WY-14643 Regulates CYP1B1 Expression through Peroxisome Proliferator-Activated Receptor α-Mediated Signaling in Human Breast Cancer Cells
Abstract
:1. Introduction
2. Results
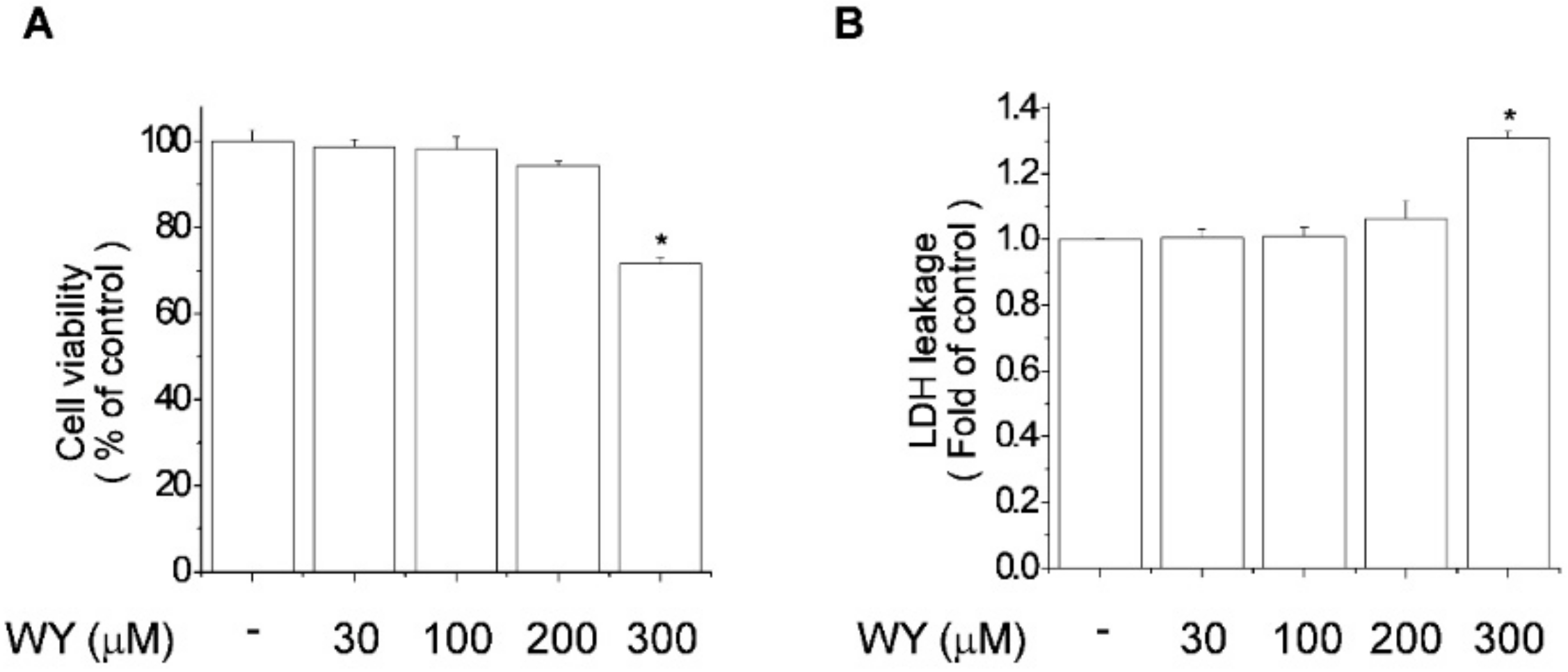
2.1. Cytotoxicity of WY-14643 in MCF-7 Breast Cancer Cells
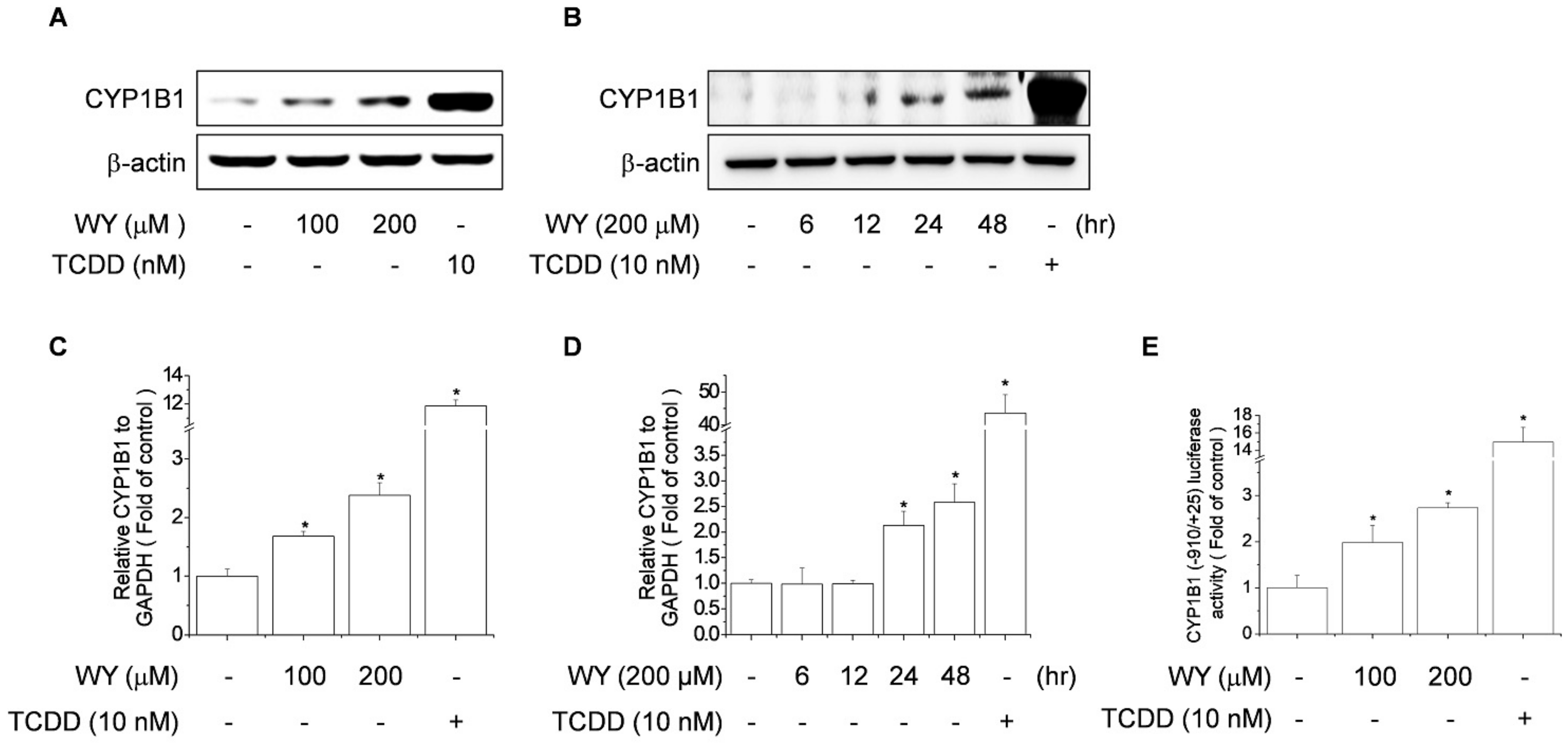
2.2. WY-14643 Induced CYP1B1 Expression, Activity, and Promoter Activity in MCF-7 Cells
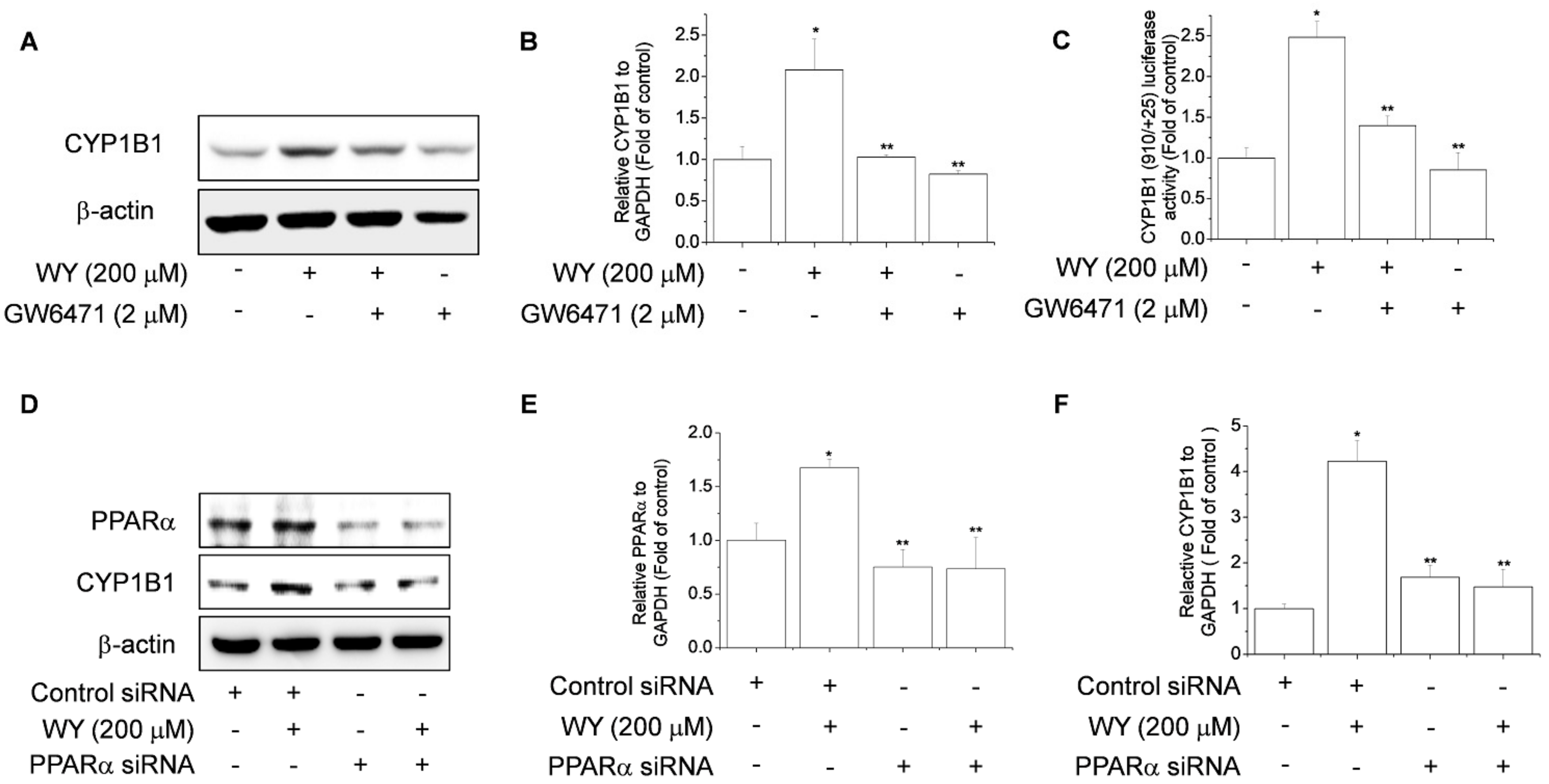
2.3. WY-14643 Induces CYP1B1 Expression in MCF-7 Cells Via a PPARα-Dependent Mechanism
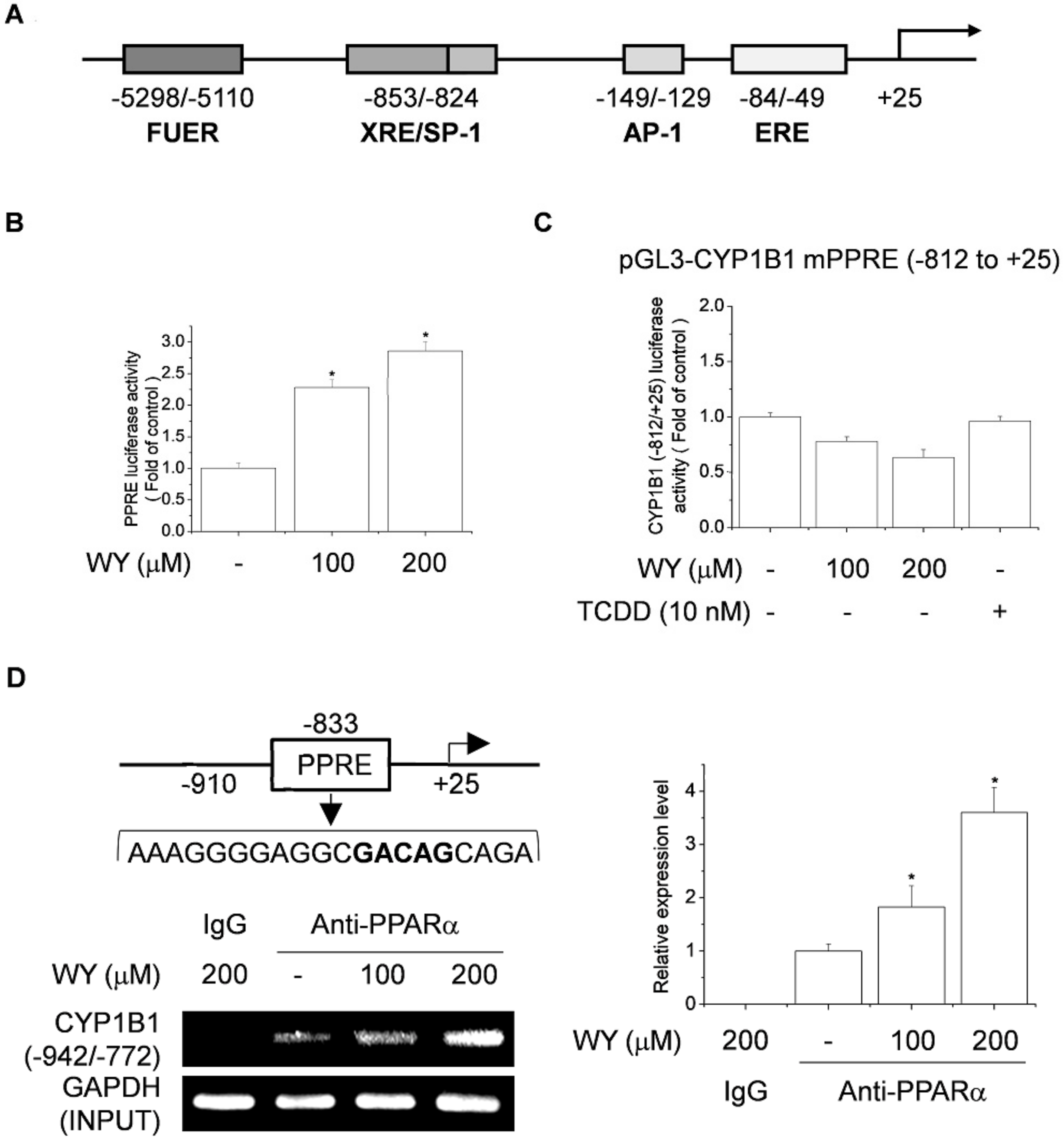
2.4. WY-14643 Upregulates CYP1B1 Expression by Activating the PPRE-Binding Site
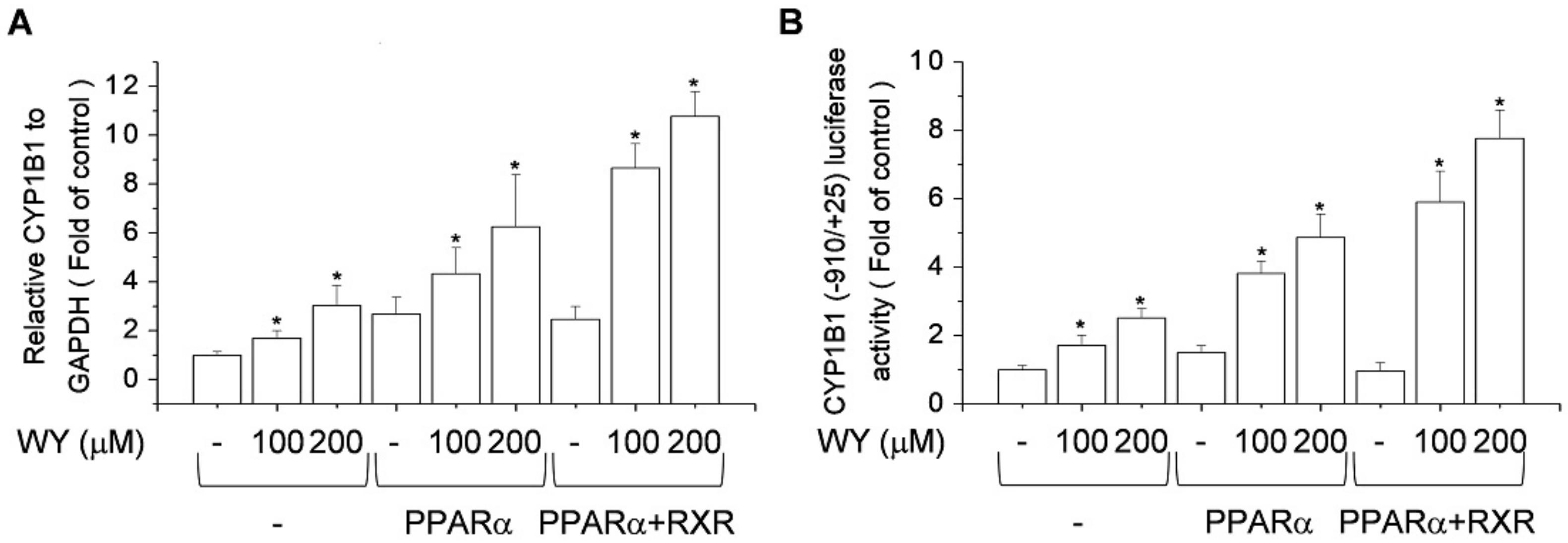
2.5. Effect of Activation of PPARα and RXR by WY-14643 on Induction of CYP1B1 Expression
3. Discussion
4. Materials and Methods
4.1. Chemicals and Reagents
4.2. Cell Culture and Treatment
4.3. Measurement of Cell Viability and Cytotoxicity
4.4. Plasmids
4.5. Transfection and Luciferase Assays
4.6. Western Blotting
4.7. Quantitative Real-Time Reverse Transcription-PCR
4.8. siRNA Transfection
4.9. Chromatin Immunoprecipitation Assay
4.10. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| AhR | Aryl hydrocarbon receptor |
| CYPs | Cytochrome P450 |
| CYP1B1 | Cytochrome P450 1B1 |
| E2 | 17 β-Estradiol |
| MTT | 3-(4,5-Dimethylthiazol-2-yl)- 2,5-diphenyltetrazolium bromide |
| 4-OHE2 | 4-Hydroxyestradiol |
| PCR | Polymerase chain reaction |
| PPARα | Peroxisome proliferator-activated receptor α |
| PPRE | Peroxisome proliferator response element |
| RXR | Retinoid X receptor |
| TCDD | 2,3,7,8-Tetrachlorodibenzo-p-dioxin |
References
- De Santis, C.E.; Bray, F.; Ferlay, J.; Lortet-Tieulent, J.; Anderson, B.O.; Jemal, A. International variation in female breast cancer incidence and mortality rates. Cancer Epidemiol. Biomarkers Prev. 2015, 24, 1495–1506. [Google Scholar] [CrossRef] [PubMed]
- Bugano, D.D.; Conforti-Froes, N.; Yamaguchi, N.H.; Baracat, E.C. Genetic polymorphisms, the metabolism of estrogens and breast cancer: A review. Eur. J. Gynaecol. Oncol. 2008, 29, 313–320. [Google Scholar] [PubMed]
- Muskhelishvili, L.; Thompson, P.A.; Kusewitt, D.F.; Wang, C.; Kadlubar, F.F. In situ hybridization and immunohistochemical analysis of cytochrome P450 1B1 expression in human normal tissues. J. Histochem. Cytochem. 2001, 49, 229–236. [Google Scholar] [CrossRef] [PubMed]
- Blackburn, H.L.; Ellsworth, D.L.; Shriver, C.D.; Ellsworth, R.E. Role of cytochrome P450 genes in breast cancer etiology and treatment: Effects on estrogen biosynthesis, metabolism, and response to endocrine therapy. Cancer Causes Control 2015, 26, 319–332. [Google Scholar] [CrossRef]
- Cui, J.; Meng, Q.; Zhang, X.; Cui, Q.; Zhou, W.; Li, S. Design and synthesis of new alpha-naphthoflavones as cytochrome P450 (CYP) 1B1 inhibitors to overcome docetaxel-resistance associated with CYP1B1 overexpression. J. Med. Chem. 2015, 58, 3534–3547. [Google Scholar] [CrossRef]
- Cavalieri, E.L.; Rogan, E.G.; Zahid, M. Critical depurinating DNA adducts: Estrogen adducts in the etiology and prevention of cancer and dopamine adducts in the etiology and prevention of Parkinson’s disease. Int. J. Cancer 2017, 141, 1078–1090. [Google Scholar] [CrossRef]
- Sissung, T.M.; Price, D.K.; Sparreboom, A.; Figg, W.D. Pharmacogenetics and regulation of human cytochrome P450 1B1: Implications in hormone-mediated tumor metabolism and a novel target for therapeutic intervention. Mol. Cancer Res. 2006, 4, 135–150. [Google Scholar] [CrossRef]
- Tsuchiya, Y.; Nakajima, M.; Yokoi, T. Cytochrome P450- mediated metabolism of estrogens and its regulation in human. Cancer Lett. 2005, 227, 115–124. [Google Scholar] [CrossRef]
- Larsen, M.C.; Bushkofsky, J.R.; Gorman, T.; Adhami, V.; Mukhtar, H.; Wang, S.; Reeder, S.B.; Sheibani, N.; Jefcoate, C.R. Cytochrome P450 1B1: An unexpected modulator of liver fatty acid homeostasis. Arch. Biochem. Biophys. 2015, 571, 21–39. [Google Scholar] [CrossRef]
- Agarwal, S.; Yadav, A.; Chaturvedi, R.K. Peroxisome proliferator-activated receptors (PPARs) as therapeutic target in neurodegenerative disorders. Biochem. Biophys. Res. Commun. 2017, 483, 1166–1177. [Google Scholar] [CrossRef]
- Guan, Y.; Breyer, M.D. Peroxisome proliferator-activated receptors (PPARs): Novel therapeutic targets in renal disease. Kidney Int. 2001, 60, 14–30. [Google Scholar] [CrossRef] [PubMed]
- Tyagi, S.; Gupta, P.; Saini, A.S.; Kaushal, C.; Sharma, S. The peroxisome proliferator-activated receptor: A family of nuclear receptors role in various diseases. J. Adv. Pharm. Technol. Res. 2011, 2, 236–240. [Google Scholar] [CrossRef] [PubMed]
- Mattijssen, F.; Georgiadi, A.; Andasarie, T.; Szalowska, E.; Zota, A.; Krones-Herzig, A.; Heier, C.; Ratman, D.; De Bosscher, K.; Qi, L.; et al. Hypoxia-inducible lipid droplet-associated (HILPDA) is a novel peroxisome proliferator-activated receptor (PPAR) target involved in hepatic triglyceride secretion. J. Biol. Chem. 2014, 289, 19279–19293. [Google Scholar] [CrossRef] [PubMed]
- Kersten, S. Integrated physiology and systems biology of PPARα. Mol. Metab. 2014, 3, 354–371. [Google Scholar] [CrossRef]
- Peters, J.M.; Shah, Y.M.; Gonzalez, F.J. The role of peroxisome proliferator-activated receptors in carcinogenesis and chemoprevention. Nat. Rev. Cancer 2012, 12, 181–195. [Google Scholar] [CrossRef]
- Suchanek, K.M.; May, F.J.; Robinson, J.A.; Lee, W.J.; Holman, N.A.; Monteith, G.R.; Roberts-Thomson, S.J. Peroxisome proliferator activated receptor alpha in the human breast cancer cell lines MCF-7 and MDA-MB-231. Mol. Carcinog. 2002, 34, 165–171. [Google Scholar] [CrossRef]
- Peters, J.M.; Cheung, C.; Gonzalez, F.J. Peroxisome proliferator-activated receptor-alpha and liver cancer: Where do we stand? J. Mol. Med. (Berl.) 2005, 83, 774–785. [Google Scholar] [CrossRef]
- Tung, S.; Shi, Y.; Wong, K.; Zhu, F.; Gorczynski, R.; Laister, R.C.; Minden, M.; Blechert, A.K.; Genzel, Y.; Reichl, U.; et al. PPARα and fatty acid oxidation mediate glucocorticoid resistance in chronic lymphocytic leukemia. Blood 2013, 122, 969–980. [Google Scholar] [CrossRef]
- Laurenti, G.; Benedetti, E.; D’Angelo, B.; Cristiano, L.; Cinque, B.; Raysi, S.; Alecci, M.; Cerù, M.P.; Cifone, M.G.; Galzio, R.; et al. Hypoxia induces peroxisome proliferator-activated receptor α (PPARα) and lipid metabolism peroxisomal enzymes in human glioblastoma cells. J. Cell. Biochem. 2011, 112, 3891–3901. [Google Scholar] [CrossRef]
- Benedetti, E.; Galzio, R.; Laurenti, G.; D’Angelo, B.; Melchiorre, E.; Cifone, M.G.; Fanelli, F.; Muzi, P.; Coletti, G.; Alecci, M.; et al. Lipid metabolism impairment in human gliomas: Expression of peroxisomal proteins in human gliomas at different grades of malignancy. Int. J. Immunopathol. Pharmacol. 2010, 23, 235–246. [Google Scholar] [CrossRef]
- Aboud, O.A.; Wettersten, H.I.; Weiss, R.H. Inhibition of PPARα induces cell cycle arrest and apoptosis, and synergizes with glycolysis inhibition in kidney cancer cells. PLoS ONE 2013, 8, e71115. [Google Scholar] [CrossRef] [PubMed]
- Peters, J.M.; Cattley, R.C.; Gonzalez, F.J. Role of PPAR alpha in the mechanism of action of the nongenotoxic carcinogen and peroxisome proliferator Wy-14,643. Carcinogenesis 1997, 18, 2029–2033. [Google Scholar] [CrossRef] [PubMed]
- Kaipainen, A.; Kieran, M.W.; Huang, S.; Butterfield, C.; Bielenberg, D.; Mostoslavsky, G.; Mulligan, R.; Folkman, J.; Panigrahy, D. PPARalpha deficiency in inflammatory cells suppresses tumor growth. PLoS ONE 2007, 2, e260. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.Z.; Ward, K.W. WY-14643, a Selective PPARα Agonist, Induces Proinflammatory and Proangiogenic Responses in Human Ocular Cells. Int. J. Toxicol. 2010, 29, 496–504. [Google Scholar] [CrossRef]
- Conzen, S. Nuclear receptors and breast cancer. Mol. Endocrinol. 2008, 22, 2215–2228. [Google Scholar] [CrossRef]
- Swedenborg, E.; Rüegg, J.; Hillenweck, A.; Rehnmark, S.; Faulds, M.H.; Zalko, D.; Pongratz, I.; Pettersson, K. 3-Methylcholanthrene displays dual effects on estrogen receptor (ER) alpha and ER beta signaling in a cell-type specific fashion. Mol. Pharmacol. 2008, 73, 575–586. [Google Scholar] [CrossRef]
- Tsuchiya, Y.; Nakajima, M.; Kyo, S.; Kanaya, T.; Inoue, M.; Yokoi, T. Human CYP1B1 is regulated by estradiol via estrogen receptor. Cancer Res. 2004, 64, 3119–3125. [Google Scholar] [CrossRef]
- Cocci, P.; Mosconi, G.; Arukwe, A.; Mozzicafreddo, M.; Angeletti, M.; Aretusi, G.; Palermo, F.A. Effects of Diisodecyl Phthalate on PPAR:RXR-Dependent Gene Expression Pathways in Sea Bream Hepatocytes. Chem. Res. Toxicol. 2015, 28, 935–947. [Google Scholar] [CrossRef]
- Li, F.; Zhu, W.; Gonzalez, F.J. Potential role of CYP1B1 in the development and treatment of metabolic diseases. Pharmacol. Ther. 2017, 178, 18–30. [Google Scholar] [CrossRef]
- Murray, G.I.; Taylor, M.C.; McFadyen, M.C.; McKay, J.A.; Greenlee, W.F.; Burke, M.D.; Melvin, W.T. Tumor-specific expression of cytochrome P450 CYP1B1. Cancer Res. 1997, 57, 3026–3031. [Google Scholar]
- Lee, A.J.; Cai, M.X.; Thomas, P.E.; Conney, A.H.; Zhu, B.T. Characterization of the oxidative metabolites of 17beta-estradiol and estrone formed by 15 selectively expressed human cytochrome p450 isoforms. Endocrinology 2013, 144, 3382–3398. [Google Scholar] [CrossRef] [PubMed]
- Spink, D.C.; Spink, B.C.; Cao, J.Q.; DePasquale, J.A.; Pentecost, B.T.; Fasco, M.J.; Li, Y.; Sutter, T.R. Differential expression of CYP1A1 and CYP1B1 in human breast epithelial cells and breast tumor cells. Carcinogenesis 1998, 19, 291–298. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Spivack, S.D.; Hurteau, G.J.; Reilly, A.A.; Aldous, K.M.; Ding, X.; Kaminsky, L.S. CYP1B1 expression in human lung. Drug Metab. Dispos. 2001, 29, 916–922. [Google Scholar] [PubMed]
- Dutour, R.; Poirier, D. Inhibitors of cytochrome P450 (CYP) 1B1. Eur. J. Med. Chem. 2017, 135, 296–306. [Google Scholar] [CrossRef]
- Sérée, E.; Villard, P.H.; Pascussi, J.M.; Pineau, T.; Maurel, P.; Nguyen, Q.B.; Fallone, F.; Martin, P.M.; Champion, S.; Lacarelle, B.; et al. Evidence for a new human CYP1A1 regulation pathway involving PPAR-alpha and 2 PPRE sites. Gastroenterology 2004, 127, 1436–1445. [Google Scholar] [CrossRef]
- Papi, A.; Guarnieri, T.; Storci, G.; Santini, D.; Ceccarelli, C.; Taffurelli, M.; De Carolis, S.; Avenia, N.; Sanguinetti, A.; Sidoni, A.; et al. Nuclear receptors agonists exert opposing effects on the inflammation dependent survival of breast cancer stem cells. Cell Death Differ. 2012, 19, 1208–1219. [Google Scholar] [CrossRef] [Green Version]
- Villard, P.H.; Caverni, S.; Baanannou, A.; Khalil, A.; Martin, P.G.; Penel, C.; Pineau, T.; Seree, E.; Barra, Y. PPARalpha transcriptionally induces AhR expression in Caco-2, but represses AhR pro-inflammatory effects. Biochem. Biophys. Res. Commun. 2007, 364, 896–901. [Google Scholar] [CrossRef]
- Coumoul, X.; Diry, M.; Robillot, C.; Barouki, R. Differential regulation of cytochrome P450 1A1 and 1B1 by a combination of dioxin and pesticides in the breast tumor cell line MCF-7. Cancer Res. 2001, 61, 3942–3948. [Google Scholar]
- Han, E.H.; Kim, H.G.; Hwang, Y.P.; Song, G.Y.; Jeong, H.G. Prostaglandin E2 induces CYP1B1 expression via ligand-independent activation of the ERalpha pathway in human breast cancer cells. Toxicol. Sci. 2010, 114, 204–216. [Google Scholar] [CrossRef] [Green Version]
- Khanal, T.; Kim, H.G.; Do, M.T.; Choi, J.H.; Won, S.S.; Kang, W.; Chung, Y.C.; Jeong, T.C.; Jeong, H.G. Leptin induces CYP1B1 expression in MCF-7 cells through ligand-independent activation of the ERα pathway. Toxicol. Appl. Pharmacol. 2014, 277, 39–48. [Google Scholar] [CrossRef]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hwang, Y.P.; Won, S.S.; Jin, S.W.; Lee, G.H.; Pham, T.H.; Choi, J.H.; Kang, K.W.; Jeong, H.G. WY-14643 Regulates CYP1B1 Expression through Peroxisome Proliferator-Activated Receptor α-Mediated Signaling in Human Breast Cancer Cells. Int. J. Mol. Sci. 2019, 20, 5928. https://doi.org/10.3390/ijms20235928
Hwang YP, Won SS, Jin SW, Lee GH, Pham TH, Choi JH, Kang KW, Jeong HG. WY-14643 Regulates CYP1B1 Expression through Peroxisome Proliferator-Activated Receptor α-Mediated Signaling in Human Breast Cancer Cells. International Journal of Molecular Sciences. 2019; 20(23):5928. https://doi.org/10.3390/ijms20235928
Chicago/Turabian StyleHwang, Yong Pil, Seong Su Won, Sun Woo Jin, Gi Ho Lee, Thi Hoa Pham, Jae Ho Choi, Keon Wook Kang, and Hye Gwang Jeong. 2019. "WY-14643 Regulates CYP1B1 Expression through Peroxisome Proliferator-Activated Receptor α-Mediated Signaling in Human Breast Cancer Cells" International Journal of Molecular Sciences 20, no. 23: 5928. https://doi.org/10.3390/ijms20235928
APA StyleHwang, Y. P., Won, S. S., Jin, S. W., Lee, G. H., Pham, T. H., Choi, J. H., Kang, K. W., & Jeong, H. G. (2019). WY-14643 Regulates CYP1B1 Expression through Peroxisome Proliferator-Activated Receptor α-Mediated Signaling in Human Breast Cancer Cells. International Journal of Molecular Sciences, 20(23), 5928. https://doi.org/10.3390/ijms20235928





