The Circadian tau Mutation in Casein Kinase 1 Is Part of a Larger Domain That Can Be Mutated to Shorten Circadian Period
Abstract
1. Introduction
2. Results
2.1. Mutations of Lysines that Are Part of or Close to the Proposed Phosphate-Binding Triad and DBT NLS Cause Short Periods When Expressed with the timGAL4 Driver
2.2. Mutations that Affected Arginines Localized Internally Lengthened Period
2.3. All of the K/N or R/N Mutant Proteins are Expressed at Comparable Levels, While the G215E Protein is not Detectably Expressed
2.4. The DBT NLS and R192N Mutant Proteins are Defective in Interactions with BDBT
3. Discussion
4. Materials and Methods
4.1. Site-Directed Mutagenesis
- DBT R192N Forward, 5′-GGGCATTGAGCAATCGAACCGTGACGACCTGGAGTC;
- DBT R192N Reverse, 5′-GACTCCAGGTCGTCACGGTTCGATTGCTCAATGCCC;
- DBT R193N Forward, 5′-GCATTGAGCAATCGCGTAACGACGACCTGGAGTCCCTG;
- DBT R193N Reverse, 5′-CAGGGACTCCAGGTCGTCGTTACGCGATTGCTCAATGC;
- DBT G215E Forward, 5′-CGCCTTGCCCTGGCAGGAATTAAAGGCAGCCAACAAG;
- DBT G215E Reverse, 5’-CTTGTTGGCTGCCTTTAATTCCTGCCAGGGCAAGGCG;
- DBT K217N Forward, 5’-CTGGCAGGGCTTAAACGCAGCCAACAAGAGG;
- DBT K217N Reverse, 5’-CCTCTTGTTGGCTGCGTTTAAGCCCTGCCAG;
- DBT NLS2 Forward, 5’-GCTTAAAGGCAGCCAACAACAACCAAAAGTACGAGAGGATC;
- DBT NLS2 Reverse, 5’-GATCCTCTCGTACTTTTGGTTGTTGTTGGCTGCCTTTAAGC;
- DBT NLS1 Forward, 5’-GCTTAAAGGCAGCCAACAACAATCAAAACTACGAGAGGATCTCG;
- DBT NLS1 Reverse, 5’-CCGAGATCCTCTCGTAGTTTTGATTGTTGTTGGCTGCCTTTAAGC.
4.2. Generation of Transgenic Flies with Mutant UAS-DBT Proteins Expressed in Circadian Cells
4.3. Analysis of Circadian Locomotor Activity Rhythms
4.4. Analysis of Transgenic and Endogeonous DBT Levels
4.5. Immunoprecipitations to Measure DBT/BDBT Interactions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Pittendrigh, C.S. Circadian rhythms and the circadian organization of living systems. Cold Spring Harbor Symp. Quant. Biol. 1960, 25, 159–184. [Google Scholar] [CrossRef] [PubMed]
- Siwicki, K.K.; Hardin, P.E.; Price, J.L. Reflections on contributing to “big discoveries” about the fly clock: Our fortunate paths as post-docs with 2017 Nobel laureates Jeff Hall, Michael Rosbash, and Mike Young. Neurobiol. Sleep Circadian Rhythms 2018, 5, 58–67. [Google Scholar] [CrossRef]
- Edery, I.; Zwiebel, L.J.; Dembinska, M.E.; Rosbash, M. Temporal phosphorylation of the Drosophila period protein. Proc. Natl. Acad. Sci. USA 1994, 91, 2260–2264. [Google Scholar] [CrossRef] [PubMed]
- Kloss, B.; Price, J.L.; Saez, L.; Blau, J.; Rothenfluh, A.; Wesley, C.S.; Young, M.W. The Drosophila clock gene double-time encodes a protein closely related to human casein kinase Iepsilon. Cell 1998, 94, 97–107. [Google Scholar] [CrossRef]
- Price, J.L.; Blau, J.; Rothenfluh, A.; Abodeely, M.; Kloss, B.; Young, M.W. Double-time is a novel Drosophila clock gene that regulates PERIOD protein accumulation. Cell 1998, 94, 83–95. [Google Scholar] [CrossRef]
- Rothenfluh, A.; Abodeely, M.; Young, M.W. Short-period mutations of per affect a double-time-dependent step in the Drosophila circadian clock. Curr. Biol. 2000, 10, 1399–1402. [Google Scholar] [CrossRef]
- Suri, V.; Hall, J.C.; Rosbash, M. Two novel doubletime mutants alter circadian properties and eliminate the delay between RNA and protein in Drosophila. J. Neurosci. 2000, 20, 7547–7555. [Google Scholar] [CrossRef] [PubMed]
- Bao, S.; Rihel, J.; Bjes, E.; Fan, J.Y.; Price, J.L. The Drosophila double-timeS mutation delays the nuclear accumulation of period protein and affects the feedback regulation of period mRNA. J. Neurosci. 2001, 21, 7117–7126. [Google Scholar] [CrossRef]
- Kloss, B.; Rothenfluh, A.; Young, M.W.; Saez, L. Phosphorylation of period is influenced by cycling physical associations of double-time, period, and timeless in the Drosophila clock. Neuron 2001, 30, 699–706. [Google Scholar] [CrossRef]
- Muskus, M.J.; Preuss, F.; Fan, J.Y.; Bjes, E.S.; Price, J.L. Drosophila DBT lacking protein kinase activity produces long-period and arrhythmic circadian behavioral and molecular rhythms. Mol. Cell. Biol. 2007, 27, 8049–8064. [Google Scholar] [CrossRef] [PubMed]
- Martinek, S.; Inonog, S.; Manoukian, A.S.; Young, M.W. A role for the segment polarity gene shaggy/GSK-3 in the Drosophila circadian clock. Cell 2001, 105, 769–779. [Google Scholar] [CrossRef]
- Lin, J.M.; Kilman, V.L.; Keegan, K.; Paddock, B.; Emery-Le, M.; Rosbash, M.; Allada, R. A role for casein kinase 2alpha in the Drosophila circadian clock. Nature 2002, 420, 816–820. [Google Scholar] [CrossRef] [PubMed]
- Akten, B.; Jauch, E.; Genova, G.K.; Kim, E.Y.; Edery, I.; Raabe, T.; Jackson, F.R. A role for CK2 in the Drosophila circadian oscillator. Nat. Neurosci. 2003, 6, 251–257. [Google Scholar] [CrossRef] [PubMed]
- Lin, J.M.; Schroeder, A.; Allada, R. In vivo circadian function of casein kinase 2 phosphorylation sites in Drosophila PERIOD. J. Neurosci. 2005, 25, 11175–11183. [Google Scholar] [CrossRef] [PubMed]
- Kim, E.Y.; Edery, I. Balance between DBT/CKIepsilon kinase and protein phosphatase activities regulate phosphorylation and stability of Drosophila CLOCK protein. Proc. Natl. Acad. Sci. USA 2006, 103, 6178–6183. [Google Scholar] [CrossRef] [PubMed]
- Yu, W.; Zheng, H.; Houl, J.H.; Dauwalder, B.; Hardin, P.E. PER-dependent rhythms in CLK phosphorylation and E-box binding regulate circadian transcription. Genes Dev. 2006, 20, 723–733. [Google Scholar] [CrossRef] [PubMed]
- Sathyanarayanan, S.; Zheng, X.; Xiao, R.; Sehgal, A. Posttranslational regulation of Drosophila PERIOD protein by protein phosphatase 2A. Cell 2004, 116, 603–615. [Google Scholar] [CrossRef]
- Fang, Y.; Sathyanarayanan, S.; Sehgal, A. Post-translational regulation of the Drosophila circadian clock requires protein phosphatase 1 (PP1). Genes Dev. 2007, 21, 1506–1518. [Google Scholar] [CrossRef]
- Lowrey, P.L.; Shimomura, K.; Antoch, M.P.; Yamazaki, S.; Zemenides, P.D.; Ralph, M.R.; Menaker, M.; Takahashi, J.S. Positional syntenic cloning and functional characterization of the mammalian circadian mutation tau. Science 2000, 288, 483–492. [Google Scholar] [CrossRef]
- Xu, Y.; Padiath, Q.S.; Shapiro, R.E.; Jones, C.R.; Wu, S.C.; Saigoh, N.; Saigoh, K.; Ptacek, L.J.; Fu, Y.H. Functional consequences of a CKIdelta mutation causing familial advanced sleep phase syndrome. Nature 2005, 434, 640–644. [Google Scholar] [CrossRef]
- Fan, J.Y.; Preuss, F.; Muskus, M.J.; Bjes, E.S.; Price, J.L. Drosophila and vertebrate casein kinase Idelta exhibits evolutionary conservation of circadian function. Genetics 2009, 181, 139–152. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.; Chen, R.; Lee, Y.; Yoo, S.; Lee, C. Essential roles of CKIdelta and CKIepsilon in the mammalian circadian clock. Proc. Natl. Acad. Sci. USA 2009, 106, 21359–21364. [Google Scholar] [CrossRef] [PubMed]
- Meng, Q.J.; Maywood, E.S.; Bechtold, D.A.; Lu, W.Q.; Li, J.; Gibbs, J.E.; Dupre, S.M.; Chesham, J.E.; Rajamohan, F.; Knafels, J.; et al. Entrainment of disrupted circadian behavior through inhibition of casein kinase 1 (CK1) enzymes. Proc. Natl. Acad. Sci. USA 2010, 107, 15240–15245. [Google Scholar] [CrossRef] [PubMed]
- Preuss, F.; Fan, J.Y.; Kalive, M.; Bao, S.; Schuenemann, E.; Bjes, E.S.; Price, J.L. Drosophila doubletime mutations which either shorten or lengthen the period of circadian rhythms decrease the protein kinase activity of casein kinase I. Mol. Cell. Biol. 2004, 24, 886–898. [Google Scholar] [CrossRef] [PubMed]
- Kivimae, S.; Saez, L.; Young, M.W. Activating PER repressor through a DBT-directed phosphorylation switch. PLoS Biol. 2008, 6, e183. [Google Scholar] [CrossRef] [PubMed]
- Gallego, M.; Eide, E.J.; Woolf, M.F.; Virshup, D.M.; Forger, D.B. An opposite role for tau in circadian rhythms revealed by mathematical modeling. Proc. Natl. Acad. Sci. USA 2006, 103, 10618–10623. [Google Scholar] [CrossRef] [PubMed]
- Meng, Q.J.; Logunova, L.; Maywood, E.S.; Gallego, M.; Lebiecki, J.; Brown, T.M.; Sladek, M.; Semikhodskii, A.S.; Glossop, N.R.; Piggins, H.D.; et al. Setting clock speed in mammals: The CK1 epsilon tau mutation in mice accelerates circadian pacemakers by selectively destabilizing PERIOD proteins. Neuron 2008, 58, 78–88. [Google Scholar] [CrossRef] [PubMed]
- Vanselow, K.; Vanselow, J.T.; Westermark, P.O.; Reischl, S.; Maier, B.; Korte, T.; Herrmann, A.; Herzel, H.; Schlosser, A.; Kramer, A. Differential effects of PER2 phosphorylation: Molecular basis for the human familial advanced sleep phase syndrome (FASPS). Genes Dev. 2006, 20, 2660–2672. [Google Scholar] [CrossRef] [PubMed]
- Venkatesan, A.; Fan, J.Y.; Nauman, C.; Price, J.L. A Doubletime Nuclear Localization Signal Mediates an Interaction with Bride of Doubletime to Promote Circadian Function. J. Biol. Rhythms 2015, 30, 302–317. [Google Scholar] [CrossRef] [PubMed]
- Longenecker, K.L.; Roach, P.J.; Hurley, T.D. Three-dimensional structure of mammalian casein kinase I: Molecular basis for phosphate recognition. J. Mol. Biol. 1996, 257, 618–631. [Google Scholar] [CrossRef]
- Goddard, T.D.; Huang, C.C.; Meng, E.C.; Pettersen, E.F.; Couch, G.S.; Morris, J.H.; Ferrin, T.E. UCSF ChimeraX: Meeting modern challenges in visualization and analysis. Protein Sci. 2018, 27, 14–25. [Google Scholar] [CrossRef] [PubMed]
- Top, D.; O’Neil, J.L.; Merz, G.E.; Dusad, K.; Crane, B.R.; Young, M.W. CK1/Doubletime activity delays transcription activation in the circadian clock. eLife 2018, 7, e32679. [Google Scholar] [CrossRef] [PubMed]
- Syed, S.; Saez, L.; Young, M.W. Kinetics of doubletime kinase-dependent degradation of the Drosophila period protein. J. Biol. Chem. 2011, 286, 27654–27662. [Google Scholar] [CrossRef] [PubMed]
- Cyran, S.A.; Yiannoulos, G.; Buchsbaum, A.M.; Saez, L.; Young, M.W.; Blau, J. The double-time protein kinase regulates the subcellular localization of the Drosophila clock protein period. J. Neurosci. 2005, 25, 5430–5437. [Google Scholar] [CrossRef] [PubMed]
- Nawathean, P.; Rosbash, M. The doubletime and CKII kinases collaborate to potentiate Drosophila PER transcriptional repressor activity. Mol. Cell 2004, 13, 213–223. [Google Scholar] [CrossRef]
- Yu, W.; Zheng, H.; Price, J.L.; Hardin, P.E. DOUBLETIME plays a noncatalytic role to mediate CLOCK phosphorylation and repress CLOCK-dependent transcription within the Drosophila circadian clock. Mol. Cell. Biol. 2009, 29, 1452–1458. [Google Scholar] [CrossRef] [PubMed]
- Kim, E.Y.; Ko, H.W.; Yu, W.; Hardin, P.E.; Edery, I. A DOUBLETIME kinase binding domain on the Drosophila PERIOD protein is essential for its hyperphosphorylation, transcriptional repression, and circadian clock function. Mol. Cell. Biol. 2007, 27, 5014–5028. [Google Scholar] [CrossRef] [PubMed]
- Fan, J.Y.; Agyekum, B.; Venkatesan, A.; Hall, D.R.; Keightley, A.; Bjes, E.S.; Bouyain, S.; Price, J.L. Noncanonical FK506-binding protein BDBT binds DBT to enhance its circadian function and forms foci at night. Neuron 2013, 80, 984–996. [Google Scholar] [CrossRef] [PubMed]
- Means, J.C.; Venkatesan, A.; Gerdes, B.; Fan, J.Y.; Bjes, E.S.; Price, J.L. Drosophila spaghetti and doubletime link the circadian clock and light to caspases, apoptosis and tauopathy. PLoS Genet. 2015, 11, e1005171. [Google Scholar] [CrossRef] [PubMed]
- Xing, L.; An, Y.; Shi, G.; Yan, J.; Xie, P.; Qu, Z.; Zhang, Z.; Liu, Z.; Pan, D.; Xu, Y. Correlated evolution between CK1delta Protein and the Serine-rich Motif Contributes to Regulating the Mammalian Circadian Clock. J. Biol. Chem. 2017, 292, 161–171. [Google Scholar] [CrossRef] [PubMed]
- Bischof, J.; Maeda, R.; Hediger, M.; Karch, F.; Basler, K. An optimized transgenesis system for Drosophila using germ-line-specific φC31 integrases. Proc. Natl. Acad. Sci. USA 2007, 104, 3312–3317. [Google Scholar] [CrossRef] [PubMed]
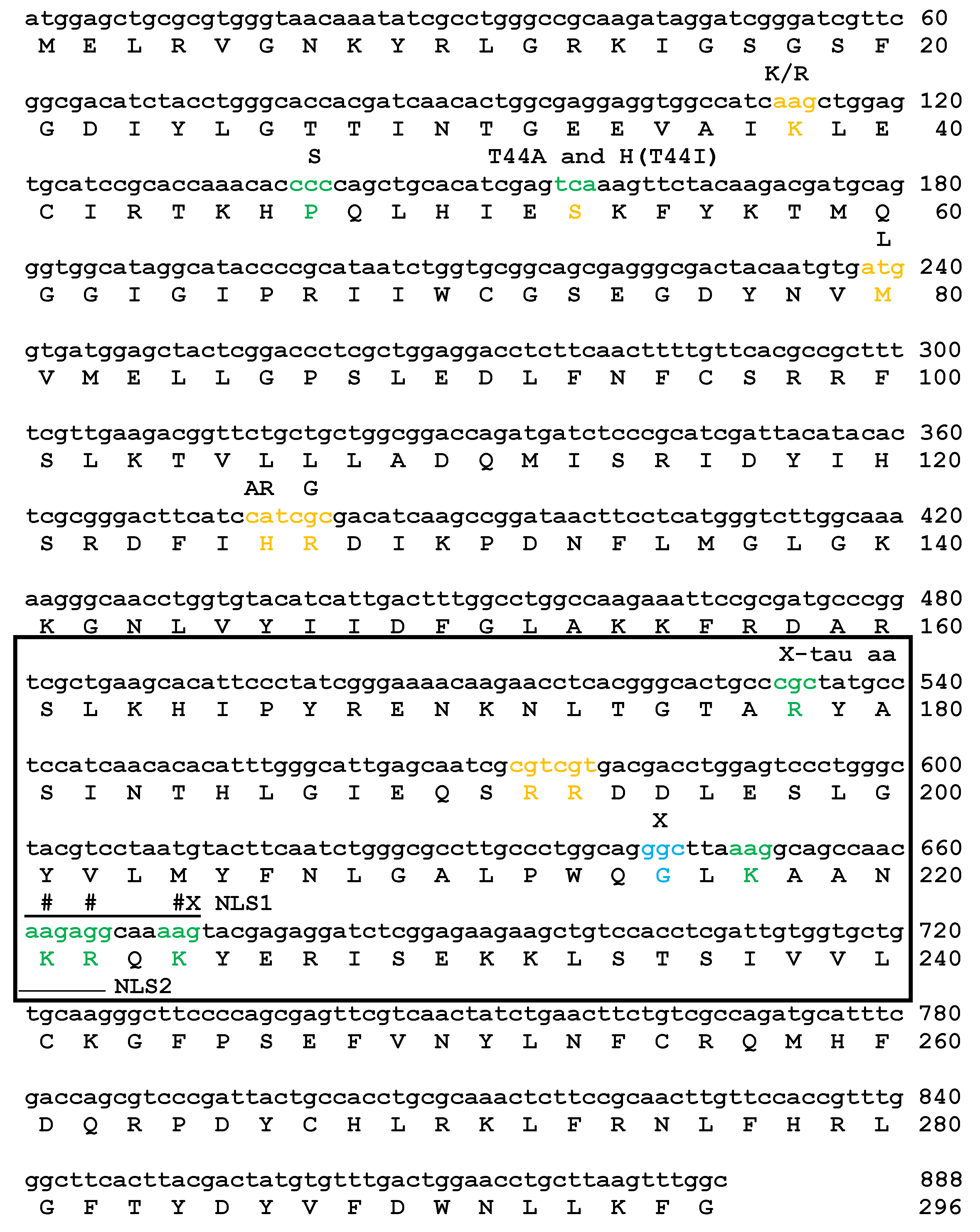
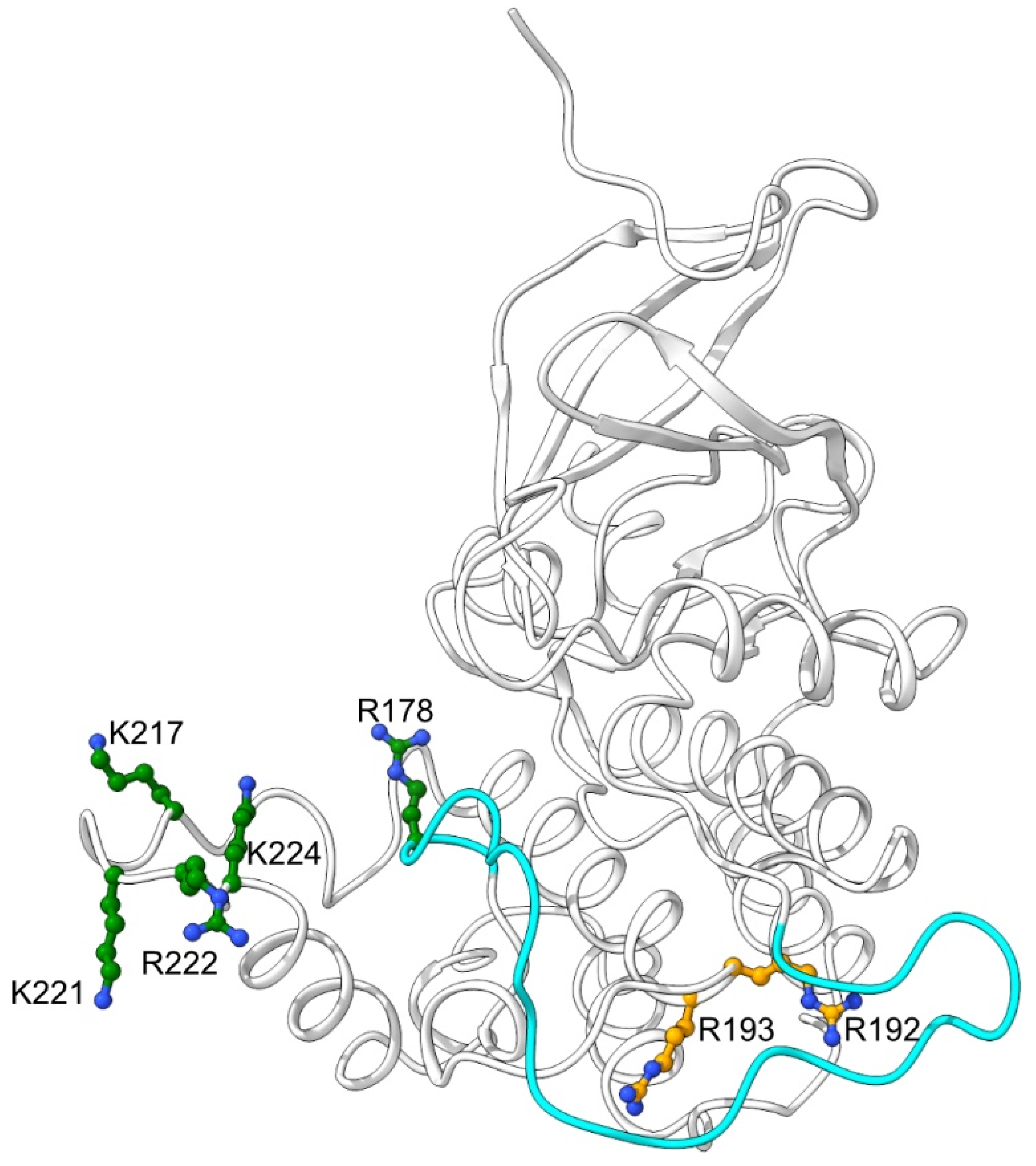
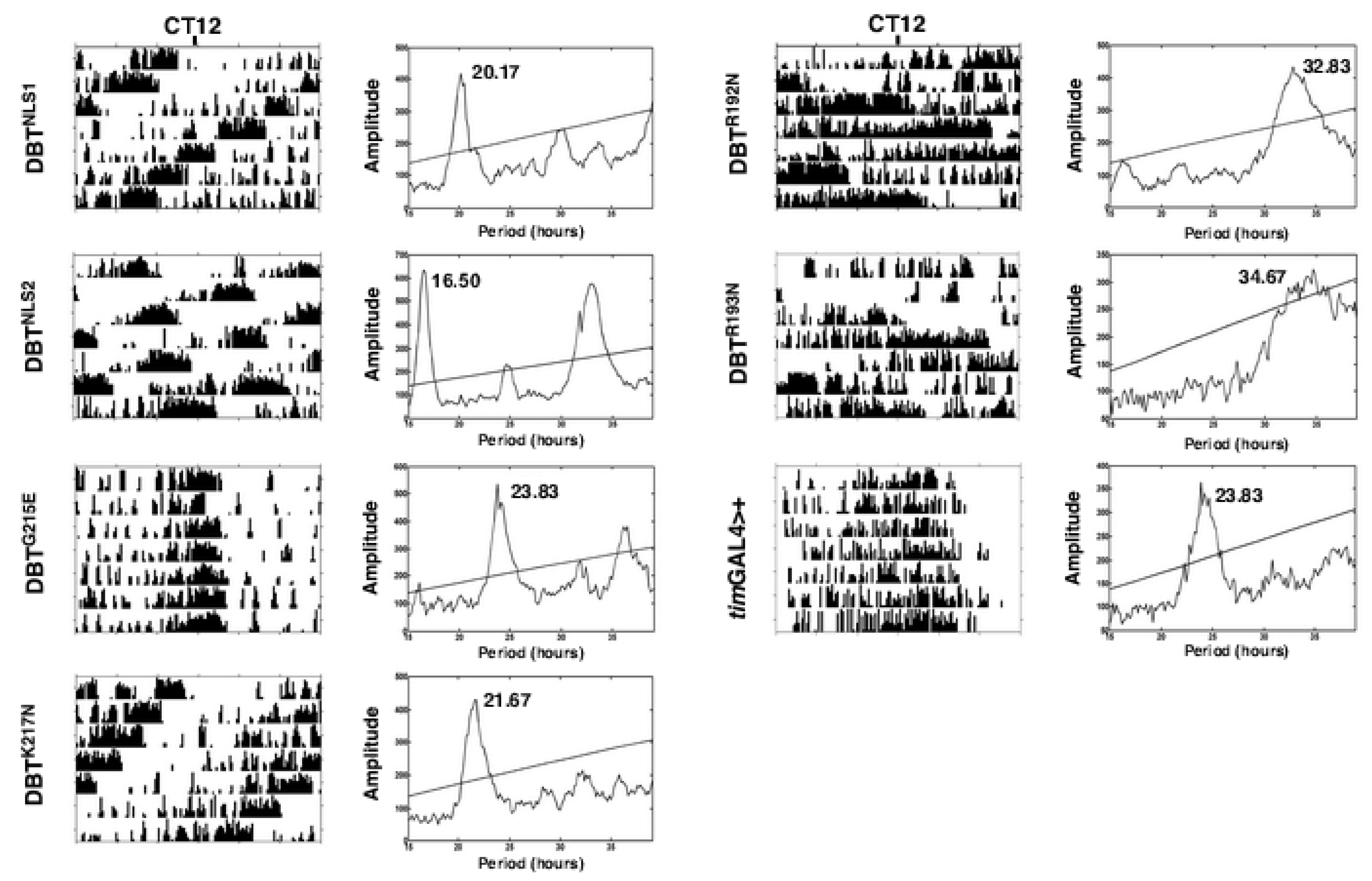
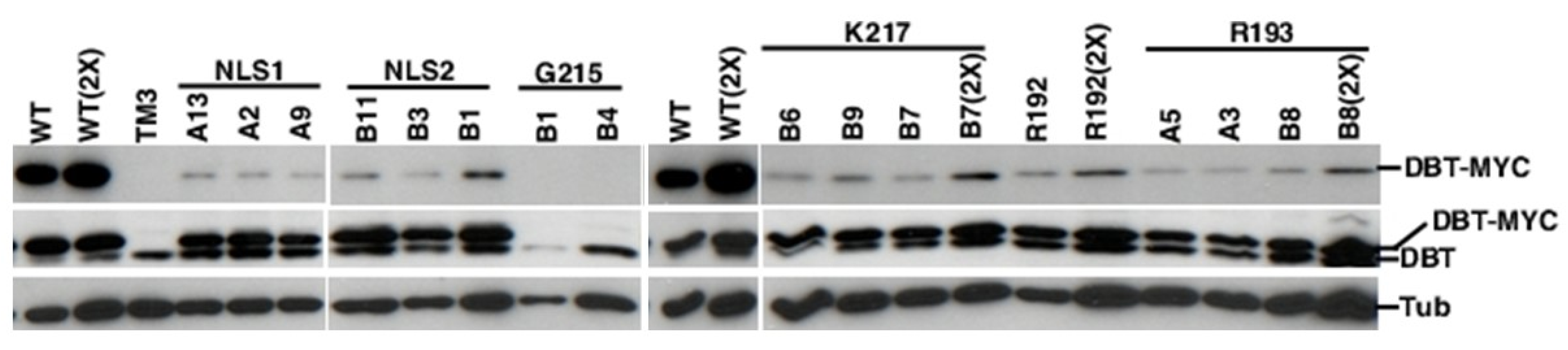

| DBT | Line | Genotype | Avg. Period (h)+SEM | %Rhythmicity (N) |
|---|---|---|---|---|
| DBTNLS1 | A2 | timGAL4>UAS-dbt | 20.3 ± 0.1 a | 93 (14) |
| A9 | timGAL4>UAS-dbt | 22.4 ± 0.2 b | 93 (14) | |
| A13 | timGAL4>UAS-dbt | 19.6 ± 0.2 a | 83 (12) | |
| DBTNLS2 | B1 | timGAL4>UAS-dbt | 16.6 ± 0.05 c | 100 (15) |
| B3 | timGAL4>UAS-dbt | 16.5 ± 0.08 c | 100 (16) | |
| B11 | timGAL4>UAS-dbt | 17.0 ± 0.07 c | 87.5 (16) | |
| B13 | timGAL4>UAS-dbt | 16.6 ± 0.03 c | 94 (16) | |
| DBTG215E | B1 | timGAL4>UAS-dbt | 23.9 ± 0.1d | 100 (25) |
| B4 | timGAL4>UAS-dbt | 24.0 ± 0.2d | 100 (12) | |
| DBTK217N | B6 | timGAL4>UAS-dbt | 21.6 ± 0.1 b | 100 (15) |
| B7 | timGAL4>UAS-dbt | 22.0 ± 0.2 b | 93 (15) | |
| B9 | timGAL4>UAS-dbt | 21.9 ± 0.08 b | 100 (15) | |
| DBTR192N | timGAL4>UAS-dbt | 33.2 ± 0.02 | 80 (15) | |
| DBTR193N | A3 | timGAL4>UAS-dbt | 34.0 ± 0.4 e | 87.5 (16) |
| A5 | timGAL4>UAS-dbt | 34.9 ± 0.5 e | 100 (7) | |
| B8 | timGAL4>UAS-dbt | 34.1 ± 0.3 e | 94 (16) | |
| DBT+ | timGAL4>+ | 24.3 ± 0.2 d | 93 (14) |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Venkatesan, A.; Fan, J.-Y.; Bouyain, S.; Price, J.L. The Circadian tau Mutation in Casein Kinase 1 Is Part of a Larger Domain That Can Be Mutated to Shorten Circadian Period. Int. J. Mol. Sci. 2019, 20, 813. https://doi.org/10.3390/ijms20040813
Venkatesan A, Fan J-Y, Bouyain S, Price JL. The Circadian tau Mutation in Casein Kinase 1 Is Part of a Larger Domain That Can Be Mutated to Shorten Circadian Period. International Journal of Molecular Sciences. 2019; 20(4):813. https://doi.org/10.3390/ijms20040813
Chicago/Turabian StyleVenkatesan, Anandakrishnan, Jin-Yuan Fan, Samuel Bouyain, and Jeffrey L. Price. 2019. "The Circadian tau Mutation in Casein Kinase 1 Is Part of a Larger Domain That Can Be Mutated to Shorten Circadian Period" International Journal of Molecular Sciences 20, no. 4: 813. https://doi.org/10.3390/ijms20040813
APA StyleVenkatesan, A., Fan, J.-Y., Bouyain, S., & Price, J. L. (2019). The Circadian tau Mutation in Casein Kinase 1 Is Part of a Larger Domain That Can Be Mutated to Shorten Circadian Period. International Journal of Molecular Sciences, 20(4), 813. https://doi.org/10.3390/ijms20040813




