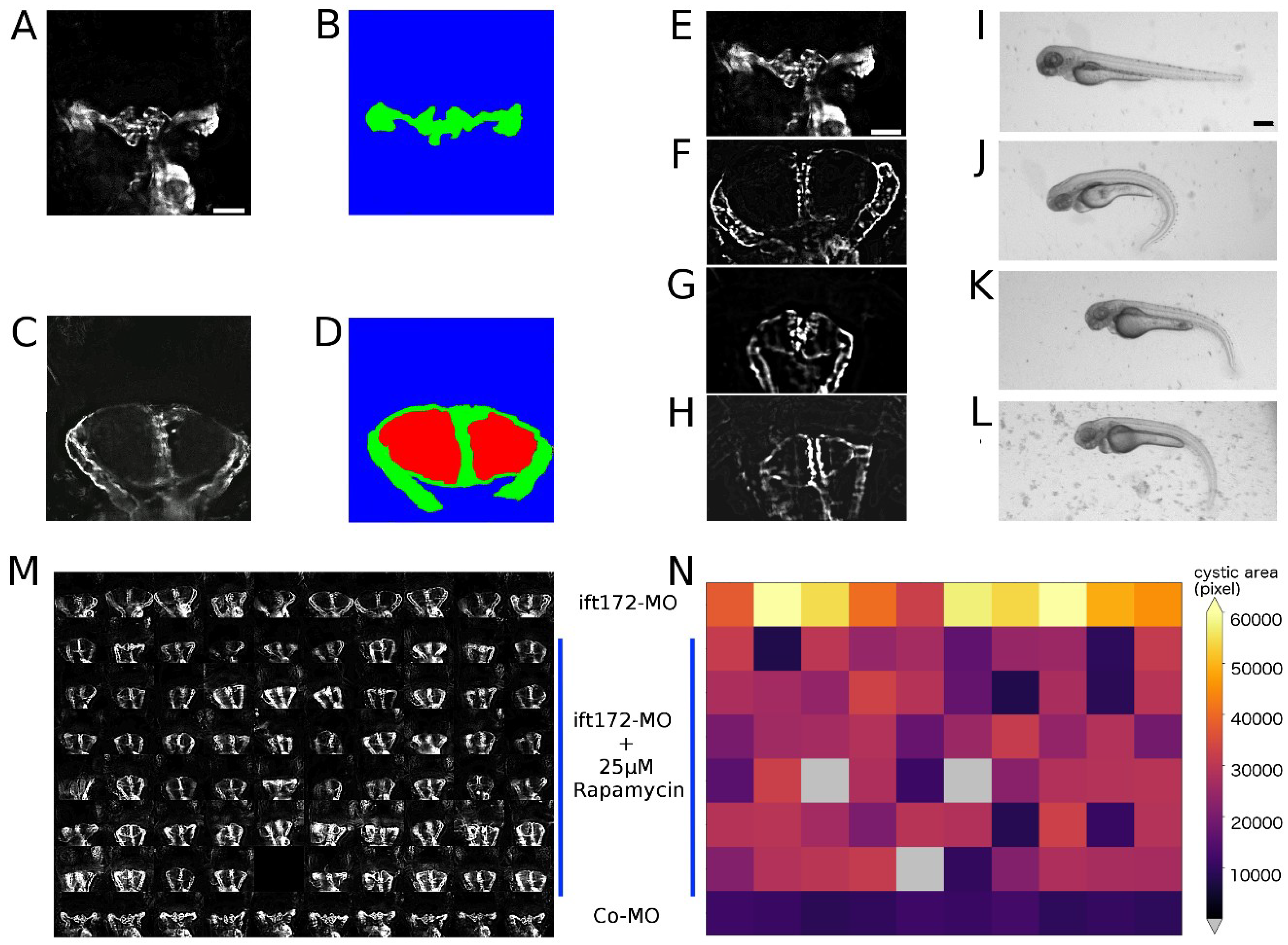
2.1. Establishment of Disease Model and Screening Assay
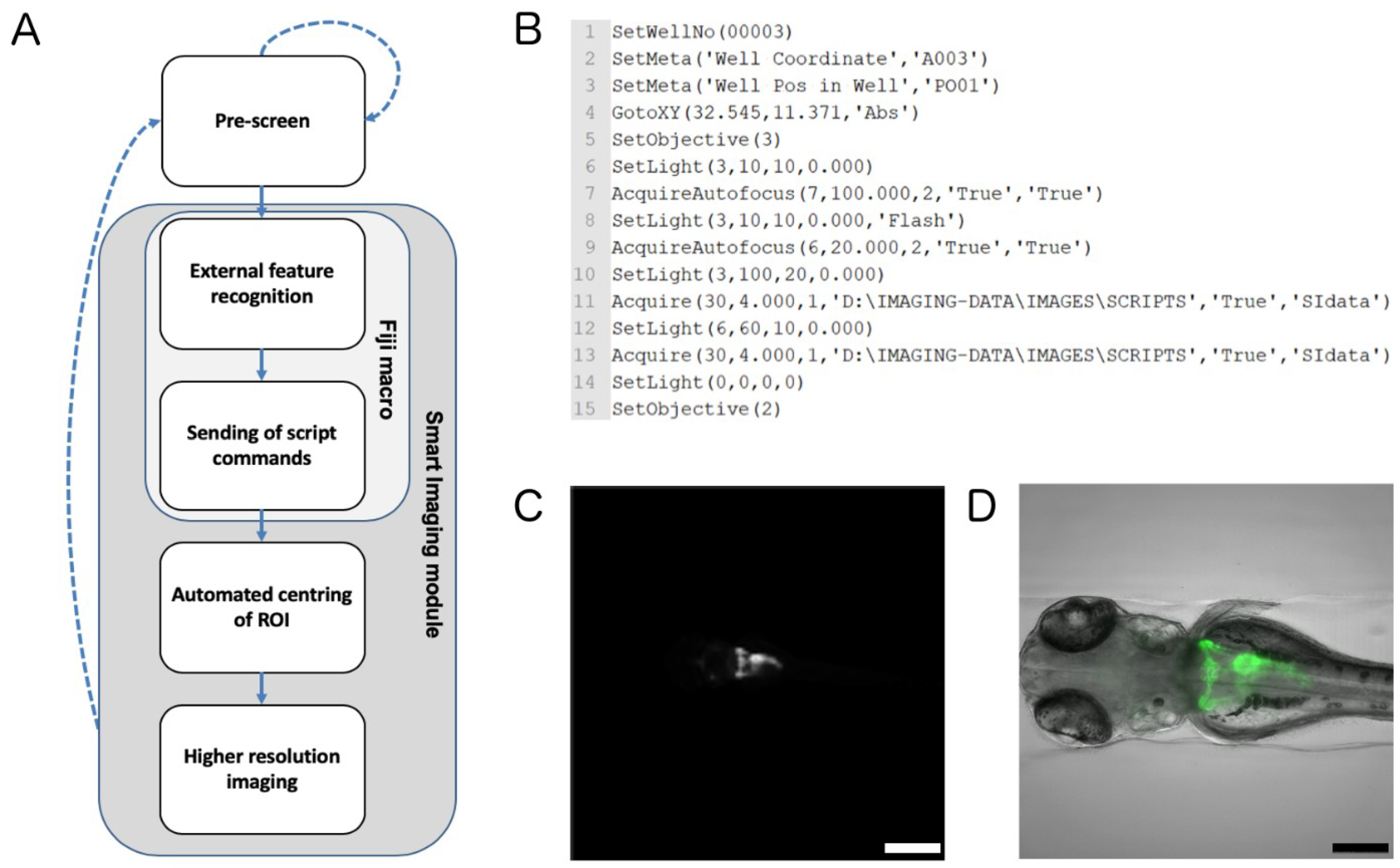
2.2. Automated Smart Microscopy of Kidney Regions
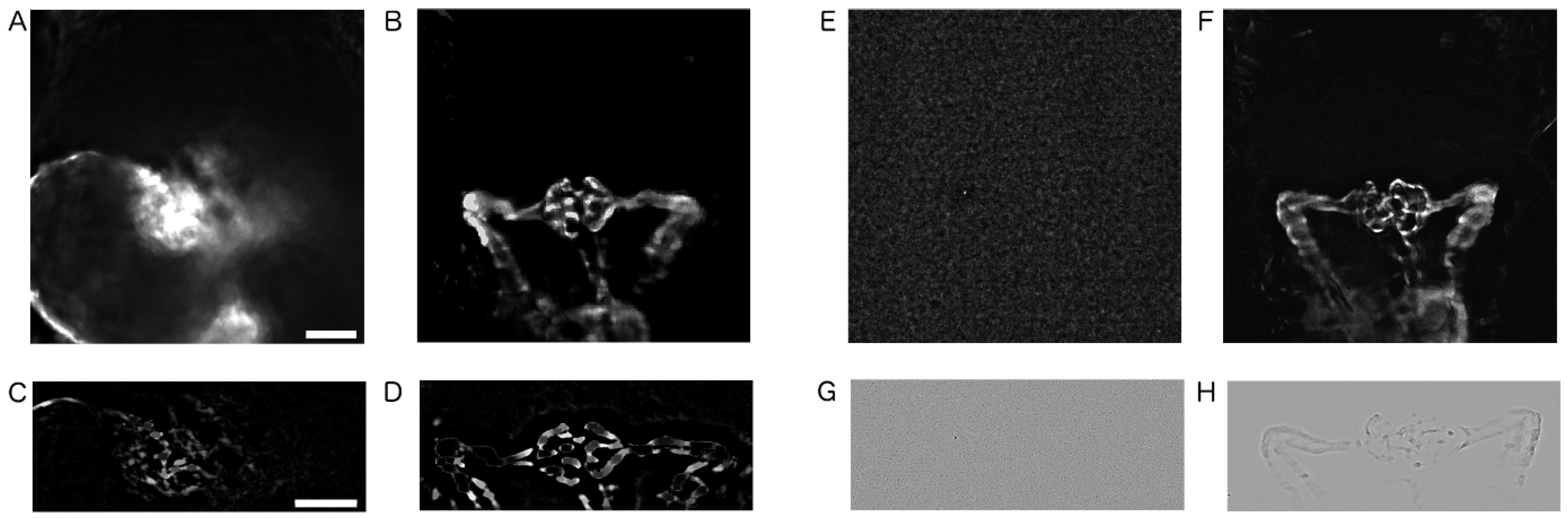
2.3. Image Pre-Processing
2.4. Image Quality Check and Phenotype Categorisation
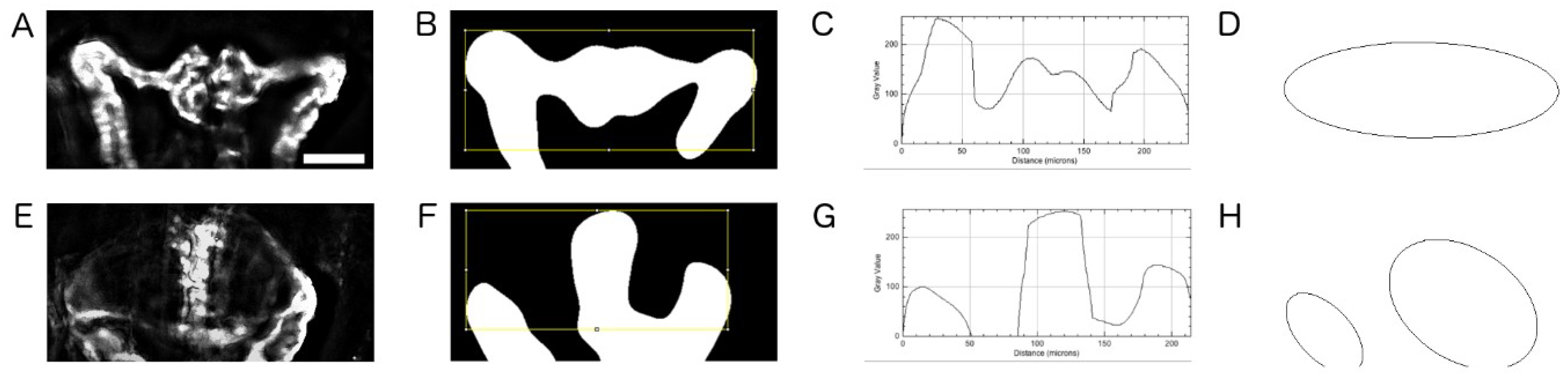
2.5. Quantification of Cystic Areas
3. Discussion
4. Materials and Methods
4.1. Ethics Statement
4.2. Fish Keeping and Embryo Handling
4.3. Morpholino Injections
4.4. Drug Treatment of Embryos
4.5. Preparation of Agarose-Filled 96-Well Plates and Embryo Positioning
4.6. Automated Image Acquisition, Smart Imaging and Data Handling
4.7. Image Pre-Processing
4.8. Image Categorisation and Image Quality Control
4.9. Image Based Quantification of Kidney Cyst Areas
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Lieschke, G.J.; Currie, P.D. Animal models of human disease: Zebrafish swim into view. Nat. Rev. Genet. 2007, 8, 353–367. [Google Scholar] [CrossRef] [PubMed]
- Santoriello, C.; Zon, L.I. Hooked! Modeling human disease in zebrafish. J. Clin. Investig. 2012, 122, 2337–2343. [Google Scholar] [CrossRef] [PubMed]
- Phillips, J.B.; Westerfield, M. Zebrafish models in translational research: Tipping the scales toward advancements in human health. Dis. Models Mech. 2014, 7, 739–743. [Google Scholar] [CrossRef] [PubMed]
- Bradford, Y.M.; Toro, S.; Ramachandran, S.; Ruzicka, L.; Howe, D.G.; Eagle, A.; Kalita, P.; Martin, R.; Taylor Moxon, S.A.; Schaper, K.; et al. Zebrafish models of human disease: Gaining insight into human disease at zfin. ILAR J. 2017, 58, 4–16. [Google Scholar] [CrossRef] [PubMed]
- Davis, E.E.; Frangakis, S.; Katsanis, N. Interpreting human genetic variation with in vivo zebrafish assays. Biochim. Biophys. Acta 2014, 1842, 1960–1970. [Google Scholar] [CrossRef] [PubMed]
- Idilli, A.I.; Precazzini, F.; Mione, M.C.; Anelli, V. Zebrafish in translational cancer research: Insight into leukemia, melanoma, glioma and endocrine tumor biology. Genes 2017, 8, 236. [Google Scholar] [CrossRef]
- Masud, S.; Torraca, V.; Meijer, A.H. Modeling infectious diseases in the context of a developing immune system. Curr. Top. Dev. Biol. 2017, 124, 277–329. [Google Scholar] [PubMed]
- Seth, A.; Stemple, D.L.; Barroso, I. The emerging use of zebrafish to model metabolic disease. Dis. Models Mech. 2013, 6, 1080–1088. [Google Scholar] [CrossRef]
- Rennekamp, A.J.; Peterson, R.T. 15 years of zebrafish chemical screening. Curr. Opin. Chem. Biol. 2015, 24, 58–70. [Google Scholar] [CrossRef]
- Garcia, G.R.; Noyes, P.D.; Tanguay, R.L. Advancements in zebrafish applications for 21st century toxicology. Pharmacol. Ther. 2016, 161, 11–21. [Google Scholar] [CrossRef]
- Esner, M.; Meyenhofer, F.; Bickle, M. Live-cell high content screening in drug development. Methods Mol. Biol. 2018, 1683, 149–164. [Google Scholar] [PubMed]
- Peterson, R.T.; Fishman, M.C. Designing zebrafish chemical screens. Methods Cell Biol. 2011, 105, 525–541. [Google Scholar] [PubMed]
- Gehrig, J.; Pandey, G.; Westhoff, J.H. Zebrafish as a model for drug screening in genetic kidney diseases. Front Pediatr. 2018, 6, 183. [Google Scholar] [CrossRef]
- Pardo-Martin, C.; Chang, T.Y.; Koo, B.K.; Gilleland, C.L.; Wasserman, S.C.; Yanik, M.F. High-throughput in vivo vertebrate screening. Nat. Methods 2010, 7, 634–636. [Google Scholar] [CrossRef] [PubMed]
- Wittbrodt, J.N.; Liebel, U.; Gehrig, J. Generation of orientation tools for automated zebrafish screening assays using desktop 3d printing. BMC Biotechnol. 2014, 14, 36. [Google Scholar] [CrossRef] [PubMed]
- Megason, S.G. In toto imaging of embryogenesis with confocal time-lapse microscopy. Methods Mol. Biol. 2009, 546, 317–332. [Google Scholar] [PubMed]
- Alessandri, K.; Andrique, L.; Feyeux, M.; Bikfalvi, A.; Nassoy, P.; Recher, G. All-in-one 3d printed microscopy chamber for multidimensional imaging, the universlide. Sci. Rep. 2017, 7, 42378. [Google Scholar] [CrossRef] [PubMed]
- Westhoff, J.H.; Giselbrecht, S.; Schmidts, M.; Schindler, S.; Beales, P.L.; Tonshoff, B.; Liebel, U.; Gehrig, J. Development of an automated imaging pipeline for the analysis of the zebrafish larval kidney. PLoS ONE 2013, 8, e82137. [Google Scholar] [CrossRef]
- Peravali, R.; Gehrig, J.; Giselbrecht, S.; Lutjohann, D.S.; Hadzhiev, Y.; Muller, F.; Liebel, U. Automated feature detection and imaging for high-resolution screening of zebrafish embryos. Biotechniques 2011, 50, 319–324. [Google Scholar] [CrossRef] [PubMed]
- Gunkel, M.; Eberle, J.P.; Erfle, H. Fluorescence-based high-throughput and targeted image acquisition and analysis for phenotypic screening. Methods Mol. Biol. 2017, 1563, 269–280. [Google Scholar] [PubMed]
- Tischer, C.; Hilsenstein, V.; Hanson, K.; Pepperkok, R. Adaptive fluorescence microscopy by online feedback image analysis. Methods Cell Biol. 2014, 123, 489–503. [Google Scholar]
- Mikut, R.; Dickmeis, T.; Driever, W.; Geurts, P.; Hamprecht, F.A.; Kausler, B.X.; Ledesma-Carbayo, M.J.; Maree, R.; Mikula, K.; Pantazis, P.; et al. Automated processing of zebrafish imaging data: A survey. Zebrafish 2013, 10, 401–421. [Google Scholar] [CrossRef]
- Eliceiri, K.W.; Berthold, M.R.; Goldberg, I.G.; Ibanez, L.; Manjunath, B.S.; Martone, M.E.; Murphy, R.F.; Peng, H.; Plant, A.L.; Roysam, B.; et al. Biological imaging software tools. Nat. Methods 2012, 9, 697–710. [Google Scholar] [CrossRef]
- Caicedo, J.C.; Cooper, S.; Heigwer, F.; Warchal, S.; Qiu, P.; Molnar, C.; Vasilevich, A.S.; Barry, J.D.; Bansal, H.S.; Kraus, O.; et al. Data-analysis strategies for image-based cell profiling. Nat. Methods 2017, 14, 849–863. [Google Scholar] [CrossRef] [PubMed]
- Halbritter, J.; Bizet, A.A.; Schmidts, M.; Porath, J.D.; Braun, D.A.; Gee, H.Y.; McInerney-Leo, A.M.; Krug, P.; Filhol, E.; Davis, E.E.; et al. Defects in the ift-b component ift172 cause jeune and mainzer-saldino syndromes in humans. Am. J. Hum. Genet. 2013, 93, 915–925. [Google Scholar] [CrossRef]
- Perner, B.; Englert, C.; Bollig, F. The wilms tumor genes wt1a and wt1b control different steps during formation of the zebrafish pronephros. Dev. Biol. 2007, 309, 87–96. [Google Scholar] [CrossRef]
- Schindelin, J.; Arganda-Carreras, I.; Frise, E.; Kaynig, V.; Longair, M.; Pietzsch, T.; Preibisch, S.; Rueden, C.; Saalfeld, S.; Schmid, B.; et al. Fiji: An open-source platform for biological-image analysis. Nat. Methods 2012, 9, 676–682. [Google Scholar] [CrossRef] [PubMed]
- Hostetter, C.L.; Sullivan-Brown, J.L.; Burdine, R.D. Zebrafish pronephros: A model for understanding cystic kidney disease. Dev. Dyn. 2003, 228, 514–522. [Google Scholar] [CrossRef] [PubMed]
- Sun, Z.; Amsterdam, A.; Pazour, G.J.; Cole, D.G.; Miller, M.S.; Hopkins, N. A genetic screen in zebrafish identifies cilia genes as a principal cause of cystic kidney. Development 2004, 131, 4085–4093. [Google Scholar] [CrossRef] [PubMed]
- Sukumaran, S.; Perkins, B.D. Early defects in photoreceptor outer segment morphogenesis in zebrafish ift57, ift88 and ift172 intraflagellar transport mutants. Vision Res. 2009, 49, 479–489. [Google Scholar] [CrossRef]
- Pandey, G. Heidelberg University: Heidelberg, Germany, Unpublished work. 2019.
- Tobin, J.L.; Beales, P.L. Restoration of renal function in zebrafish models of ciliopathies. Pediatr. Nephrol. 2008, 23, 2095–2099. [Google Scholar] [CrossRef] [PubMed]
- Rossi, A.; Kontarakis, Z.; Gerri, C.; Nolte, H.; Holper, S.; Kruger, M.; Stainier, D.Y. Genetic compensation induced by deleterious mutations but not gene knockdowns. Nature 2015, 524, 230–233. [Google Scholar] [CrossRef] [PubMed]
- Stainier, D.Y.R.; Raz, E.; Lawson, N.D.; Ekker, S.C.; Burdine, R.D.; Eisen, J.S.; Ingham, P.W.; Schulte-Merker, S.; Yelon, D.; Weinstein, B.M.; et al. Guidelines for morpholino use in zebrafish. PLoS Genet. 2017, 13, e1007000. [Google Scholar] [CrossRef] [PubMed]
- Kok, F.O.; Shin, M.; Ni, C.W.; Gupta, A.; Grosse, A.S.; van Impel, A.; Kirchmaier, B.C.; Peterson-Maduro, J.; Kourkoulis, G.; Male, I.; et al. Reverse genetic screening reveals poor correlation between morpholino-induced and mutant phenotypes in zebrafish. Dev. Cell 2015, 32, 97–108. [Google Scholar] [CrossRef] [PubMed]
- Ryan, S.; Willer, J.; Marjoram, L.; Bagwell, J.; Mankiewicz, J.; Leshchiner, I.; Goessling, W.; Bagnat, M.; Katsanis, N. Rapid identification of kidney cyst mutations by whole exome sequencing in zebrafish. Development 2013, 140, 4445–4451. [Google Scholar] [CrossRef] [PubMed]
- Spaink, H.P.; Cui, C.; Wiweger, M.I.; Jansen, H.J.; Veneman, W.J.; Marin-Juez, R.; de Sonneville, J.; Ordas, A.; Torraca, V.; van der Ent, W.; et al. Robotic injection of zebrafish embryos for high-throughput screening in disease models. Methods 2013, 62, 246–254. [Google Scholar] [CrossRef] [PubMed]
- Cordero-Maldonado, M.L.; Perathoner, S.; van der Kolk, K.J.; Boland, R.; Heins-Marroquin, U.; Spaink, H.P.; Meijer, A.H.; Crawford, A.D.; de Sonneville, J. Deep learning image recognition enables efficient genome editing in zebrafish by automated injections. PLoS ONE 2019, 14, e0202377. [Google Scholar] [CrossRef] [PubMed]
- Jarque, S.; Fetter, E.; Veneman, W.J.; Spaink, H.P.; Peravali, R.; Strahle, U.; Scholz, S. An automated screening method for detecting compounds with goitrogenic activity using transgenic zebrafish embryos. PLoS ONE 2018, 13, e0203087. [Google Scholar] [CrossRef] [PubMed]
- Early, J.J.; Cole, K.L.; Williamson, J.M.; Swire, M.; Kamadurai, H.; Muskavitch, M.; Lyons, D.A. An automated high-resolution in vivo screen in zebrafish to identify chemical regulators of myelination. Elife 2018, 7, e35136. [Google Scholar] [CrossRef]
- Scheeder, C.; Heigwer, F.; Boutros, M. Machine learning and image-based profiling in drug discovery. Curr. Opin. Syst. Biol. 2018, 10, 43–52. [Google Scholar] [CrossRef]
- Westerfield, M. The Zebrafish Book. A Guide for the Laboratory Use of Zebrafish (Danio Rerio), 4th ed.; University of Oregon Press: Eugene, OR, USA, 2000. [Google Scholar]
- Gehrig, J.; Reischl, M.; Kalmar, E.; Ferg, M.; Hadzhiev, Y.; Zaucker, A.; Song, C.; Schindler, S.; Liebel, U.; Muller, F. Automated high-throughput mapping of promoter-enhancer interactions in zebrafish embryos. Nat. Methods 2009, 6, 911–916. [Google Scholar] [CrossRef] [PubMed]
- Haralick, R.M.; Shapiro, L.G. Computer and Robot Vision; Addison-Wesley: Reading, MA, USA, 1993. [Google Scholar]
- Gong, Y.; Sbalzarini, I.F. Curvature filters efficiently reduce certain variational energies. IEEE Trans. Image Process. 2017, 26, 1786–1798. [Google Scholar] [CrossRef] [PubMed]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pandey, G.; Westhoff, J.H.; Schaefer, F.; Gehrig, J. A Smart Imaging Workflow for Organ-Specific Screening in a Cystic Kidney Zebrafish Disease Model. Int. J. Mol. Sci. 2019, 20, 1290. https://doi.org/10.3390/ijms20061290
Pandey G, Westhoff JH, Schaefer F, Gehrig J. A Smart Imaging Workflow for Organ-Specific Screening in a Cystic Kidney Zebrafish Disease Model. International Journal of Molecular Sciences. 2019; 20(6):1290. https://doi.org/10.3390/ijms20061290
Chicago/Turabian StylePandey, Gunjan, Jens H. Westhoff, Franz Schaefer, and Jochen Gehrig. 2019. "A Smart Imaging Workflow for Organ-Specific Screening in a Cystic Kidney Zebrafish Disease Model" International Journal of Molecular Sciences 20, no. 6: 1290. https://doi.org/10.3390/ijms20061290
APA StylePandey, G., Westhoff, J. H., Schaefer, F., & Gehrig, J. (2019). A Smart Imaging Workflow for Organ-Specific Screening in a Cystic Kidney Zebrafish Disease Model. International Journal of Molecular Sciences, 20(6), 1290. https://doi.org/10.3390/ijms20061290




