Deregulation of Biologically Significant Genes and Associated Molecular Pathways in the Oral Epithelium of Electronic Cigarette Users
Abstract
1. Introduction
2. Results
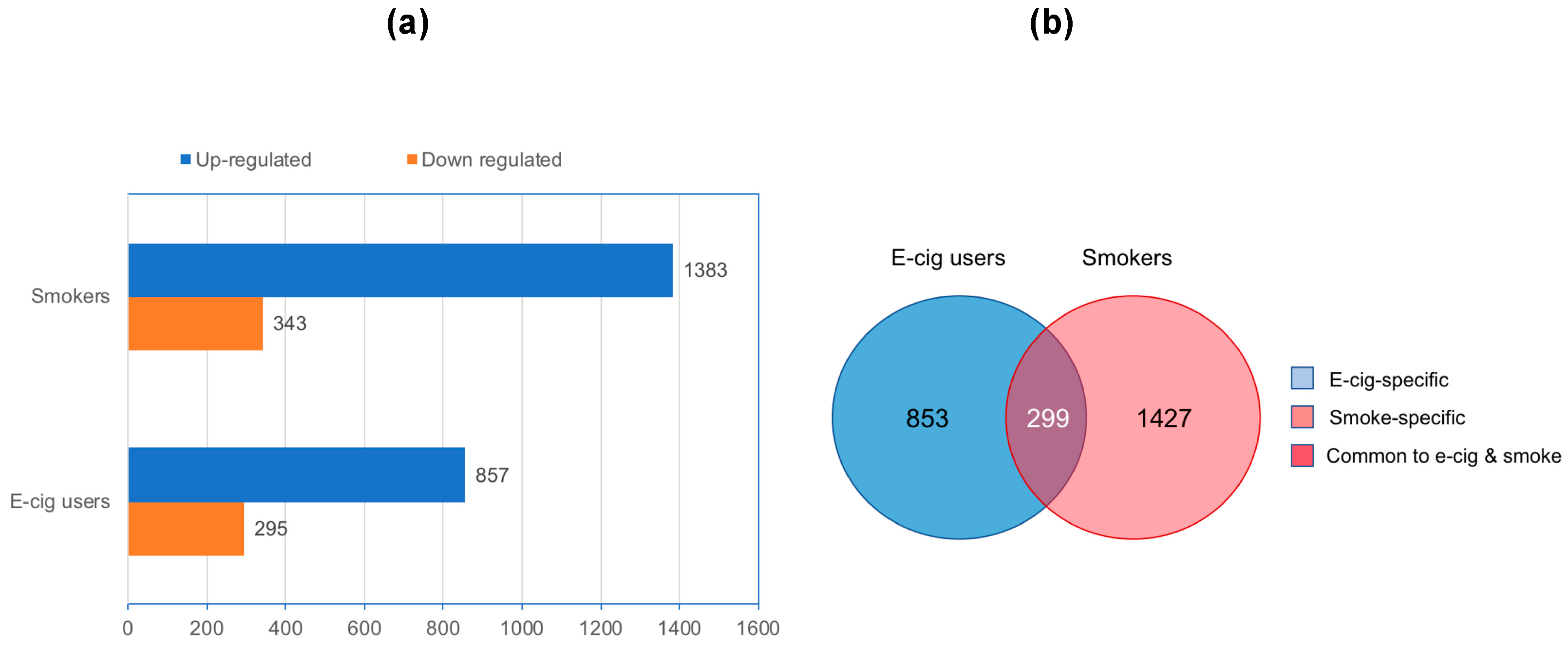
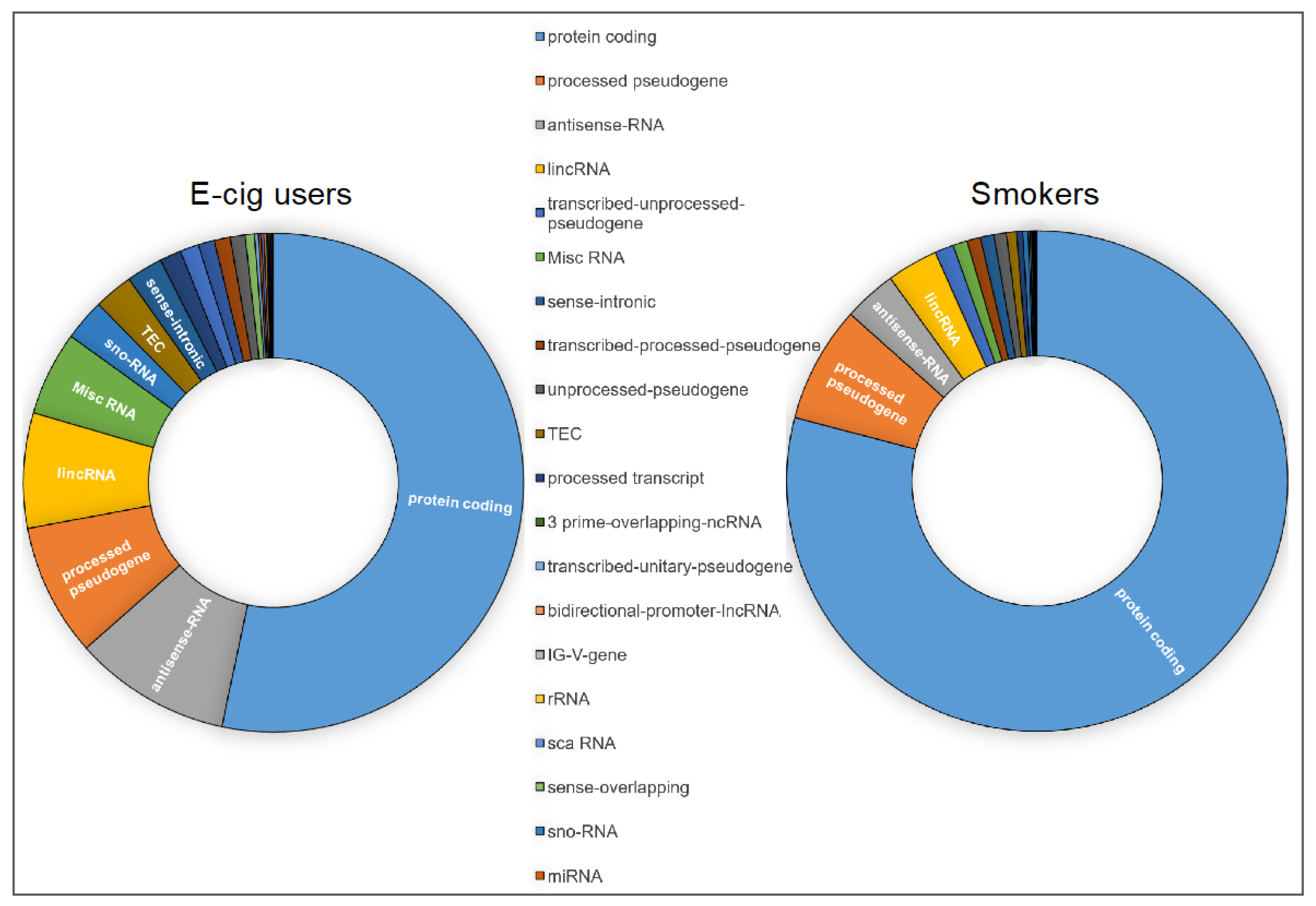
2.1. Genome-Wide Gene-Expression Analysis
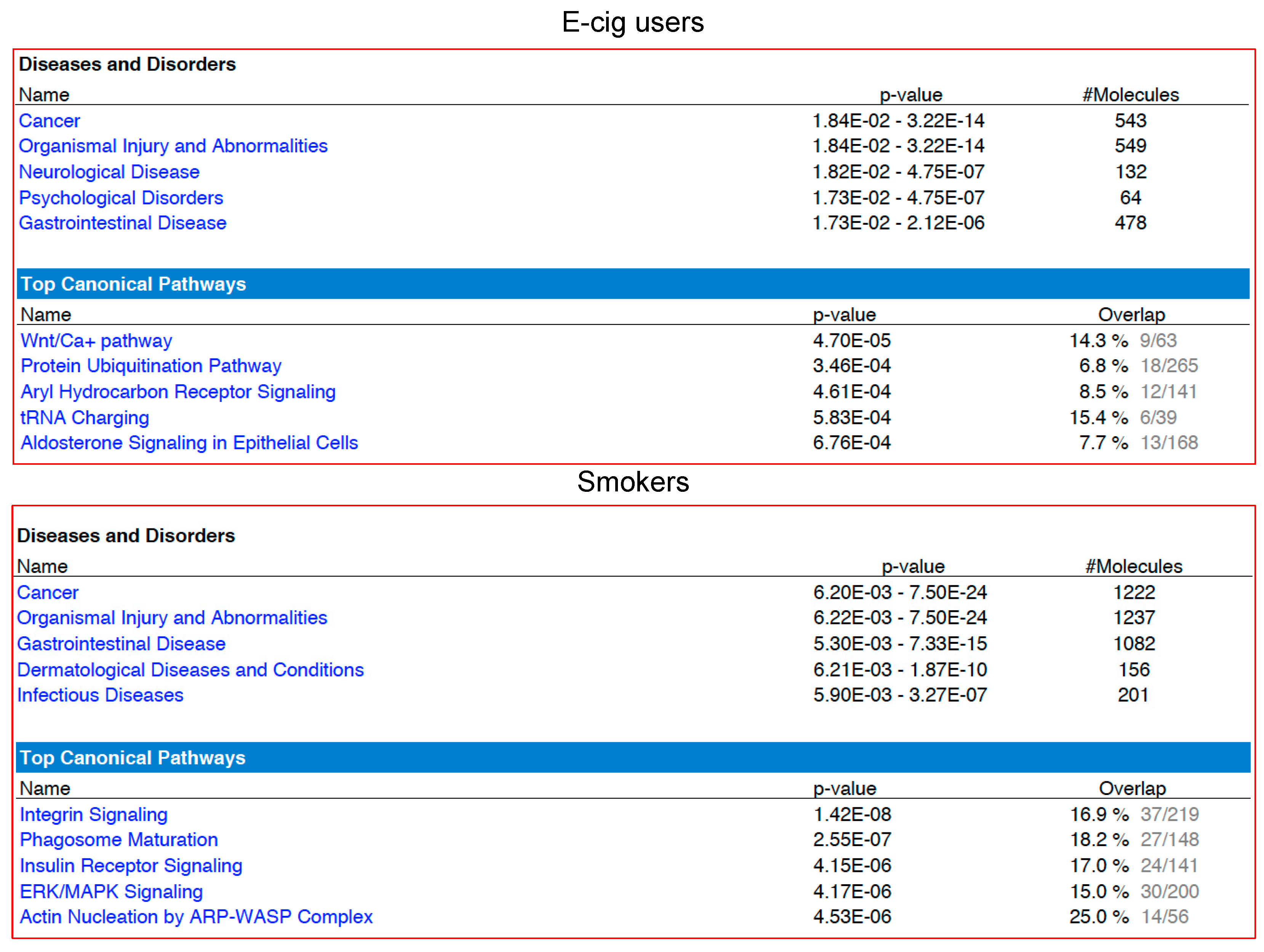
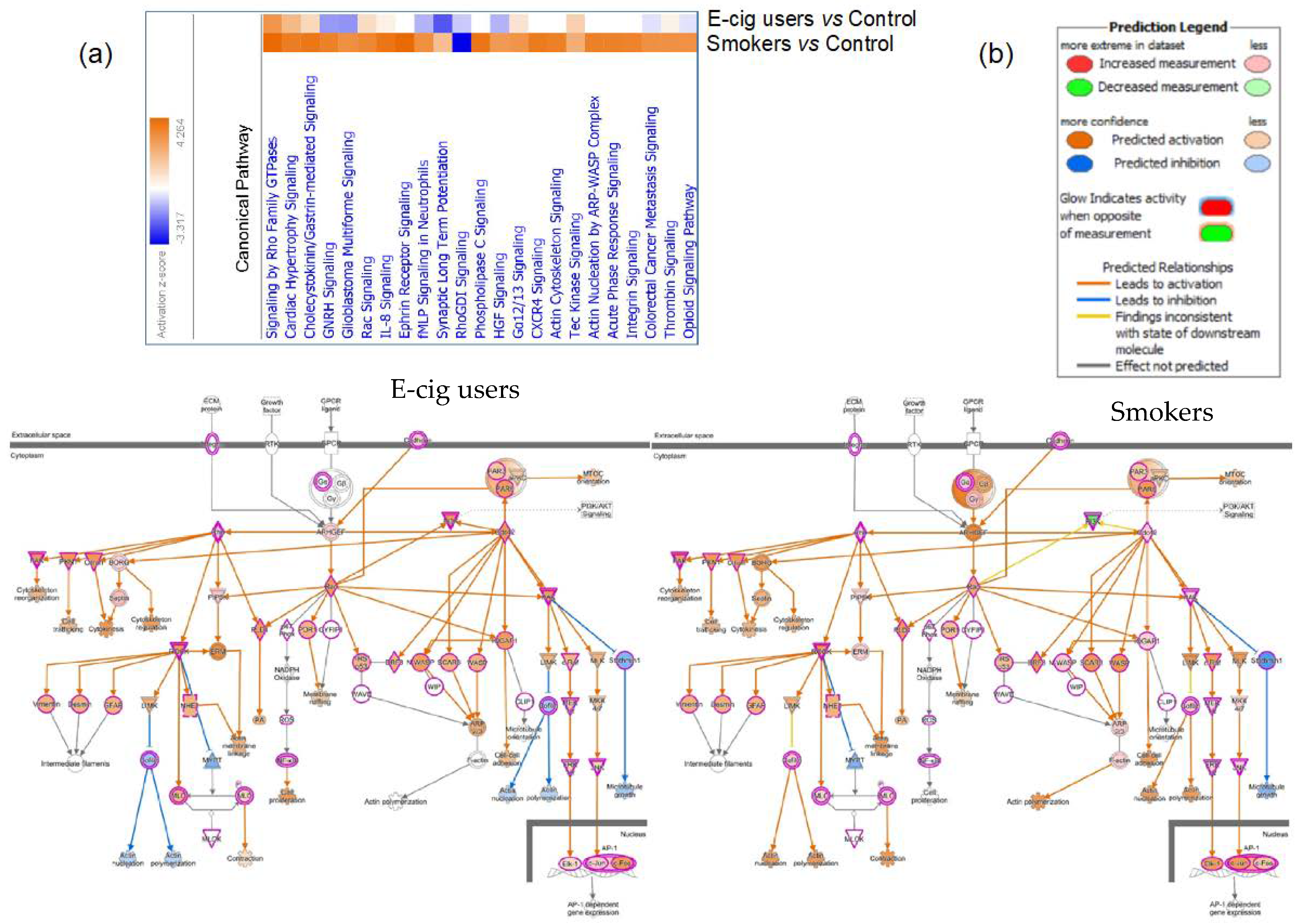
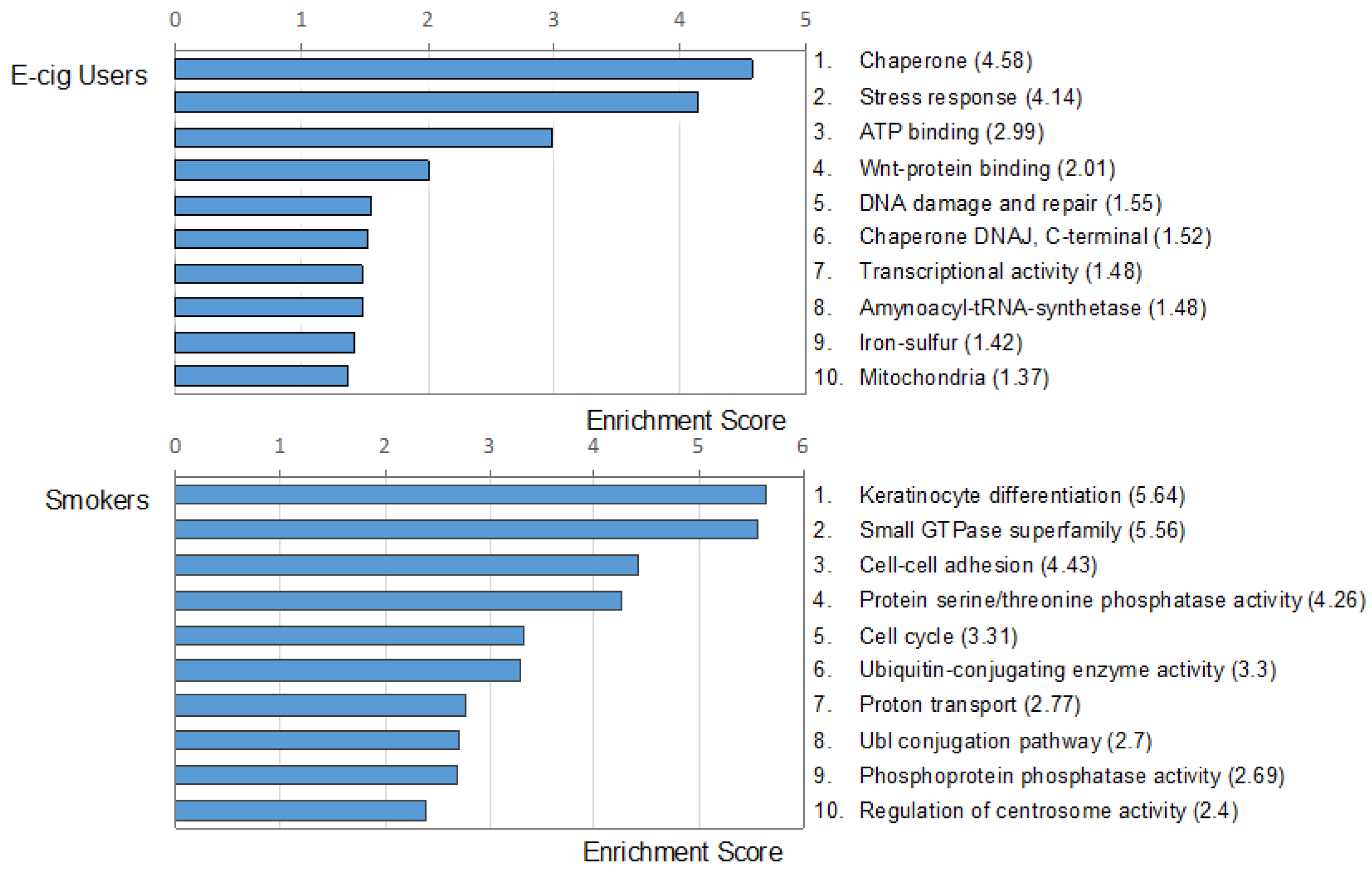
2.2. Gene Ontology and Molecular Pathway and Functional Network Analyses
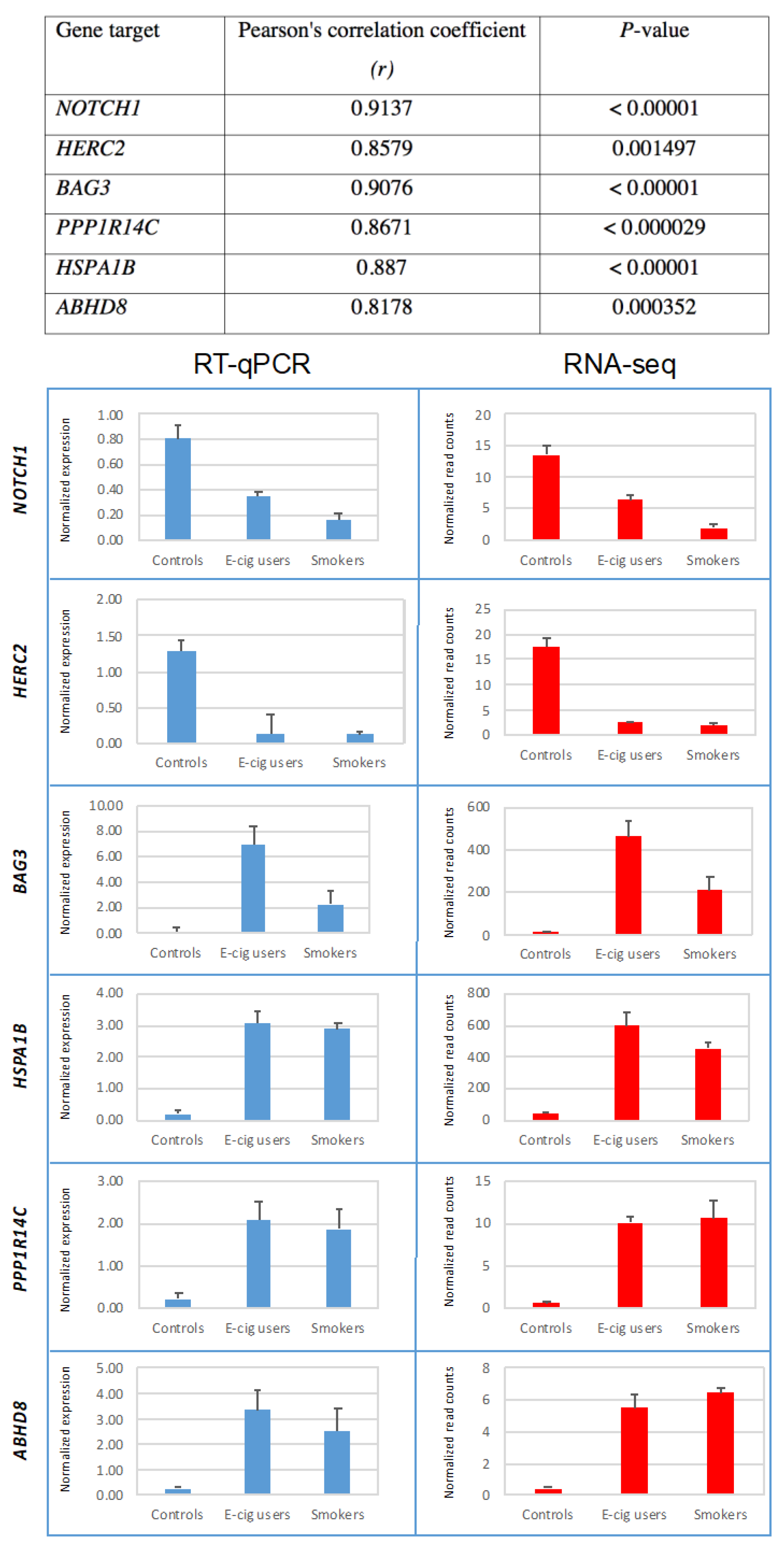
2.3. Validation of Gene-Expression Data by Real-Time Reverse Transcription Quantitative PCR (RT-qPCR)
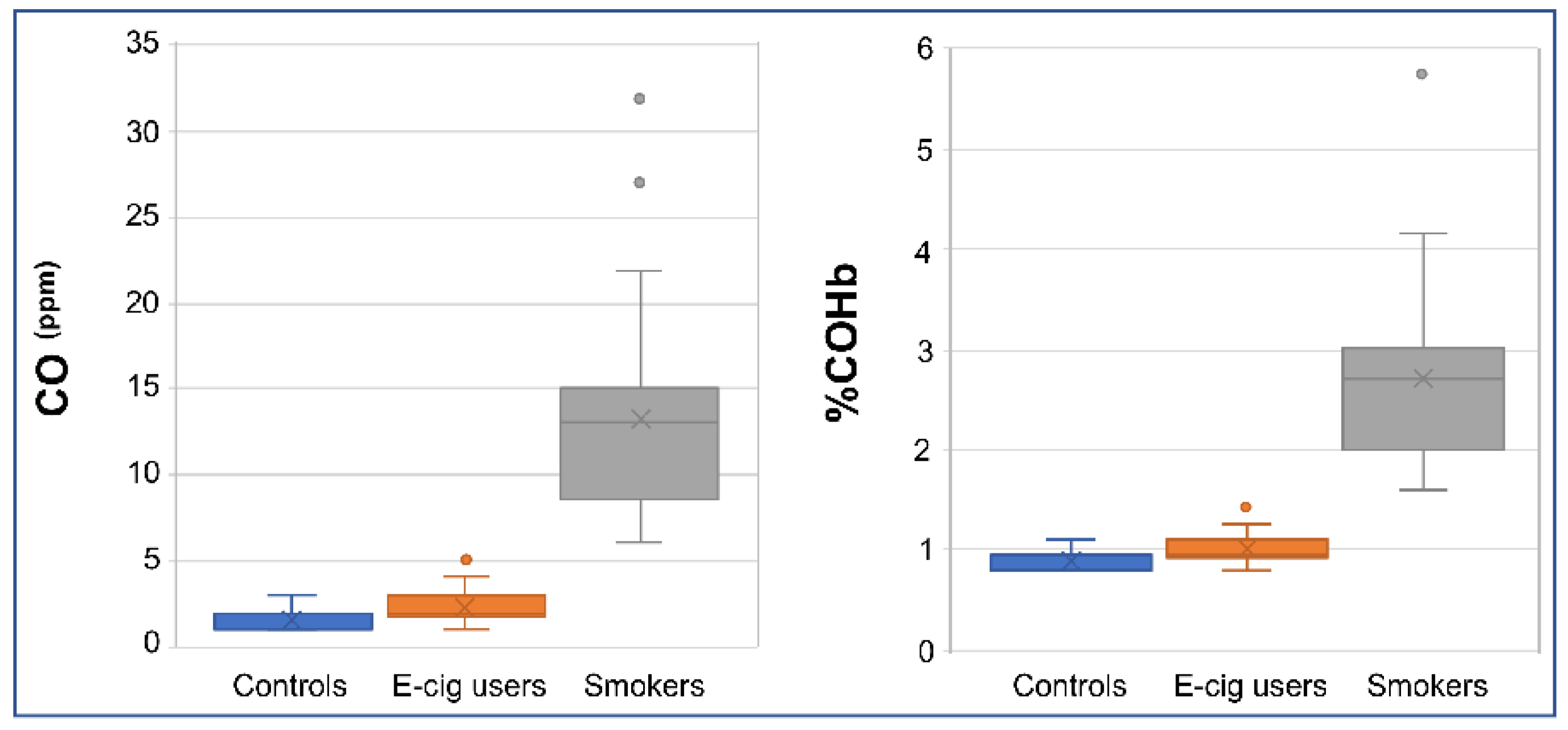
2.4. Verification of Smoking/Vaping Status
3. Discussion
4. Materials and Methods
4.1. Subject Recruitment and Enrollment
4.2. Inclusion and Exclusion Criteria for the Study
4.3. Personal Interview
4.4. Study Population
4.5. Sampling and Processing of Oral Epithelial Cells
4.6. Sampling of Peripheral Blood
4.7. RNA-Seq Analysis
4.8. Gene Ontology and Canonical Pathways Analyses
4.9. Reverse Transcription Quantitative PCR (RT-qPCR)
4.10. Cotinine Measurement
4.11. Measurement of Carbon Monoxide in Breath and Determination of %Carboxyhemoglobin
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Dedication
Conflicts of Interest
References
- National Academies of Sciences, Engineering, and Medicine. Public Health Consequences of e-cigarettes; National Academies Press: Washington, DC, USA, 2018. [Google Scholar] [CrossRef]
- Besaratinia, A.; Tommasi, S. Electronic cigarettes: The road ahead. Prev. Med. 2014, 66, 65–67. [Google Scholar] [CrossRef]
- Besaratinia, A.; Tommasi, S. An opportune and unique research to evaluate the public health impact of electronic cigarettes. Cancer Causes Control 2017, 28, 1167–1171. [Google Scholar] [CrossRef] [PubMed]
- Schoenborn, C.A.; Gindi, R.M. Electronic Cigarette Use Among Adults: United States; Department of Health and Human Services, Centers for Disease Control and Prevention, National Center for Health Statistics: Atlanta, GA, USA, 2014; pp. 1–8.
- Singh, T.; Arrazola, R.A.; Corey, C.G.; Husten, C.G.; Neff, L.J.; Homa, D.M.; King, B.A. Tobacco Use Among Middle and High School Students—United States, 2011–2015. MMWR Morb. Mortal. Wkly. Rep. 2016, 65, 361–367. [Google Scholar] [CrossRef] [PubMed]
- Centers for Disease Control and Prevention. Tobacco Use Among Middle and High School Students—United States, 2011–2016. MMWR Morb. Mortal. Wkly. Rep. 2017, 66, 597–603. [Google Scholar] [CrossRef]
- Shiffman, S.; Sembower, M.A.; Pillitteri, J.L.; Gerlach, K.K.; Gitchell, J.G. The impact of flavor descriptors on nonsmoking teens’ and adult smokers’ interest in electronic cigarettes. Nicotine Tob. Res. 2015, 17, 1255–1262. [Google Scholar] [CrossRef] [PubMed]
- Coleman, B.N.; Apelberg, B.J.; Ambrose, B.K.; Green, K.M.; Choiniere, C.J.; Bunnell, R.; King, B.A. Association Between Electronic Cigarette Use and Openness to Cigarette Smoking Among US Young Adults. Nicotine Tob. Res. 2015, 17, 212–218. [Google Scholar] [CrossRef] [PubMed]
- Schick, S.F.; Blount, B.C.; Jacob, P.R.; Saliba, N.A.; Bernert, J.T.; El Hellani, A.; Jatlow, P.; Pappas, R.S.; Wang, L.; Foulds, J.; et al. Biomarkers of exposure to new and emerging tobacco delivery products. Am. J. Physiol. Lung Cell. Mol. Physiol. 2017, 313, L425–L452. [Google Scholar] [CrossRef]
- Timp, W.; Feinberg, A.P. Cancer as a dysregulated epigenome allowing cellular growth advantage at the expense of the host. Nat. Rev. Cancer 2013, 13, 497–510. [Google Scholar] [CrossRef]
- Liu, F.; Wang, L.; Perna, F.; Nimer, S.D. Beyond transcription factors: How oncogenic signalling reshapes the epigenetic landscape. Nat. Rev. Cancer 2016, 16, 359–372. [Google Scholar] [CrossRef]
- Brody, J.S.; Steiling, K. Interaction of cigarette exposure and airway epithelial cell gene expression. Annu. Rev. Physiol. 2011, 73, 437–456. [Google Scholar] [CrossRef]
- Brothers, J.F.; Hijazi, K.; Mascaux, C.; El-Zein, R.A.; Spitz, M.R.; Spira, A. Bridging the clinical gaps: Genetic, epigenetic and transcriptomic biomarkers for the early detection of lung cancer in the post-National Lung Screening Trial era. BMC Med. 2013, 11, 168. [Google Scholar] [CrossRef] [PubMed]
- Sridhar, S.; Schembri, F.; Zeskind, J.; Shah, V.; Gustafson, A.M.; Steiling, K.; Liu, G.; Dumas, Y.M.; Zhang, X.; Brody, J.S.; et al. Smoking-induced gene expression changes in the bronchial airway are reflected in nasal and buccal epithelium. BMC Genom. 2008, 9, 259. [Google Scholar] [CrossRef] [PubMed]
- Kupfer, D.M.; White, V.L.; Jenkins, M.C.; Burian, D. Examining smoking-induced differential gene expression changes in buccal mucosa. BMC Med. Genom. 2010, 3, 24. [Google Scholar] [CrossRef] [PubMed]
- Silverman, S. Oral Cancer, 5th ed.; BC Decker, Inc.: Hamilton, ON, Canada, 2003. [Google Scholar]
- Proia, N.K.; Paszkiewicz, G.M.; Nasca, M.A.; Franke, G.E.; Pauly, J.L. Smoking and smokeless tobacco-associated human buccal cell mutations and their association with oral cancer—A review. Cancer Epidemiol. Biomark. Prev. 2006, 15, 1061–1077. [Google Scholar] [CrossRef] [PubMed]
- Van Schooten, F.J.; Besaratinia, A.; de Flora, S.; D’Agostini, F.; Izzotti, A.; Camoirano, A.; Balm, A.J.; Dallinga, J.W.; Bast, A.; Haenen, G.R.; et al. Effects of oral administration of N-acetyl-L-cysteine: A multi-biomarker study in smokers. Cancer Epidemiol. Biomark. Prev. 2002, 11, 167–175. [Google Scholar]
- Cancer Genome Atlas Research Network. Comprehensive genomic characterization of head and neck squamous cell carcinomas. Nature 2015, 517, 576–582. [Google Scholar] [CrossRef] [PubMed]
- Sakamoto, K. Notch signaling in oral squamous neoplasia. Pathol. Int. 2016, 66, 609–617. [Google Scholar] [CrossRef] [PubMed]
- Sanchez-Tena, S.; Cubillos-Rojas, M.; Schneider, T.; Rosa, J.L. Functional and pathological relevance of HERC family proteins: A decade later. Cell. Mol. Life Sci. 2016, 73, 1955–1968. [Google Scholar] [CrossRef]
- Benowitz, N.L.; Hukkanen, J.; Jacob, P. Nicotine Chemistry, Metabolism, Kinetics and Biomarkers, 3rd ed.; Handbook of Experimental Pharmacology; Springer: Berlin/Heidelberg, Germany, 2009; pp. 29–60. [Google Scholar]
- Emery, R.L.; Levine, M.D. Optimal Carbon Monoxide Criteria to Confirm Smoking Status Among Postpartum Women. Nicotine Tob. Res. 2016, 18, 966–970. [Google Scholar] [CrossRef]
- Ripoll, J.; Girauta, H.; Ramos, M.; Medina-Bombardo, D.; Pastor, A.; Alvarez-Ossorio, C.; Gorreto, L.; Esteva, M.; Garcia, E.; Urendez, A.; et al. Clinical trial on the efficacy of exhaled carbon monoxide measurement in smoking cessation in primary health care. BMC Public Health 2012, 12, 322. [Google Scholar] [CrossRef]
- Blumenthal, I. Carbon monoxide poisoning. J. R. Soc. Med. 2001, 94, 270–272. [Google Scholar] [CrossRef] [PubMed]
- Cairns, J. Mutation selection and the natural history of cancer. Nature 1975, 255, 197–200. [Google Scholar] [CrossRef]
- Holland, N.; Bolognesi, C.; Kirsch-Volders, M.; Bonassi, S.; Zeiger, E.; Knasmueller, S.; Fenech, M. The micronucleus assay in human buccal cells as a tool for biomonitoring DNA damage: The HUMN project perspective on current status and knowledge gaps. Mutat. Res. 2008, 659, 93–108. [Google Scholar] [CrossRef] [PubMed]
- Fenech, M.; Holland, N.; Zeiger, E.; Chang, W.P.; Burgaz, S.; Thomas, P.; Bolognesi, C.; Knasmueller, S.; Kirsch-Volders, M.; Bonassi, S. The HUMN and HUMNxL international collaboration projects on human micronucleus assays in lymphocytes and buccal cells—Past, present and future. Mutagenesis 2011, 26, 239–245. [Google Scholar] [CrossRef]
- Autrup, H.; Seremet, T.; Arenholt, D.; Dragsted, L.; Jepsen, A. Metabolism of benzo[a]pyrene by cultured rat and human buccal mucosa cells. Carcinogenesis 1985, 6, 1761–1765. [Google Scholar] [CrossRef] [PubMed]
- Spivack, S.D.; Hurteau, G.J.; Jain, R.; Kumar, S.V.; Aldous, K.M.; Gierthy, J.F.; Kaminsky, L.S. Gene-environment interaction signatures by quantitative mRNA profiling in exfoliated buccal mucosal cells. Cancer Res. 2004, 64, 6805–6813. [Google Scholar] [CrossRef]
- Zhang, Y.; Chen, S.Y.; Hsu, T.; Santella, R.M. Immunohistochemical detection of malondialdehyde-DNA adducts in human oral mucosa cells. Carcinogenesis 2002, 23, 207–211. [Google Scholar] [CrossRef]
- Schwartz, J.L.; Muscat, J.E.; Baker, V.; Larios, E.; Stephenson, G.D.; Guo, W.; Xie, T.; Gu, X.; Chung, F.L. Oral cytology assessment by flow cytometry of DNA adducts, aneuploidy, proliferation and apoptosis shows differences between smokers and non-smokers. Oral Oncol. 2003, 39, 842–854. [Google Scholar] [CrossRef]
- Shani, T.; Onn, A.; Kabha, A.; Ben-Dov, I.; Adam, I.; Amariglio, N.; Yahalom, R.; Rechavi, G.; Trakhtenbrot, L.; Hirshberg, A. Chromosomal numerical aberrations in apparently normal oral mucosa of heavy smokers affected by lung cancer. Oral Oncol. 2010, 46, 96–99. [Google Scholar] [CrossRef]
- Bonassi, S.; Coskun, E.; Ceppi, M.; Lando, C.; Bolognesi, C.; Burgaz, S.; Holland, N.; Kirsh-Volders, M.; Knasmueller, S.; Zeiger, E.; et al. The HUman MicroNucleus project on eXfoLiated buccal cells (HUMN(XL)): The role of life-style, host factors, occupational exposures, health status, and assay protocol. Mutat. Res. 2011, 728, 88–97. [Google Scholar] [CrossRef]
- Shah, V.; Sridhar, S.; Beane, J.; Brody, J.S.; Spira, A. SIEGE: Smoking Induced Epithelial Gene Expression Database. Nucleic Acids Res. 2005, 33, D573–D579. [Google Scholar] [CrossRef] [PubMed]
- Boyle, J.O.; Gumus, Z.H.; Kacker, A.; Choksi, V.L.; Bocker, J.M.; Zhou, X.K.; Yantiss, R.K.; Hughes, D.B.; Du, B.; Judson, B.L. Effects of cigarette smoke on the human oral mucosal transcriptome. Cancer Prev. Res. 2010, 3, 266–278. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Closas, M.; Egan, K.M.; Abruzzo, J.; Newcomb, P.A.; Titus-Ernstoff, L.; Franklin, T.; Bender, P.K.; Beck, J.C.; Le Marchand, L.; Lum, A.; et al. Collection of genomic DNA from adults in epidemiological studies by buccal cytobrush and mouthwash. Cancer Epidemiol. Biomark. Prev. 2001, 10, 687–696. [Google Scholar]
- Huang, B.; Zhang, R. Regulatory non-coding RNAs: Revolutionizing the RNA world. Mol. Biol. Rep. 2014, 41, 3915–3923. [Google Scholar] [CrossRef] [PubMed]
- Maute, R.L.; Dalla-Favera, R.; Basso, K. RNAs with multiple personalities. Wiley Interdiscip. Rev. RNA 2014, 5, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Song, W.; Sun, Y.; Lin, J.; Bi, X. Current research on head and neck cancer-associated long noncoding RNAs. Oncotarget 2018, 9, 1403–1425. [Google Scholar] [CrossRef] [PubMed]
- Zou, A.E.; Ku, J.; Honda, T.K.; Yu, V.; Kuo, S.Z.; Zheng, H.; Xuan, Y.; Saad, M.A.; Hinton, A.; Brumund, K.T.; et al. Transcriptome sequencing uncovers novel long noncoding and small nucleolar RNAs dysregulated in head and neck squamous cell carcinoma. RNA 2015, 21, 1122–1134. [Google Scholar] [CrossRef]
- Leemans, C.R.; Snijders, P.J.F.; Brakenhoff, R.H. The molecular landscape of head and neck cancer. Nat. Rev. Cancer 2018, 18, 269–282. [Google Scholar] [CrossRef]
- De, A. Wnt/Ca2+ signaling pathway: A brief overview. Acta Biochim. Biophys. Sin. 2011, 43, 745–756. [Google Scholar] [CrossRef]
- Guo, W.; Giancotti, F.G. Integrin signalling during tumour progression. Nat. Rev. Mol. Cell Biol. 2004, 5, 816–826. [Google Scholar] [CrossRef]
- Fritz, G.; Henninger, C. Rho GTPases: Novel Players in the Regulation of the DNA Damage Response? Biomolecules 2015, 5, 2417–2434. [Google Scholar] [CrossRef] [PubMed]
- Stransky, N.; Egloff, A.M.; Tward, A.D.; Kostic, A.D.; Cibulskis, K.; Sivachenko, A.; Kryukov, G.V.; Lawrence, M.S.; Sougnez, C.; McKenna, A.; et al. The mutational landscape of head and neck squamous cell carcinoma. Science 2011, 333, 1157–1160. [Google Scholar] [CrossRef]
- Sturner, E.; Behl, C. The Role of the Multifunctional BAG3 Protein in Cellular Protein Quality Control and in Disease. Front. Mol. Neurosci. 2017, 10, 177. [Google Scholar] [CrossRef] [PubMed]
- Eto, M. Regulation of cellular protein phosphatase-1 (PP1) by phosphorylation of the CPI-17 family, C-kinase-activated PP1 inhibitors. J. Biol. Chem. 2009, 284, 35273–35277. [Google Scholar] [CrossRef] [PubMed]
- Huang da, W.; Sherman, B.T.; Lempicki, R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009, 4, 44–57. [Google Scholar] [CrossRef] [PubMed]
- Tommasi, S.; Zheng, A.; Weninger, A.; Bates, S.E.; Li, X.A.; Wu, X.; Hollstein, M.; Besaratinia, A. Mammalian cells acquire epigenetic hallmarks of human cancer during immortalization. Nucleic Acids Res. 2013, 41, 182–195. [Google Scholar] [CrossRef]
| E-cig Users (n = 42) | Smokers (n = 24) | Controls (n = 27) | ||
|---|---|---|---|---|
| Age * | 28 ± 1.3 | 42 ± 2.8 | 24 ± 1.7 | |
| Gender | Male | 34 (81.0%) | 19 (79.2%) | 16 (59.3%) |
| Female | 8 (19.0%) | 5 (20.8%) | 11 (40.7%) | |
| Race | White | 16 (38.1%) | 5 (20.8%) | 5 (18.5%) |
| Hispanic | 12 (28.6%) | 1 (4.2%) | 5 (18.5%) | |
| African American | 5 (11.9%) | 9 (37.5%) | 2 (7.4%) | |
| Asian | 7 (16.7%) | 4 (16.7%) | 12 (44.4%) | |
| Other | 2 (4.8%) | 5 (20.8%) | 3 (11.1%) | |
| Education | Less than high school | 0 (0%) | 3 (12.5%) | 0 (0%) |
| High school diploma or GED | 11 (26.2%) | 2 (8.3%) | 0 (0%) | |
| Some college completed or currently enrolled in college | 19 (45.2%) | 8 (33.3%) | 1 (3.7%) | |
| College degree or higher | 12 (28.6%) | 11 (45.8%) | 26 (96.3%) | |
| Marital status | Married | 6 (14.3%) | 3 (12.5%) | 0 (0%) |
| Currently living with someone | 2 (4.8%) | 0 (0%) | 0 (0%) | |
| Widowed | 1 (2.4%) | 0 (0%) | 0 (0%) | |
| Separated | 1 (2.4%) | 1 (4.2%) | 0 (0%) | |
| Divorced | 5 (11.9%) | 2 (8.3%) | 3 (11.1%) | |
| Single and never married | 27 (64.3%) | 18 (75.0%) | 24 (88.9%) | |
| Employment status | Full time | 26 (61.9%) | 12 (50%) | 19 (70.4%) |
| Part time | 9 (21.4%) | 4 (16.7%) | 6 (22.2%) | |
| Retired or disability | 1 (2.4%) | 3 (12.5%) | 0 (0%) | |
| Unemployed | 6 (14.3%) | 5 (20.8%) | 2 (7.4%) | |
| Pre-tax-annual income | <$15,000 | 4 (9.5%) | 9 (37.5%) | 9 (33.3%) |
| ≥$15,000 to <$30,000 | 12 (28.6%) | 5 (20.8%) | 5 (18.5%) | |
| ≥$30,000 to <$45,000 | 9 (21.4%) | 4 (16.7%) | 4 (14.8%) | |
| ≥$45,000 to <$60,000 | 6 (14.3%) | 1 (4.2%) | 2 (7.4%) | |
| ≥$60,000 to <$75,000 | 3 (7.1%) | 0 (0%) | 2 (7.4%) | |
| ≥$75,000 to <$90,000 | 3 (7.1%) | 1 (4.2%) | 1 (3.7%) | |
| ≥$90,000 to <$105,000 | 1 (2.4%) | 1 (4.2%) | 1 (3.7%) | |
| ≥$105,000 to <$120,000 | 0 (0%) | 1 (4.2%) | 0 (0%) | |
| ≥$120,000 | 4 (9.5%) | 2 (8.3%) | 3 (11.1%) | |
| BMI *,† | 27.9 ± 1.1 | 27.2 ± 0.9 | 25.0 ± 1.3 | |
| Pack Year *,‡ | NA | 12.3 ± 2.5 | NA | |
| Cumulative e-liquid (mL) *,¶ | 5369.5 ± 3038.5 | NA | NA | |
| Cumulative e-nicotine (mg) *,¶¶ | 20,859.7 ± 10,346.6 | NA | NA | |
| Plasma cotinine (ng/mL) * | 115.0 ± 8.5 | 122.0 ± 10.8 | 2.5 ± 0.1 | |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tommasi, S.; Caliri, A.W.; Caceres, A.; Moreno, D.E.; Li, M.; Chen, Y.; Siegmund, K.D.; Besaratinia, A. Deregulation of Biologically Significant Genes and Associated Molecular Pathways in the Oral Epithelium of Electronic Cigarette Users. Int. J. Mol. Sci. 2019, 20, 738. https://doi.org/10.3390/ijms20030738
Tommasi S, Caliri AW, Caceres A, Moreno DE, Li M, Chen Y, Siegmund KD, Besaratinia A. Deregulation of Biologically Significant Genes and Associated Molecular Pathways in the Oral Epithelium of Electronic Cigarette Users. International Journal of Molecular Sciences. 2019; 20(3):738. https://doi.org/10.3390/ijms20030738
Chicago/Turabian StyleTommasi, Stella, Andrew W. Caliri, Amanda Caceres, Debra E. Moreno, Meng Li, Yibu Chen, Kimberly D. Siegmund, and Ahmad Besaratinia. 2019. "Deregulation of Biologically Significant Genes and Associated Molecular Pathways in the Oral Epithelium of Electronic Cigarette Users" International Journal of Molecular Sciences 20, no. 3: 738. https://doi.org/10.3390/ijms20030738
APA StyleTommasi, S., Caliri, A. W., Caceres, A., Moreno, D. E., Li, M., Chen, Y., Siegmund, K. D., & Besaratinia, A. (2019). Deregulation of Biologically Significant Genes and Associated Molecular Pathways in the Oral Epithelium of Electronic Cigarette Users. International Journal of Molecular Sciences, 20(3), 738. https://doi.org/10.3390/ijms20030738







