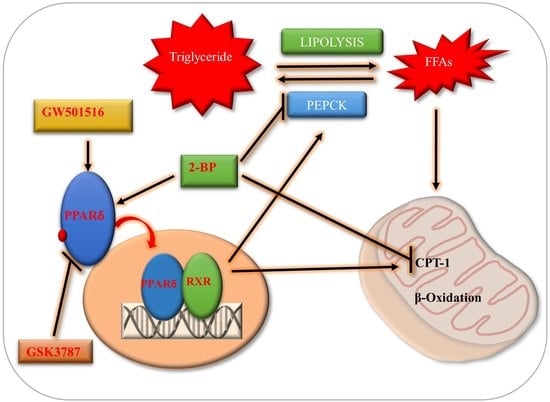
The PPARδ Agonist GW501516 Improves Lipolytic/Lipogenic Balance through CPT1 and PEPCK during the Development of Pre-Implantation Bovine Embryos
Abstract
1. Introduction
2. Results
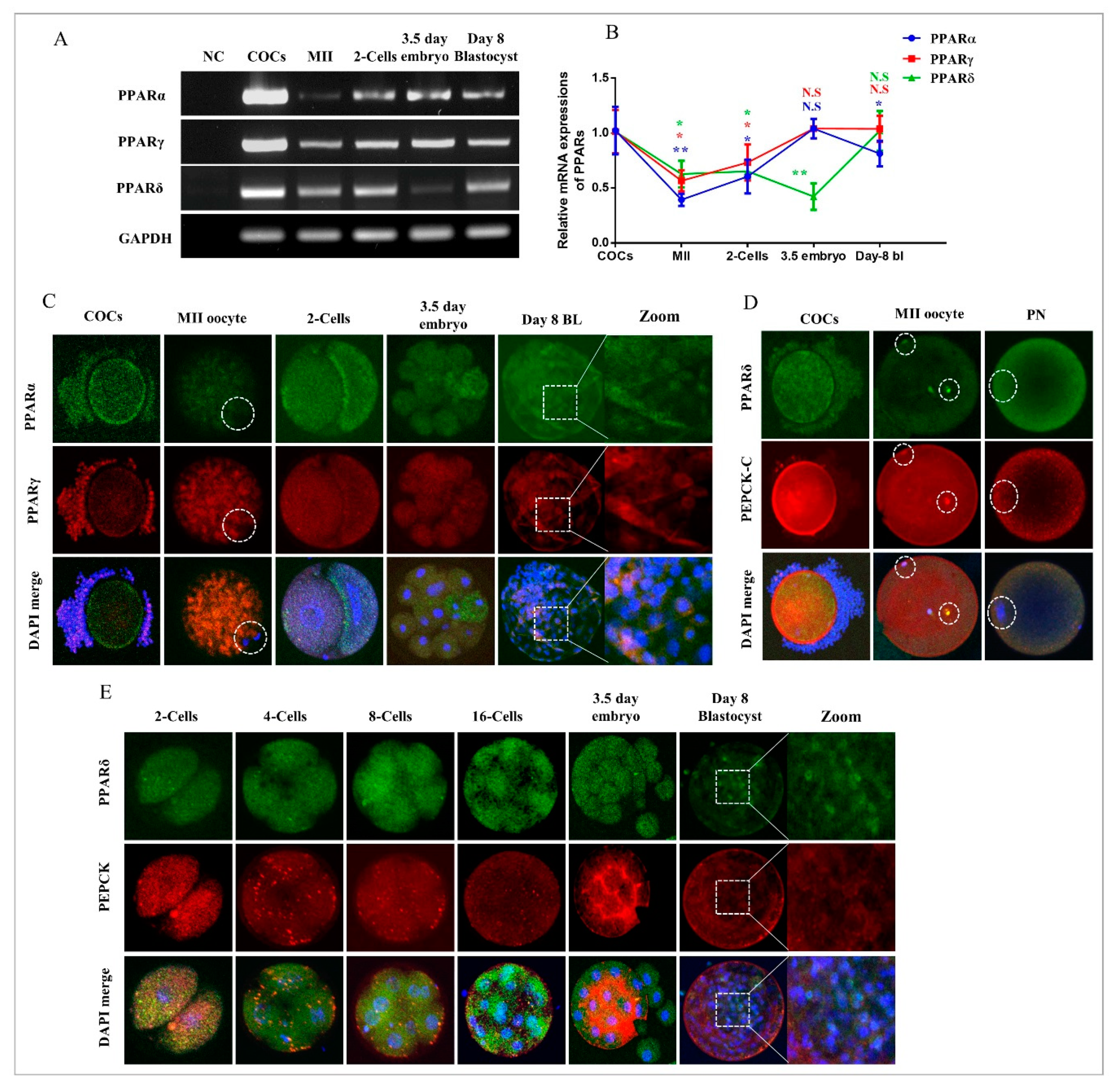
2.1. Dynamic Changes in PPARs and PEPCK Expression during In Vitro Oocyte Maturation and Embryo Development
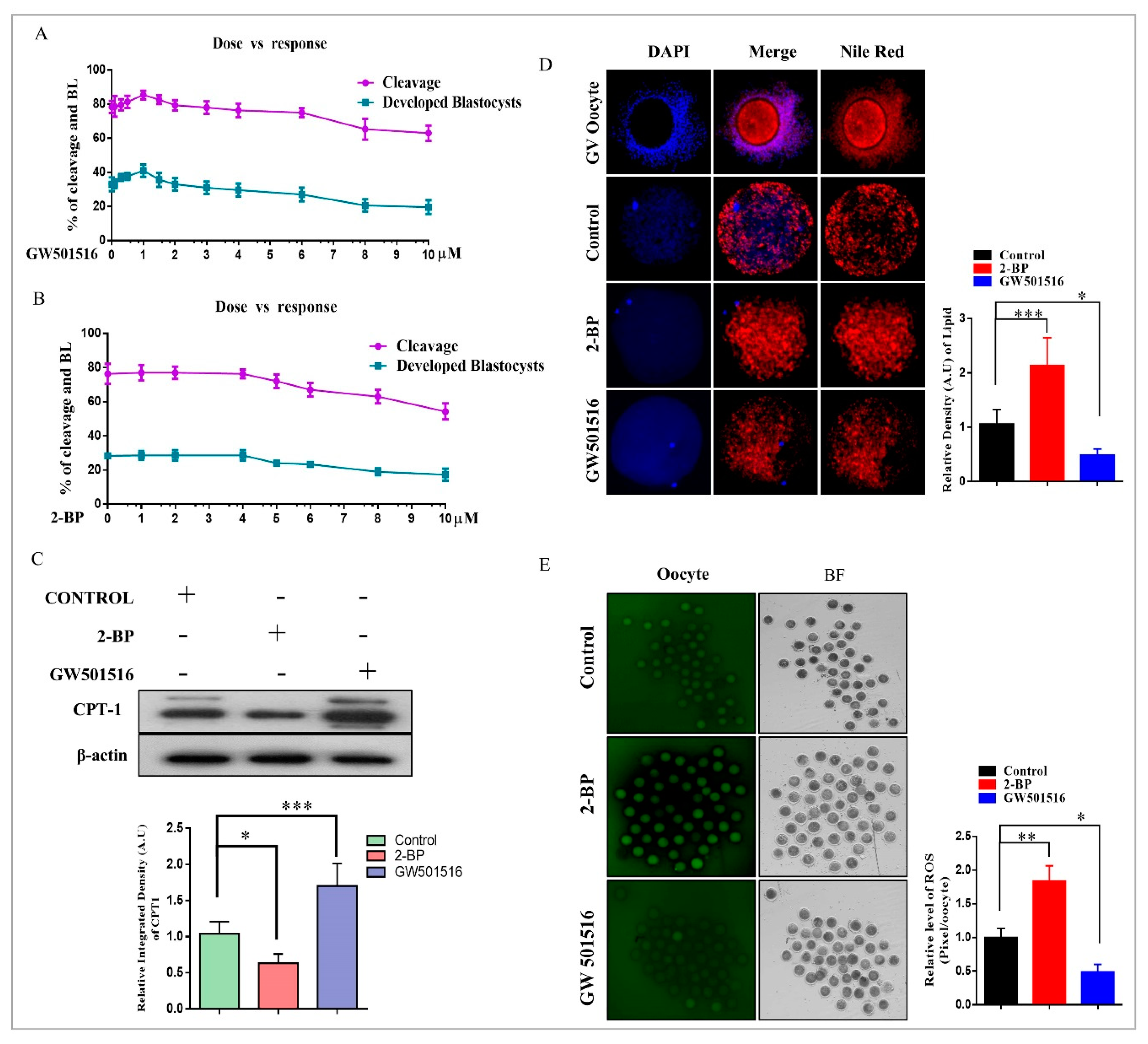
2.2. Effect of 2-BP and GW501516 on PPARs, Lipid Metabolism, and ROS Level during GVBD Induction
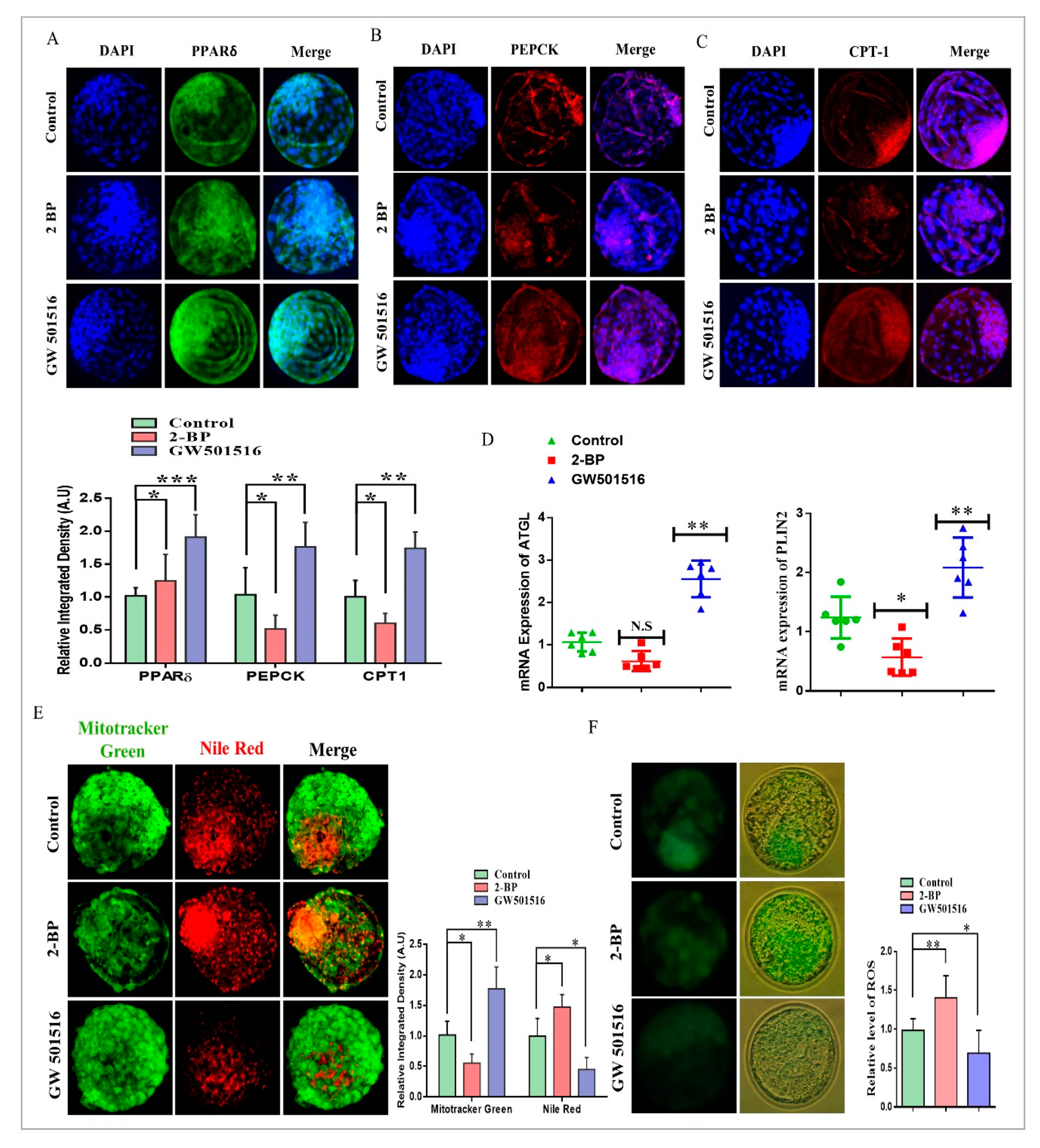
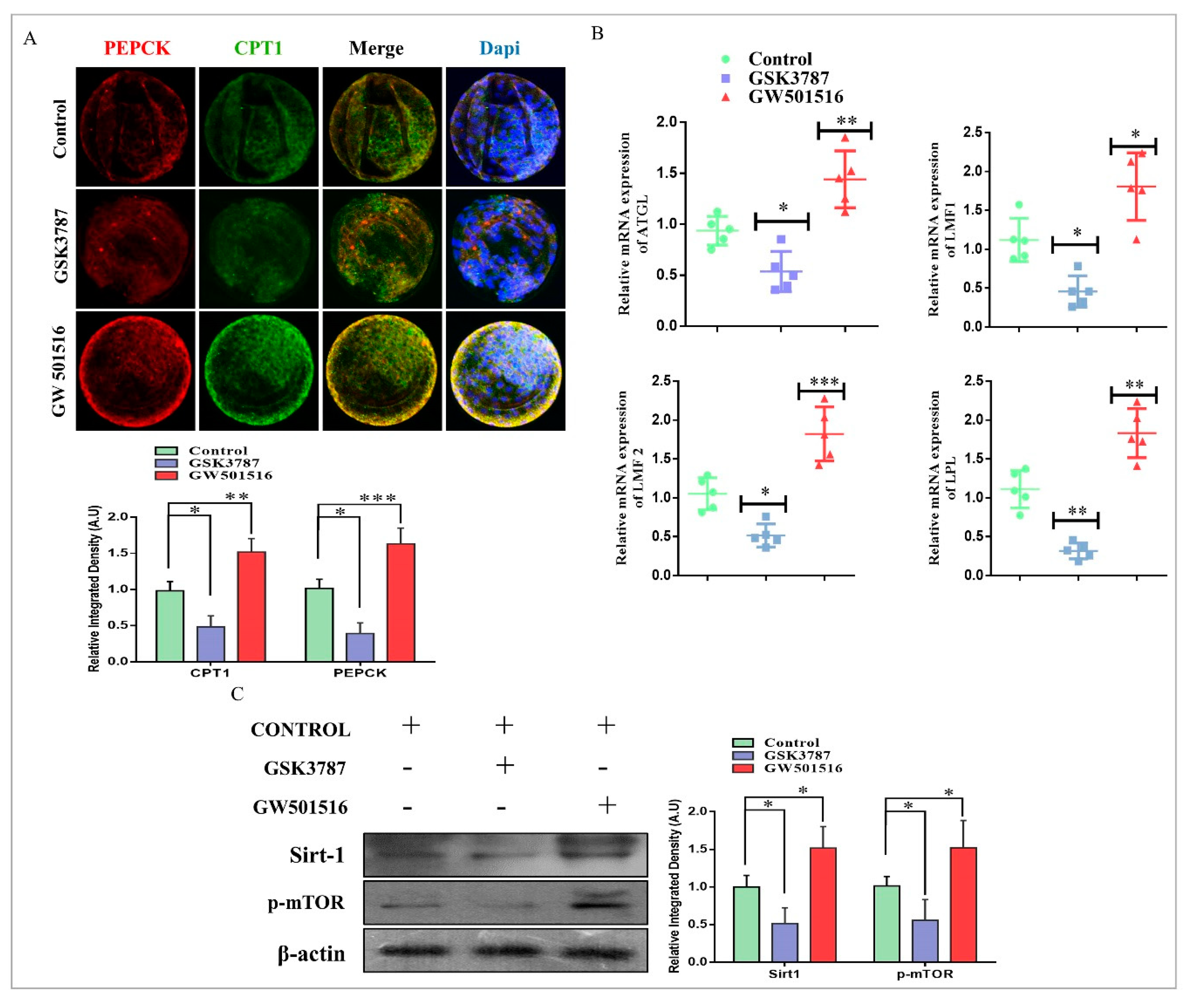
2.3. PPARδ/PEPCK Expression and Mitochondrial β-Oxidation in Bovine Embryos
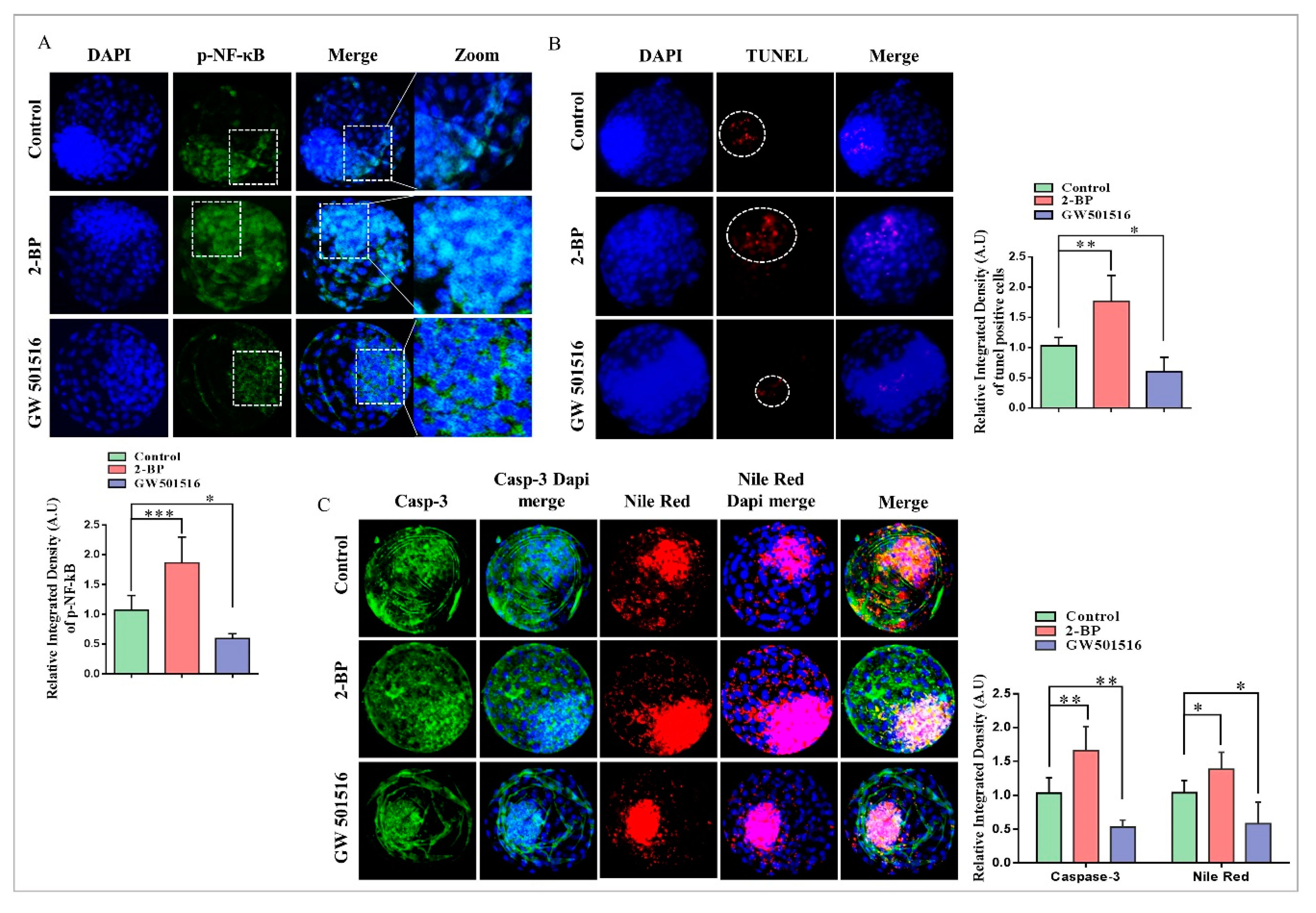
2.4. Disturbance in Lipolytic/Lipogenic Balance Enhanced Apoptosis in Bovine Blastocysts
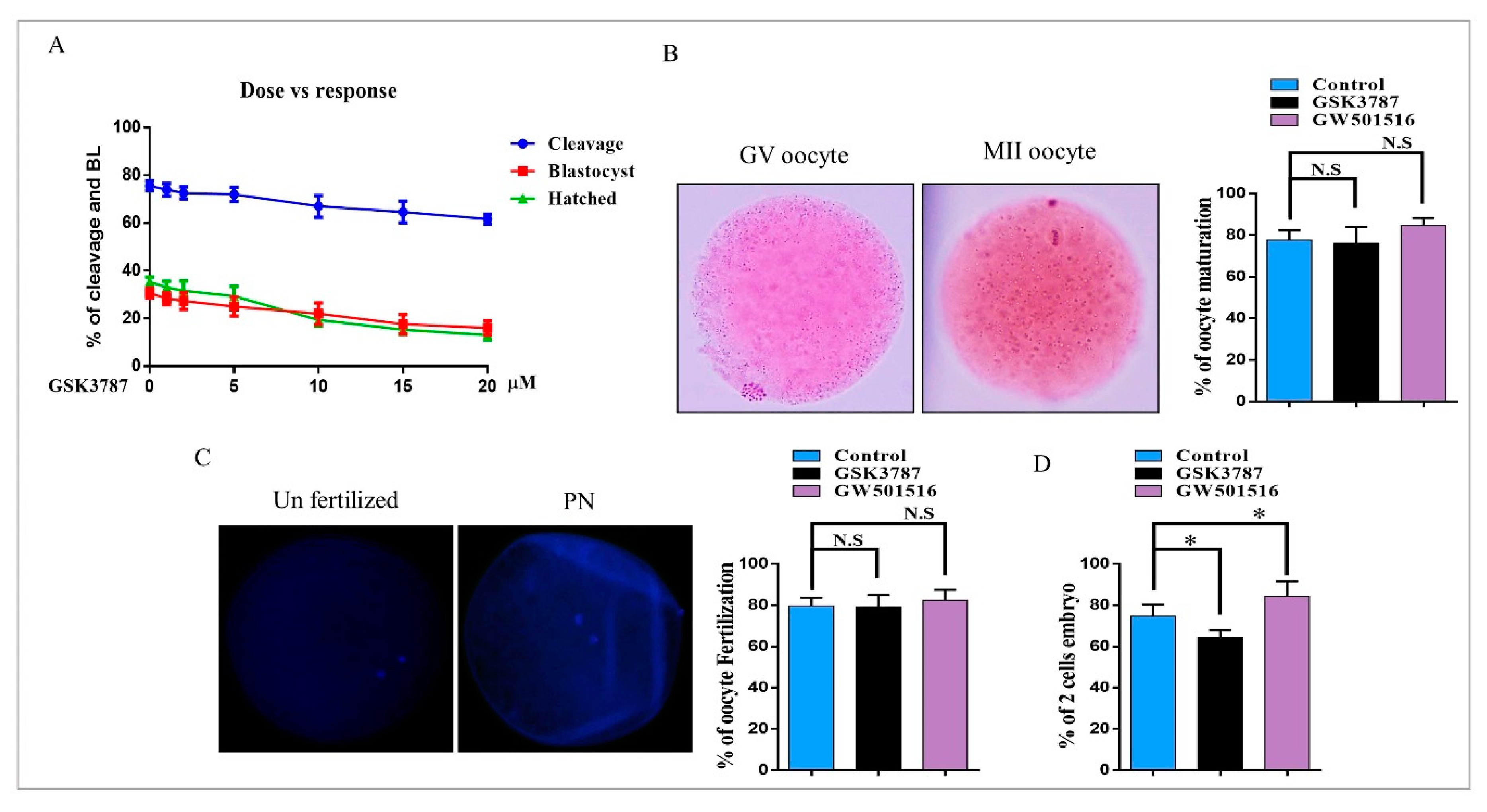
2.5. PPARδ Inhibition Reduced Embryo Development and Hatching
2.6. PPARδ Reversibility Affected Lipid Metabolism and Embryo Survival
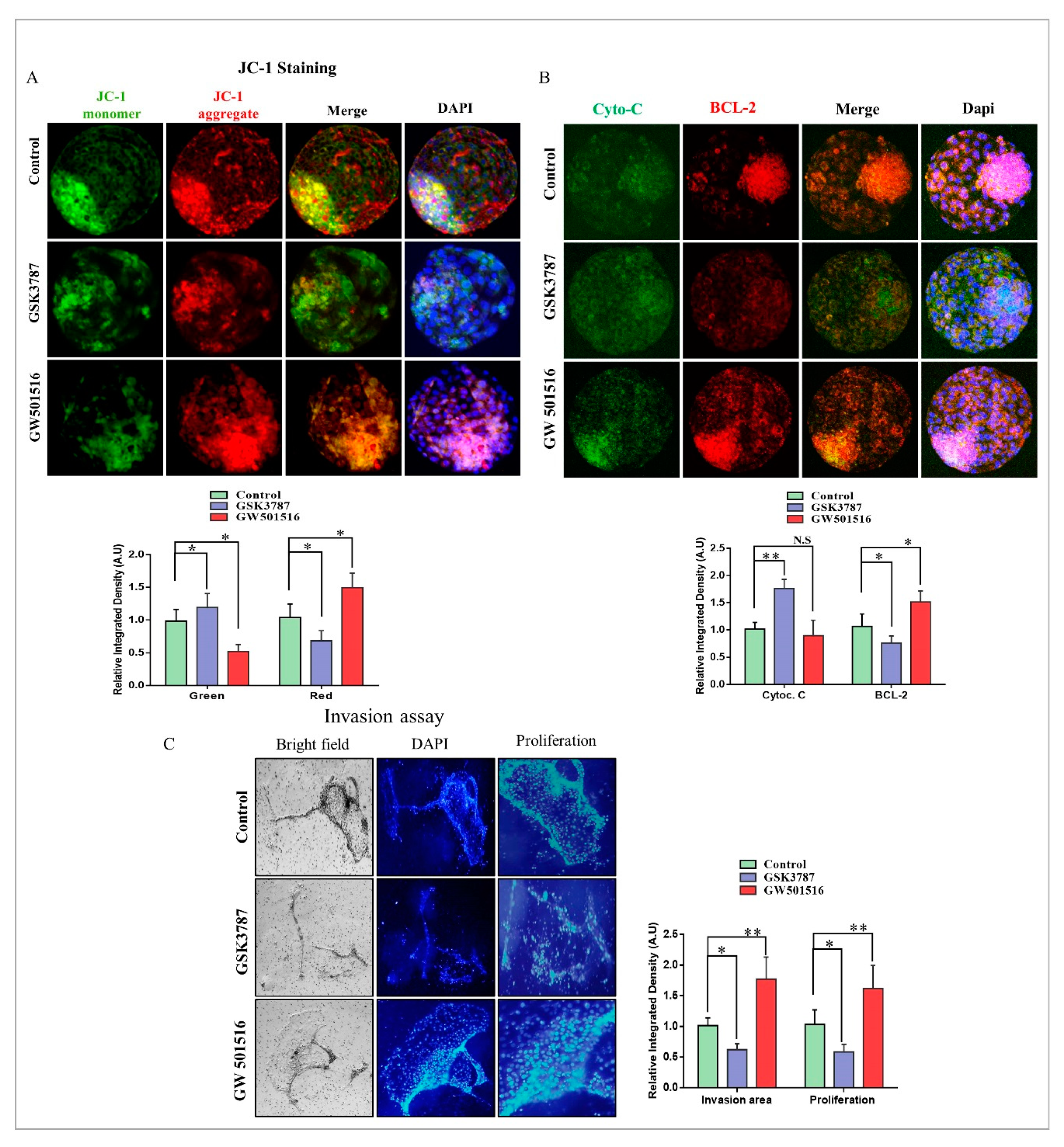
2.7. PPARδ Effects on Mitochondria and Implantation Potential of Bovine Day-8 Blastocysts
3. Discussion
4. Materials and Methods
4.1. Experimental Design
4.1.1. Experiment 1 (PPARδ Activation)
4.1.2. Experiment 2 (PPARδ Inhibition)
4.2. Oocyte Collection
4.3. In Vitro Maturation (IVM)
4.4. In Vitro Fertilization and Culture
4.5. Immunofluorescence
4.6. TUNEL Assay
4.7. H2DCFDA Assay for ROS Detection
4.8. Extraction of mRNA and cDNA Synthesis
4.9. Real-Time Polymerase Chain Reaction
4.10. Protein Extraction and Western Blot Analysis
4.11. Mitochondrial–Lipid Dual Staining
4.12. Antibodies
4.13. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| PPARδ | Peroxisome Proliferator-activated receptor δ |
| PEPCK | Phosphoenolpyruvate carboxykinase |
| CPT1 | Carnitine palmitoyltransferase I |
| p-NF-κB | Phosphorylated-nuclear factor kappa light chain enhancer of activated B cells |
| 2-BP | 2-Bromo Palmitate |
| GSK3787 | 4-chloro-N-(2-ethyl) Benz amide |
| GV oocyte | Germinal Vesicle oocyte |
| ROS | Reactive Oxygen Species |
References
- Dunning, K.R.; Russell, D.L.; Robker, R.L. Lipids and oocyte developmental competence: The role of fatty acids and beta-oxidation. Reproduction 2014, 148, 15–27. [Google Scholar] [CrossRef] [PubMed]
- Aardema, H.; Vos, P.L.; Lolicato, F.; Roelen, B.A.; Knijn, H.M.; Vaandrager, A.B.; Helms, J.B.; Gadella, B.M. Oleic acid prevents detrimental effects of saturated fatty acids on bovine oocyte developmental competence. Biol. Reprod. 2011, 85, 62–69. [Google Scholar] [CrossRef] [PubMed]
- Cetica, P.; Pintos, L.; Dalvit, G.; Beconi, M. Activity of key enzymes involved in glucose and triglyceride catabolism during bovine oocyte maturation in vitro. Reproduction 2002, 124, 675–681. [Google Scholar] [CrossRef] [PubMed]
- Prastowo, S.; Amin, A.; Rings, F.; Held, E.; Wondim, D.S.; Gad, A.; Neuhoff, C.; Tholen, E.; Looft, C.; Schellander, K.; et al. Fateful triad of reactive oxygen species, mitochondrial dysfunction and lipid accumulation is associated with expression outline of the AMP-activated protein kinase pathway in bovine blastocysts. Reprod. Fertil. Dev. 2016, 29, 890–905. [Google Scholar] [CrossRef] [PubMed]
- Abe, H.; Yamashita, S.; Satoh, T.; Hoshi, H. Accumulation of cytoplasmic lipid droplets in bovine embryos and cryotolerance of embryos developed in different culture systems using serum-free or serum-containing media. Mol. Reprod. Dev. 2002, 61, 57–66. [Google Scholar] [CrossRef] [PubMed]
- Schwarz, K.R.L.; Botigelli, R.C.; Del Collado, M.; de Castro, F.C.; Fernandes, H.; Paschoal, D.M.; Leal, C.L.V. Effects of fetal calf serum on cGMP pathway and oocyte lipid metabolism in vitro. Reprod. Fertil. Dev. 2017, 29, 1593–1601. [Google Scholar] [CrossRef]
- Luquet, S.; Gaudel, C.; Holst, D.; Lopez-Soriano, J.; Jehl-Pietri, C.; Fredenrich, A.; Grimaldi, P.A. Roles of PPAR delta in lipid absorption and metabolism: A new target for the treatment of type 2 diabetes. Biochim. Biophys. Acta 2005, 1740, 313–317. [Google Scholar] [CrossRef]
- Ravnskjaer, K.; Frigerio, F.; Boergesen, M.; Nielsen, T.; Maechler, P.; Mandrup, S. PPARdelta is a fatty acid sensor that enhances mitochondrial oxidation in insulin-secreting cells and protects against fatty acid-induced dysfunction. J. Lipid Res. 2010, 51, 1370–1379. [Google Scholar] [CrossRef]
- Liu, Y.; Colby, J.K.; Zuo, X.; Jaoude, J.; Wei, D.; Shureiqi, I. The Role of PPAR-delta in Metabolism, Inflammation, and Cancer: Many Characters of a Critical Transcription Factor. Int. J. Mol. Sci. 2018, 19, 3339. [Google Scholar] [CrossRef]
- De Lange, P.; Lombardi, A.; Silvestri, E.; Goglia, F.; Lanni, A.; Moreno, M. Peroxisome Proliferator-Activated Receptor Delta: A Conserved Director of Lipid Homeostasis through Regulation of the Oxidative Capacity of Muscle. PPAR Res. 2008, 2008, 172676. [Google Scholar] [CrossRef]
- Kramer, D.K.; Al-Khalili, L.; Guigas, B.; Leng, Y.; Garcia-Roves, P.M.; Krook, A. Role of AMP kinase and PPARdelta in the regulation of lipid and glucose metabolism in human skeletal muscle. J. Biol. Chem. 2007, 282, 19313–19320. [Google Scholar] [CrossRef] [PubMed]
- Rogers, N.H.; Perfield II, J.W.; Strissel, K.J.; Obin, M.S.; Greenberg, A.S. Loss of ovarian function in mice results in abrogated skeletal muscle PPARdelta and FoxO1-mediated gene expression. Biochem. Biophys. Res. Commun. 2010, 392, 1–3. [Google Scholar] [CrossRef] [PubMed]
- Montal, E.D.; Dewi, R.; Bhalla, K.; Ou, L.; Hwang, B.J.; Ropell, A.E.; Gordon, C.; Liu, W.J.; DeBerardinis, R.J.; Sudderth, J.; et al. PEPCK Coordinates the Regulation of Central Carbon Metabolism to Promote Cancer Cell Growth. Mol. Cell 2015, 60, 571–583. [Google Scholar] [CrossRef] [PubMed]
- Fan, W.; Waizenegger, W.; Lin, C.S.; Sorrentino, V.; He, M.X.; Wall, C.E.; Li, H.; Liddle, C.; Ruth, T.Y.; Atkins, A.R.; et al. PPARdelta Promotes Running Endurance by Preserving Glucose. Cell Metab. 2017, 25, 1186–1193. [Google Scholar] [CrossRef] [PubMed]
- Millward, C.A.; DeSantis, D.; Hsieh, C.W.; Heaney, J.D.; Pisano, S.; Olswang, Y.; Reshef, L.; Beidelschies, M.; Puchowicz, M.; Croniger, C.M. Phosphoenolpyruvate carboxykinase (Pck1) helps regulate the triglyceride/fatty acid cycle and development of insulin resistance in mice. J. Lipid Res. 2010, 51, 1452–1463. [Google Scholar] [CrossRef] [PubMed]
- Assou, S.; Boumela, I.; Haouzi, D.; Anahory, T.; Dechaud, H.; De Vos, J.; Hamamah, S. Dynamic changes in gene expression during human early embryo development: From fundamental aspects to clinical applications. Hum. Reprod. Update 2011, 17, 272–290. [Google Scholar] [CrossRef]
- Assou, S.; Haouzi, D.; Mahmoud, K.; Aouacheria, A.; Guillemin, Y.; Pantesco, V.; Reme, T.; Dechaud, H.; De Vos, J.; Hamamah, S. A non-invasive test for assessing embryo potential by gene expression profiles of human cumulus cells: A proof of concept study. Mol. Hum. Reprod. 2008, 14, 711–719. [Google Scholar] [CrossRef]
- Barak, Y.; Liao, D.; He, W.; Ong, E.S.; Nelson, M.C.; Olefsky, J.M.; Boland, R.; Evans, R.M. Effects of peroxisome proliferator-activated receptor delta on placentation, adiposity, and colorectal cancer. Proc. Natl. Acad. Sci. USA 2002, 99, 303–308. [Google Scholar] [CrossRef]
- Komar, C.M.; Braissant, O.; Wahli, W.; Curry, T.E., Jr. Expression and localization of PPARs in the rat ovary during follicular development and the periovulatory period. Endocrinology 2001, 142, 4831–4838. [Google Scholar] [CrossRef]
- MacLaren, L.A.; Guzeloglu, A.; Michel, F.; Thatcher, W.W. Peroxisome proliferator-activated receptor (PPAR) expression in cultured bovine endometrial cells and response to omega-3 fatty acid, growth hormone and agonist stimulation in relation to series 2 prostaglandin production. Domest. Anim. Endocrinol. 2006, 30, 155–169. [Google Scholar] [CrossRef]
- Huang, J.C.; Wun, W.S.; Goldsby, J.S.; Wun, I.C.; Noorhasan, D.; Wu, K.K. Stimulation of embryo hatching and implantation by prostacyclin and peroxisome proliferator-activated receptor delta activation: Implication in IVF. Hum. Reprod. 2007, 22, 807–814. [Google Scholar] [CrossRef]
- Huang, J.C. The role of peroxisome proliferator-activated receptors in the development and physiology of gametes and preimplantation embryos. PPAR Res. 2008, 2008, 732303. [Google Scholar] [CrossRef] [PubMed]
- Palkar, P.S.; Borland, M.G.; Naruhn, S.; Ferry, C.H.; Lee, C.; Sk, U.H.; Sharma, A.K.; Amin, S.; Murray, I.A.; Anderson, C.R.; et al. Cellular and pharmacological selectivity of the peroxisome proliferator-activated receptor-beta/delta antagonist GSK3787. Mol. Pharmacol. 2010, 78, 419–430. [Google Scholar] [CrossRef] [PubMed]
- Memili, E.; First, N.L. Control of gene expression at the onset of bovine embryonic development. Biol. Reprod. 1999, 61, 1198–1207. [Google Scholar] [CrossRef] [PubMed]
- Kang, H.J.; Kang, H.J.; Hwang, S.J.; Lim, H.; Song, H. Activation of peroxisome proliferators-activated receptor delta (PPARdelta) promotes blastocyst hatching in mice. Mol. Hum. Reprod. 2011, 17, 653–660. [Google Scholar] [CrossRef] [PubMed]
- Sudano, M.J.; Paschoal, D.M.; da Silva Rascado, T.; Crocomo, L.F.; Magalhães, L.C.O.; Junior, A.M.; Machado, R.; da Cruz Landim-Alvarenga, F. Crucial surviving aspects for vitrified in vitro-produced bovine embryos. Zygote 2014, 22, 124–131. [Google Scholar] [CrossRef]
- Wan, J.; Jiang, L.; Lü, Q.; Ke, L.; Li, X.; Tong, N. Activation of PPARdelta up-regulates fatty acid oxidation and energy uncoupling genes of mitochondria and reduces palmitate-induced apoptosis in pancreatic beta-cells. Biochem. Biophys. Res. Commun. 2010, 391, 1567–1572. [Google Scholar] [CrossRef]
- Somfai, T.; Kaneda, M.; Akagi, S.; Watanabe, S.; Haraguchi, S.; Mizutani, E.; Dang-Nguyen, T.Q.; Geshi, M.; Kikuchi, K.; Nagai, T. Enhancement of lipid metabolism with L-carnitine during in vitro maturation improves nuclear maturation and cleavage ability of follicular porcine oocytes. Reprod. Fertil. Dev. 2011, 23, 912–920. [Google Scholar] [CrossRef]
- Bickel, P.E.; Tansey, J.T.; Welte, M.A. PAT proteins, an ancient family of lipid droplet proteins that regulate cellular lipid stores. Biochim. Biophys. Acta 2009, 1791, 419–440. [Google Scholar] [CrossRef]
- Szkudelski, T.; Szkudelska, K. Short-term effects of palmitate and 2-bromopalmitate on the lipolytic activity of rat adipocytes. Life Sci. 2011, 89, 450–455. [Google Scholar] [CrossRef]
- Benkhalifa, M.; Ferreira, Y.J.; Chahine, H.; Louanjli, N.; Miron, P.; Merviel, P.; Copin, H. Mitochondria: Participation to infertility as source of energy and cause of senescence. Int. J. Biochem. Cell Biol. 2014, 55, 60–64. [Google Scholar] [CrossRef] [PubMed]
- Ferguson, E.M.; Leese, H.J. Triglyceride content of bovine oocytes and early embryos. J. Reprod. Fertil. 1999, 116, 373–378. [Google Scholar] [CrossRef] [PubMed]
- Jin, J.X.; Lee, S.; Taweechaipaisankul, A.; Kim, G.A.; Lee, B.C. Melatonin regulates lipid metabolism in porcine oocytes. J. Pineal. Res. 2017, 62, e12388. [Google Scholar] [CrossRef] [PubMed]
- Sadeghi, A.; Seyyed Ebrahimi, S.S.; Golestani, A.; Meshkani, R. Resveratrol Ameliorates Palmitate-Induced Inflammation in Skeletal Muscle Cells by Attenuating Oxidative Stress and JNK/NF-κB Pathway in a SIRT1-Independent Mechanism. J. Cell. Biochem. 2017, 118, 2654–2663. [Google Scholar] [CrossRef] [PubMed]
- Okazaki, M.; Iwasaki, Y.; Nishiyama, M.; Taguchi, T.; Tsugita, M.; Nakayama, S.; Kambayashi, M.; Hashimoto, K.; Terada, Y. PPARbeta/delta regulates the human SIRT1 gene transcription via Sp1. Endocr. J. 2010, 57, 403–413. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.H.; Olson, P.; Hevener, A.; Mehl, I.; Chong, L.W.; Olefsky, J.M.; Gonzalez, F.J.; Ham, J.; Kang, H.; Peters, J.M.; et al. PPARdelta regulates glucose metabolism and insulin sensitivity. Proc. Natl. Acad. Sci. USA 2006, 103, 3444–3449. [Google Scholar] [CrossRef]
- Bradley, J.; Swann, K. Mitochondria and lipid metabolism in mammalian oocytes and early embryos. Int. J. Dev. Biol. 2019, 63, 93–103. [Google Scholar] [CrossRef]
- Lolicato, F.; Brouwers, J.F.; de Lest, C.H.V.; Wubbolts, R.; Aardema, H.; Priore, P.; Roelen, B.A.; Helms, J.B.; Gadella, B.M. The cumulus cell layer protects the bovine maturing oocyte against fatty acid-induced lipotoxicity. Biol. Reprod. 2015, 92, 16. [Google Scholar] [CrossRef]
- Kim, J.Y.; Kinoshita, M.; Ohnishi, M.; Fukui, Y. Lipid and fatty acid analysis of fresh and frozen-thawed immature and in vitro matured bovine oocytes. Reproduction 2001, 122, 131–138. [Google Scholar] [CrossRef]
- Ferguson, E.M.; Leese, H.J. A potential role for triglyceride as an energy source during bovine oocyte maturation and early embryo development. Mol. Reprod. Dev. 2006, 73, 1195–1201. [Google Scholar] [CrossRef]
- Schwarz, K.R.L.; de Castro, F.C.; Schefer, L.; Botigelli, R.C.; Paschoal, D.M.; Fernandes, H.; Leal, C.L. The role of cGMP as a mediator of lipolysis in bovine oocytes and its effects on embryo development and cryopreservation. PLoS ONE 2018, 13, e0191023. [Google Scholar]
- Forman, B.M.; Chen, J.; Evans, R.M. Hypolipidemic drugs, polyunsaturated fatty acids, and eicosanoids are ligands for peroxisome proliferator-activated receptors alpha and delta. Proc. Natl. Acad. Sci. USA 1997, 94, 4312–4317. [Google Scholar] [CrossRef] [PubMed]
- Ogawa, K.; Itami, N.; Ueda, M.; Kansaku, K.; Shirasuna, K.; Kuwayama, T.; Iwata, H. Non-esterified fatty acid-associated ability of follicular fluid to support porcine oocyte maturation and development. Reprod. Med. Biol. 2018, 17, 155–163. [Google Scholar] [CrossRef] [PubMed]
- Uhde, K.; van Tol, H.T.; Stout, T.A.; Roelen, B.A. Exposure to elevated glucose concentrations alters the metabolomic profile of bovine blastocysts. PLoS ONE 2018, 13, e0199310. [Google Scholar] [CrossRef] [PubMed]
- Abe, T.; Kawahara-Miki, R.; Hara, T.; Noguchi, T.; Hayashi, T.; Shirasuna, K.; Kuwayama, T.; Iwata, H. Modification of mitochondrial function, cytoplasmic lipid content and cryosensitivity of bovine embryos by resveratrol. J. Reprod. Dev. 2017, 63, 455–461. [Google Scholar] [CrossRef] [PubMed]
- Paczkowski, M.; Silva, E.; Schoolcraft, W.B.; Krisher, R.L. Comparative importance of fatty acid beta-oxidation to nuclear maturation, gene expression, and glucose metabolism in mouse, bovine, and porcine cumulus oocyte complexes. Biol. Reprod. 2013, 88, 111. [Google Scholar] [CrossRef]
- Song, B.S.; Kim, J.S.; Koo, D.B.; Park, J.S.; Lee, K.K.; Han, Y.M. 178 Improved Developmental Competence of Bovine Embryos by Pgi2 Analog Treatment. Reprod. Fertil. Dev. 2006, 18, 197. [Google Scholar] [CrossRef]
- Elo, B.; Villano, C.M.; Govorko, D.; White, L.A. Larval zebrafish as a model for glucose metabolism: Expression of phosphoenolpyruvate carboxykinase as a marker for exposure to anti-diabetic compounds. J. Mol. Endocrinol. 2007, 38, 433–440. [Google Scholar] [CrossRef]
- Ahn, S.W.; Gang, G.T.; Tadi, S.; Nedumaran, B.; Kim, Y.D.; Park, J.H.; Kweon, G.R.; Koo, S.H.; Lee, K.; Ahn, R.S.; et al. Phosphoenolpyruvate carboxykinase and glucose-6-phosphatase are required for steroidogenesis in testicular Leydig cells. J. Biol. Chem. 2012, 287, 41875–41887. [Google Scholar] [CrossRef]
- Desmet, K.L.; Van Hoeck, V.; Gagné, D.; Fournier, E.; Thakur, A.; O’doherty, A.M.; Walsh, C.P.; Sirard, M.A.; Bols, P.E.J.; Leroy, J.L.M.R. Exposure of bovine oocytes and embryos to elevated non-esterified fatty acid concentrations: Integration of epigenetic and transcriptomic signatures in resultant blastocysts. BMC Genom. 2016, 17, 1004. [Google Scholar] [CrossRef]
- Wan, Z.; Matravadia, S.; Holloway, G.P.; Wright, D.C. FAT/CD36 regulates PEPCK expression in adipose tissue. Am. J. Physiol. Cell Physiol. 2013, 304, 478–484. [Google Scholar] [CrossRef] [PubMed]
- Yuan, Y.G.; Xu, L.; Zhang, S.; Mesalam, A.; Lee, K.L.; Liu, H.; Joo, M.D.; Idrees, M.; Kong, I.K. Polydatin and I-CBP112 protects early bovine embryo against nicotinamide-induced mitochondrial dysfunction. Theriogenology 2019, 134, 1–10. [Google Scholar] [CrossRef] [PubMed]
- El Sheikh, M.; Mesalam, A.; Mesalam, A.A.; Idrees, M.; Lee, K.L.; Kong, I.K. Melatonin Abrogates the Anti-Developmental Effect of the AKT Inhibitor SH6 in Bovine Oocytes and Embryos. Int. J. Mol. Sci. 2019, 20, 2956. [Google Scholar] [CrossRef] [PubMed]
- Idrees, M.; Xu, L.; Song, S.H.; Joo, M.D.; Lee, K.L.; Muhammad, T.; El Sheikh, M.; Sidrat, T.; Kong, I.K. PTPN11 (SHP2) Is Indispensable for Growth Factors and Cytokine Signal Transduction During Bovine Oocyte Maturation and Blastocyst Development. Cells 2019, 8, 1272. [Google Scholar] [CrossRef]
- Chowdhury, M.M.R.; Mesalam, A.; Khan, I.; Joo, M.D.; Lee, K.L.; Xu, L.; Afrin, F.; Kong, I.K. Improved developmental competence in embryos treated with lycopene during in vitro culture system. Mol. Reprod. Dev. 2018, 85, 46–61. [Google Scholar] [CrossRef]
- Gao, M.; Li, X.; He, Y.; Han, L.; Qiu, D.; Ling, L.; Liu, H.; Liu, J.; Gu, L. SIRT7 functions in redox homeostasis and cytoskeletal organization during oocyte maturation. FASEB J. 2018, 32, 6228–6238. [Google Scholar] [CrossRef]
- Milakovic, I.; Jeseta, M.; Hanulakova, S.; Knitlova, D.; Hanzalova, K.; Hulinska, P.; Machal, L.; Kempisty, B.; Antosik, P.; Machatkova, M. Energy Status Characteristics of Porcine Oocytes During In Vitro Maturation is Influenced by Their Meiotic Competence. Reprod. Domest. Anim. 2015, 50, 812–819. [Google Scholar] [CrossRef]
- Sommerfeld, V.; Niemann, H. Cryopreservation of bovine in vitro produced embryos using ethylene glycol in controlled freezing or vitrification. Cryobiology 1999, 38, 95–105. [Google Scholar] [CrossRef]
| Groups | No. of Fertilized Zygotes | No. of Cleavage Embryo (% ± SEM) | No. of Blastocysts (% ± SEM) | No. of Hatched Blastocysts (% ± SEM) |
|---|---|---|---|---|
| Control | 399 | 306 (76.6 ± 1.2) b | 133 (33.2 ± 0.9) b | 47 (35.9 ± 2.8) b |
| GW501516 | 399 | 323 (81.1 ± 1.1) b | 154 (38.8 ± 1.6) c | 72 (47.0 ± 2.5) c |
| GSK3787 | 375 | 266 (70.8 ± 1.6) a | 102 (27.3 ± 1.1) a | 20 (19.4 ± 2.3) a |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Idrees, M.; Xu, L.; El Sheikh, M.; Sidrat, T.; Song, S.-H.; Joo, M.-D.; Lee, K.-L.; Kong, I.-K. The PPARδ Agonist GW501516 Improves Lipolytic/Lipogenic Balance through CPT1 and PEPCK during the Development of Pre-Implantation Bovine Embryos. Int. J. Mol. Sci. 2019, 20, 6066. https://doi.org/10.3390/ijms20236066
Idrees M, Xu L, El Sheikh M, Sidrat T, Song S-H, Joo M-D, Lee K-L, Kong I-K. The PPARδ Agonist GW501516 Improves Lipolytic/Lipogenic Balance through CPT1 and PEPCK during the Development of Pre-Implantation Bovine Embryos. International Journal of Molecular Sciences. 2019; 20(23):6066. https://doi.org/10.3390/ijms20236066
Chicago/Turabian StyleIdrees, Muhammad, Lianguang Xu, Marwa El Sheikh, Tabinda Sidrat, Seok-Hwan Song, Myeong-Don Joo, Kyeong-Lim Lee, and Il-Keun Kong. 2019. "The PPARδ Agonist GW501516 Improves Lipolytic/Lipogenic Balance through CPT1 and PEPCK during the Development of Pre-Implantation Bovine Embryos" International Journal of Molecular Sciences 20, no. 23: 6066. https://doi.org/10.3390/ijms20236066
APA StyleIdrees, M., Xu, L., El Sheikh, M., Sidrat, T., Song, S.-H., Joo, M.-D., Lee, K.-L., & Kong, I.-K. (2019). The PPARδ Agonist GW501516 Improves Lipolytic/Lipogenic Balance through CPT1 and PEPCK during the Development of Pre-Implantation Bovine Embryos. International Journal of Molecular Sciences, 20(23), 6066. https://doi.org/10.3390/ijms20236066