Evaluation of the Reactivity and Receptor Competition of HLA-G Isoforms toward Available Antibodies: Implications of Structural Characteristics of HLA-G Isoforms
Abstract
1. Introduction
2. Results
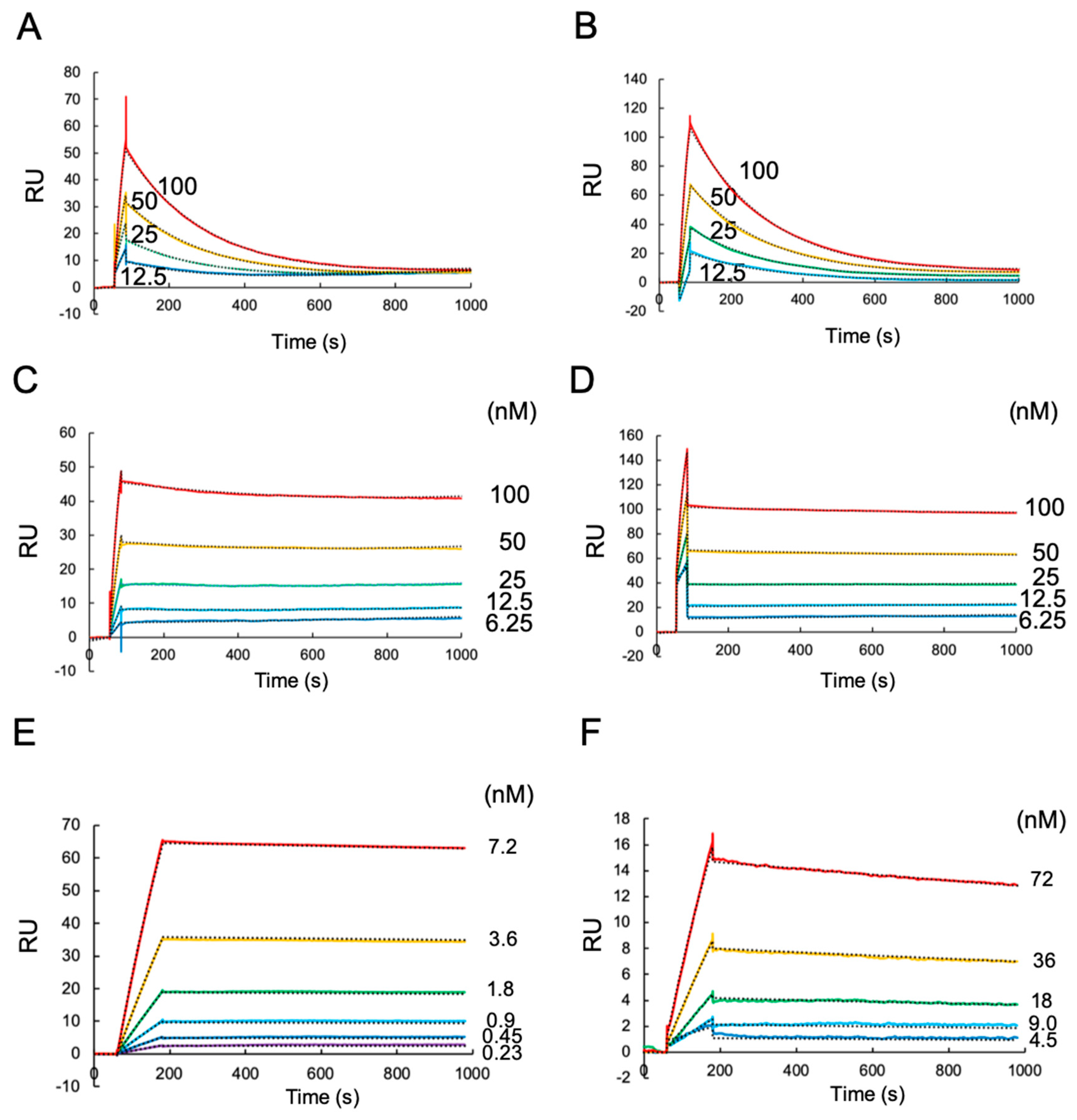
2.1. A Surface Plasmon Resonance (SPR) Analysis of Antibody Binding Towards HLA-G1 and Its Disulfide-Linked Dimer
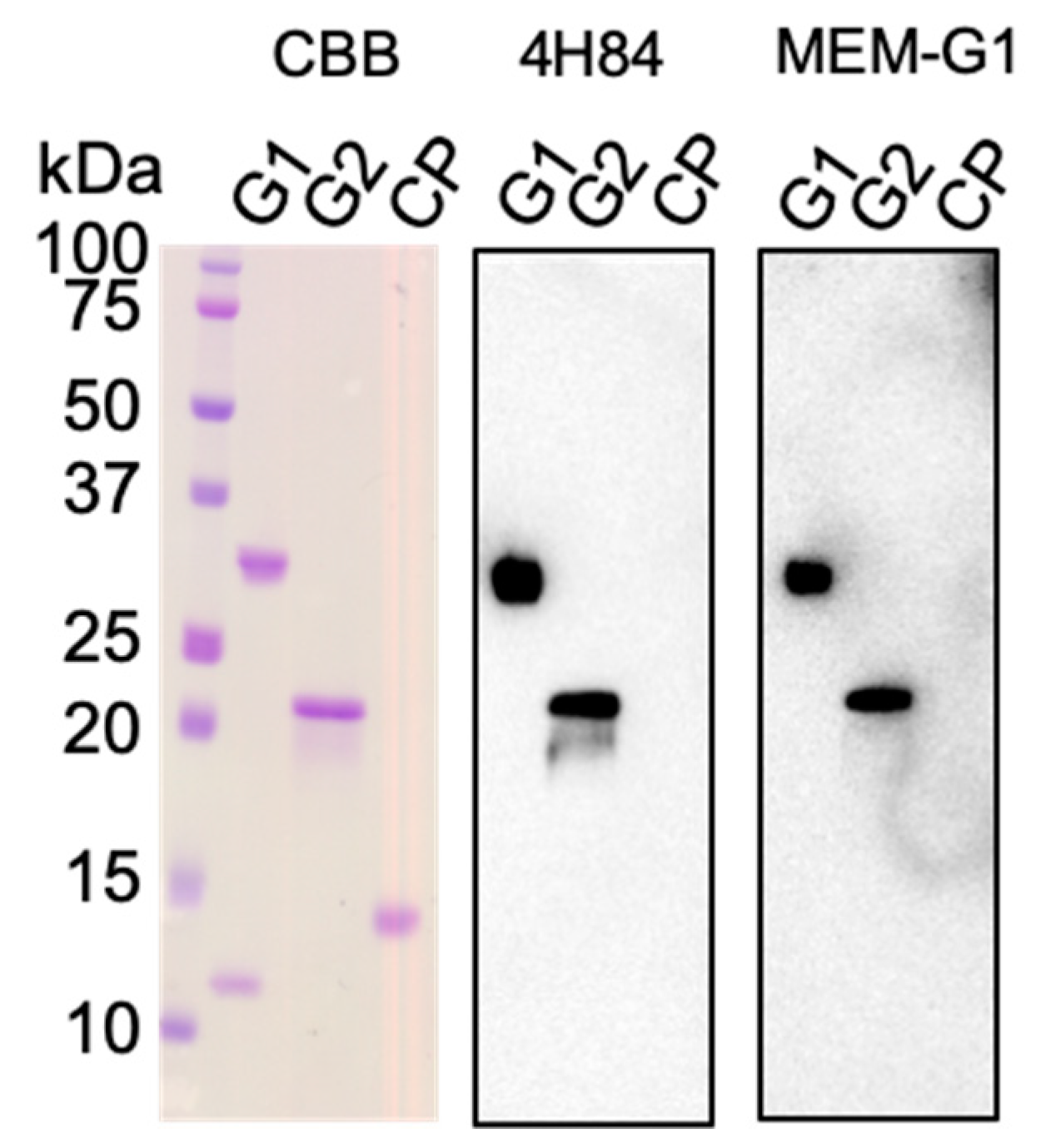
2.2. Western Blotting and SPR Interaction Analyses of HLA-G2 Using the Antibodies
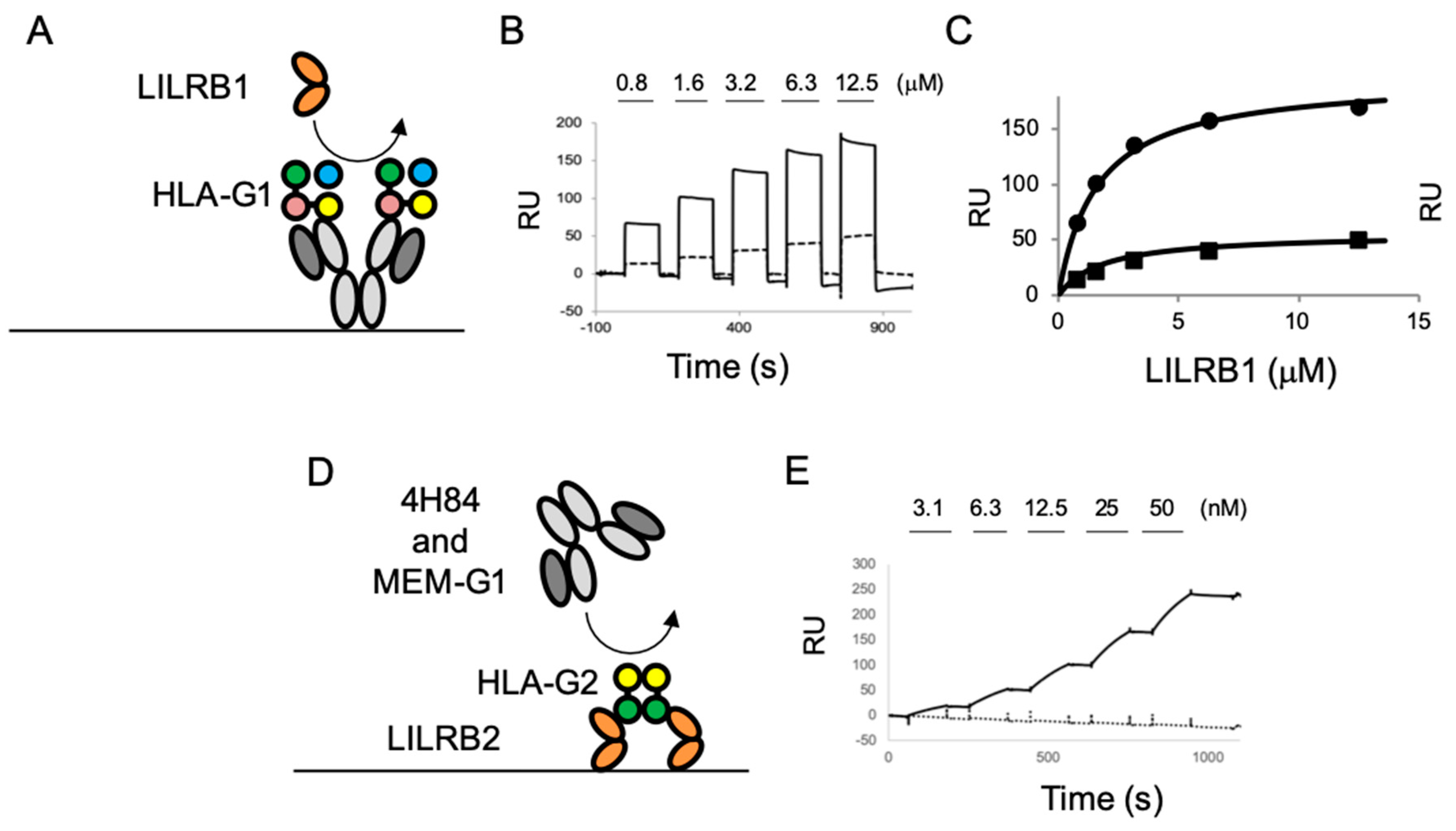
2.3. The Competition Assay for the LILR Receptor Binding of HLA-G Isoforms with Anti-HLA-G Antibodies
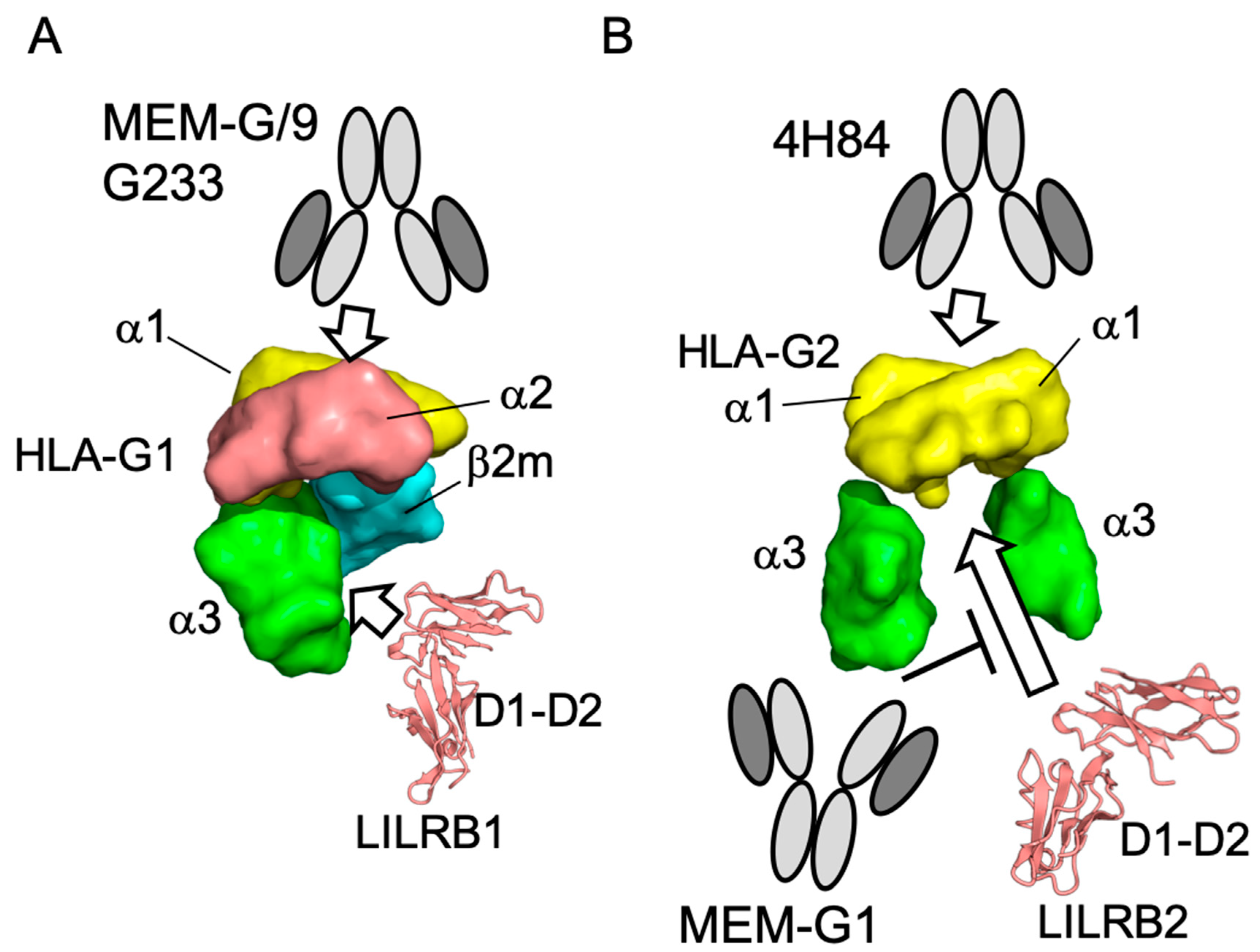
3. Discussion
4. Materials and Methods
4.1. Antibodies
4.2. Production of the Ectodomains of HLA-G1 (Monomer/Dimer), HLA-G2, and LILRB Receptors
4.3. SPR Analysis
4.4. Western Blotting
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| β2m | β2-microglobulin |
| CTLA-4 | cytotoxic cell lymphocyte antigen-4 |
| DTT | Dithiothreitol |
| ELISA | enzyme linked immunosolvent assay |
| HLA | Human Leucocyte antigen |
| HRP | horseradish peroxidase |
| LILR | Leukocyte immunoglobulin-like receptors |
| MHC | Main Histocompatibility Complex |
| PD-1 | programed cell death protein-1 |
| SDS-PAGE | sodium dodecyl sulfate polyacrylamide gel electrophoresis |
| SPR | Surface Plasmon Resonance |
References
- Foroni, I.; Couto, A.R.; Bettencourt, B.F.; Santos, M.; Lima, M.; Bruges-Armas, J. HLA-E, HLA-F and HLA-G—the non-classical side of the MHC cluster. HLA Assoc. Important Dis. 2014, 3, 61–109. [Google Scholar] [CrossRef]
- Feger, U.; Tolosa, E.; Huang, Y.H.; Waschbisch, A.; Biedermann, T.; Melms, A.; Wiendl, H. HLA-G expression defines a novel regulatory T-cell subset present in human peripheral blood and sites of inflammation. Blood 2007, 110, 568–577. [Google Scholar] [CrossRef]
- Rouas-Freiss, N.; Gonçalves, R.M.; Menier, C.; Dausset, J.; Carosella, E.D. Direct evidence to support the role of HLA-G in protecting the fetus from maternal uterine natural killer cytolysis. Proc. Natl. Acad. Sci. USA 1997, 94, 11520–11525. [Google Scholar] [CrossRef]
- Rajagopalan, S.; Long, E.O. A human histocompatibility leukocyte antigen (HLA)-G-specific receptor expressed on all natural killer cells. J. Exp. Med. 1999, 189, 1093–1100. [Google Scholar] [CrossRef]
- Rizzo, R.; Bortolotti, D.; Bolzani, S.; Fainardi, E. HLA-G Molecules in Autoimmune Diseases and Infections. Front. Immunol. 2014, 5, 592. [Google Scholar] [CrossRef]
- Verbruggen, L.A.; Rebmann, V.; Demanet, C.; De Cock, S.; Grosse-Wilde, H. Soluble HLA-G in rheumatoid arthritis. Hum. Immunol. 2006, 67, 561–567. [Google Scholar] [CrossRef]
- Prigione, I.; Penco, F.; Martini, A.; Gattorno, M.; Pistoia, V.; Morandi, F. HLA-G and HLA-E in patients with juvenile idiopathic arthritis. Rheumatology (Oxford) 2011, 50, 966–972. [Google Scholar] [CrossRef][Green Version]
- Kuroki, K.; Hirose, K.; Okabe, Y.; Fukunaga, Y.; Takahashi, A.; Shiroishi, M.; Kajikawa, M.; Tabata, S.; Nakamura, S.; Takai, T.; et al. The long-term immunosuppressive effects of disulfide-linked HLA-G dimer in mice with collagen-induced arthritis. Hum. Immunol. 2013, 74, 433–438. [Google Scholar] [CrossRef]
- Takahashi, A.; Kuroki, K.; Okabe, Y.; Kasai, Y.; Matsumoto, N.; Yamada, C.; Takai, T.; Ose, T.; Kon, S.; Matsuda, T.; et al. The immunosuppressive effect of domain-deleted dimer of HLA-G2 isoform in collagen-induced arthritis mice. Hum. Immunol. 2016. [Google Scholar] [CrossRef]
- Maeda, N.; Yamada, C.; Takahashi, A.; Kuroki, K.; Maenaka, K. Therapeutic application of human leukocyte antigen-G1 improves atopic dermatitis-like skin lesions in mice. Int. Immunopharmacol. 2017, 50, 202–207. [Google Scholar] [CrossRef]
- Amiot, L.; Ferrone, S.; Grosse-Wilde, H.; Seliger, B. Biology of HLA-G in cancer: A candidate molecule for therapeutic intervention? Cell Mol. Life Sci. 2011, 68, 417–431. [Google Scholar] [CrossRef]
- Paul, P.; Rouas-Freiss, N.; Khalil-Daher, I.; Moreau, P.; Riteau, B.; Le Gal, F.A.; Avril, M.F.; Dausset, J.; Guillet, J.G.; Carosella, E.D. HLA-G expression in melanoma: A way for tumor cells to escape from immunosurveillance. Proc. Natl. Acad. Sci. USA 1998, 95, 4510–4515. [Google Scholar] [CrossRef]
- Pardoll, D.M. The blockade of immune checkpoints in cancer immunotherapy. Nat. Rev. Cancer 2012, 12, 252–264. [Google Scholar] [CrossRef]
- Walker, L.S.; Sansom, D.M. The emerging role of CTLA4 as a cell-extrinsic regulator of T cell responses. Nat. Rev. Immunol. 2011, 11, 852–863. [Google Scholar] [CrossRef]
- Agaugué, S.; Carosella, E.D.; Rouas-Freiss, N. Role of HLA-G in tumor escape through expansion of myeloid-derived suppressor cells and cytokinic balance in favor of Th2 versus Th1/Th17. Blood 2011, 117, 7021–7031. [Google Scholar] [CrossRef]
- Carosella, E.D.; Rouas-Freiss, N.; Tronik-Le Roux, D.; Moreau, P.; LeMaoult, J. HLA-G: An Immune Checkpoint Molecule. Adv. Immunol. 2015, 127, 33–144. [Google Scholar]
- Kuroki, K.; Mio, K.; Takahashi, A.; Matsubara, H.; Kasai, Y.; Manaka, S.; Kikkawa, M.; Hamada, D.; Sato, C.; Maenaka, K. Cutting Edge: Class II-like Structural Features and Strong Receptor Binding of the Nonclassical HLA-G2 Isoform Homodimer. J. Immunol. 2017, 198, 3399–3403. [Google Scholar] [CrossRef]
- Menier, C.; Riteau, B.; Dausset, J.; Carosella, E.D.; Rouas-Freiss, N. HLA-G truncated isoforms can substitute for HLA-G1 in fetal survival. Hum. Immunol. 2000, 61, 1118–1125. [Google Scholar] [CrossRef]
- Casro, M.J.; Morales, P.; Rojo-Amigo, R.; Martinez-Laso, J.; Allende, L.; Varela, P.; Garcia-Berciano, M.; Guillen-Perales, J.; Arnaiz-Villena, A. Homozygous HLA-G*0105N healthy individuals indicate that membrane-anchored HLA-G1 molecule is not necessary for survival. Tissue Antigens 2000, 56, 232–239. [Google Scholar] [CrossRef]
- HoWangYin, K.Y.; Loustau, M.; Wu, J.; Alegre, E.; Daouya, M.; Caumartin, J.; Sousa, S.; Horuzsko, A.; Carosella, E.D.; LeMaoult, J. Multimeric structures of HLA-G isoforms function through differential binding to LILRB receptors. Cell Mol. Life Sci. 2012, 69, 4041–4049. [Google Scholar] [CrossRef]
- Paul, P.; Cabestre, F.A.; Ibrahim, E.C.; Lefebvre, S.; Khalil-Daher, I.; Vazeux, G.; Quiles, R.M.; Bermond, F.; Dausset, J.; Carosella, E.D. Identification of HLA-G7 as a new splice variant of the HLA-G mRNA and expression of soluble HLA-G5, -G6, and -G7 transcripts in human transfected cells. Hum. Immunol. 2000, 61, 1138–1149. [Google Scholar] [CrossRef]
- Boyson, J.E.; Erskine, R.; Whitman, M.C.; Chiu, M.; Lau, J.M.; Koopman, L.A.; Valter, M.M.; Angelisova, P.; Horejsi, V.; Strominger, J.L. Disulfide bond-mediated dimerization of HLA-G on the cell surface. Proc. Natl. Acad. Sci. USA 2002, 99, 16180–16185. [Google Scholar] [CrossRef] [PubMed]
- Shiroishi, M.; Kuroki, K.; Ose, T.; Rasubala, L.; Shiratori, I.; Arase, H.; Tsumoto, K.; Kumagai, I.; Kohda, D.; Maenaka, K. Efficient leukocyte Ig-like receptor signaling and crystal structure of disulfide-linked HLA-G dimer. J. Biol. Chem. 2006, 281, 10439–10447. [Google Scholar] [CrossRef] [PubMed]
- Alegre, E.; Rizzo, R.; Bortolotti, D.; Fernandez-Landázuri, S.; Fainardi, E.; González, A. Some basic aspects of HLA-G biology. J. Immunol. Res. 2014, 2014, 657625. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez, A.; Alegre, E.; Arroyo, A.; LeMaoult, J.; Echeveste, J.I. Identification of circulating nonclassic human leukocyte antigen G (HLA-G)-like molecules in exudates. Clin. Chem. 2011, 57, 1013–1022. [Google Scholar] [CrossRef] [PubMed]
- Menier, C.; Saez, B.; Horejsi, V.; Martinozzi, S.; Krawice-Radanne, I.; Bruel, S.; Le Danff, C.; Reboul, M.; Hilgert, I.; Rabreau, M.; et al. Characterization of monoclonal antibodies recognizing HLA-G or HLA-E: New tools to analyze the expression of nonclassical HLA class I molecules. Hum. Immunol. 2003, 64, 315–326. [Google Scholar] [CrossRef]
- Zhao, L.; Teklemariam, T.; Hantash, B.M. Reassessment of HLA-G isoform specificity of MEM-G/9 and 4H84 monoclonal antibodies. Tissue Antigens 2012, 80, 231–238. [Google Scholar] [CrossRef]
- Juch, H.; Blaschitz, A.; Daxböck, C.; Rueckert, C.; Kofler, K.; Dohr, G. A novel sandwich ELISA for alpha1 domain based detection of soluble HLA-G heavy chains. J. Immunol. Methods 2005, 307, 96–106. [Google Scholar] [CrossRef]
- Poláková, K.; Železníková, T.; Russ, G. HLA-G5 in the blood of leukemia patients and healthy individuals. Leuk. Res. 2013, 37, 139–145. [Google Scholar] [CrossRef]
- Spurny, C.; Kailayangiri, S.; Altvater, B.; Jamitzky, S.; Hartmann, W.; Wardelmann, E.; Ranft, A.; Dirksen, U.; Amler, S.; Hardes, J.; et al. T cell infiltration into Ewing sarcomas is associated with local expression of immune-inhibitory HLA-G. Oncotarget 2018, 9, 6536–6549. [Google Scholar] [CrossRef]
- Ouji-Sageshima, N.; Geraghty, D.E.; Ishitani, A.; Hatake, K.; Ito, T. Establishment of optimized ELISA system specific for HLA-G in body fluids. HLA 2016, 88, 293–299. [Google Scholar] [CrossRef]
- Rebmann, V.; LeMaoult, J.; Rouas-Freiss, N.; Carosella, E.D.; Grosse-Wilde, H. Quantification and identification of soluble HLA-G isoforms. Tissue Antigens 2007, 69 (Suppl. 1), 143–149. [Google Scholar] [CrossRef]
- McMaster, M.; Zhou, Y.; Shorter, S.; Kapasi, K.; Geraghty, D.; Lim, K.H.; Fisher, S. HLA-G isoforms produced by placental cytotrophoblasts and found in amniotic fluid are due to unusual glycosylation. J. Immunol 1998, 160, 5922–5928. [Google Scholar] [PubMed]
- Loke, Y.W.; King, A.; Burrows, T.; Gardner, L.; Bowen, M.; Hiby, S.; Howlett, S.; Holmes, N.; Jacobs, D. Evaluation of trophoblast HLA-G antigen with a specific monoclonal antibody. Tissue Antigens 1997, 50, 135–146. [Google Scholar] [CrossRef] [PubMed]
- Fournel, S.; Huc, X.; Aguerre-Girr, M.; Solier, C.; Legros, M.; Praud-Brethenou, C.; Moussa, M.; Chaouat, G.; Berrebi, A.; Bensussan, A.; et al. Comparative reactivity of different HLA-G monoclonal antibodies to soluble HLA-G molecules. Tissue Antigens 2000, 55, 510–518. [Google Scholar] [CrossRef] [PubMed]
- Pela, F.P.; Rustiguel, J.K.; Rodrigues, L.C.; Mendonca, J.N.; Andrade, C.D.C.; Lopes, N.P.; Rosa, J.C.; Nonato, M.C.; Favier, B.; Donadi, E.A.; et al. A soluble recombinant form of human leucocyte antigen-G 6 (srHLA-G6). Biochem. Biophys. Res. Commun. 2017, 487, 28–33. [Google Scholar] [CrossRef] [PubMed]
- Poláková, K.; Bennink, J.R.; Yewdell, J.W.; Bystrická, M.; Bandzuchová, E.; Russ, G. Mild acid treatment induces cross-reactivity of 4H84 monoclonal antibody specific to nonclassical HLA-G antigen with classical HLA class I molecules. Hum. Immunol. 2003, 64, 256–264. [Google Scholar] [CrossRef]
- Carosella, E.D.; Moreau, P.; Lemaoult, J.; Rouas-Freiss, N. HLA-G: From biology to clinical benefits. Trends Immunol. 2008, 29, 125–132. [Google Scholar] [CrossRef]
- Shiroishi, M.; Tsumoto, K.; Amano, K.; Shirakihara, Y.; Colonna, M.; Braud, V.M.; Allan, D.S.; Makadzange, A.; Rowland-Jones, S.; Willcox, B.; et al. Human inhibitory receptors Ig-like transcript 2 (ILT2) and ILT4 compete with CD8 for MHC class I binding and bind preferentially to HLA-G. Proc. Natl. Acad. Sci. USA 2003, 100, 8856–8861. [Google Scholar] [CrossRef]
- Shiroishi, M.; Kuroki, K.; Rasubala, L.; Tsumoto, K.; Kumagai, I.; Kurimoto, E.; Kato, K.; Kohda, D.; Maenaka, K. Structural basis for recognition of the nonclassical MHC molecule HLA-G by the leukocyte Ig-like receptor B2 (LILRB2/LIR2/ILT4/CD85d). Proc. Natl. Acad. Sci. USA 2006, 103, 16412–16417. [Google Scholar] [CrossRef]
- Kuroki, K.; Matsubara, H.; Kanda, R.; Miyashita, N.; Shiroishi, M.; Fukunaga, Y.; Kamishikiryo, J.; Fukunaga, A.; Fukuhara, H.; Hirose, K.; et al. Structural and Functional Basis for LILRB Immune Checkpoint Receptor Recognition of HLA-G Isoforms. J. Immunol. 2019. [Google Scholar] [CrossRef]
- Shiroishi, M.; Kuroki, K.; Tsumoto, K.; Yokota, A.; Sasaki, T.; Amano, K.; Shimojima, T.; Shirakihara, Y.; Rasubala, L.; van der Merwe, P.A.; et al. Entropically driven MHC class I recognition by human inhibitory receptor leukocyte Ig-like receptor B1 (LILRB1/ILT2/CD85j). J. Mol. Biol. 2006, 355, 237–248. [Google Scholar] [CrossRef] [PubMed]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Furukawa, A.; Meguro, M.; Yamazaki, R.; Watanabe, H.; Takahashi, A.; Kuroki, K.; Maenaka, K. Evaluation of the Reactivity and Receptor Competition of HLA-G Isoforms toward Available Antibodies: Implications of Structural Characteristics of HLA-G Isoforms. Int. J. Mol. Sci. 2019, 20, 5947. https://doi.org/10.3390/ijms20235947
Furukawa A, Meguro M, Yamazaki R, Watanabe H, Takahashi A, Kuroki K, Maenaka K. Evaluation of the Reactivity and Receptor Competition of HLA-G Isoforms toward Available Antibodies: Implications of Structural Characteristics of HLA-G Isoforms. International Journal of Molecular Sciences. 2019; 20(23):5947. https://doi.org/10.3390/ijms20235947
Chicago/Turabian StyleFurukawa, Atsushi, Manami Meguro, Rika Yamazaki, Hiroshi Watanabe, Ami Takahashi, Kimiko Kuroki, and Katsumi Maenaka. 2019. "Evaluation of the Reactivity and Receptor Competition of HLA-G Isoforms toward Available Antibodies: Implications of Structural Characteristics of HLA-G Isoforms" International Journal of Molecular Sciences 20, no. 23: 5947. https://doi.org/10.3390/ijms20235947
APA StyleFurukawa, A., Meguro, M., Yamazaki, R., Watanabe, H., Takahashi, A., Kuroki, K., & Maenaka, K. (2019). Evaluation of the Reactivity and Receptor Competition of HLA-G Isoforms toward Available Antibodies: Implications of Structural Characteristics of HLA-G Isoforms. International Journal of Molecular Sciences, 20(23), 5947. https://doi.org/10.3390/ijms20235947




