SH3Ps—Evolution and Diversity of a Family of Proteins Engaged in Plant Cytokinesis
Abstract
1. Introduction
2. Results
2.1. Inventory of SH3P-Encoding Genes in Selected Plant Lineages
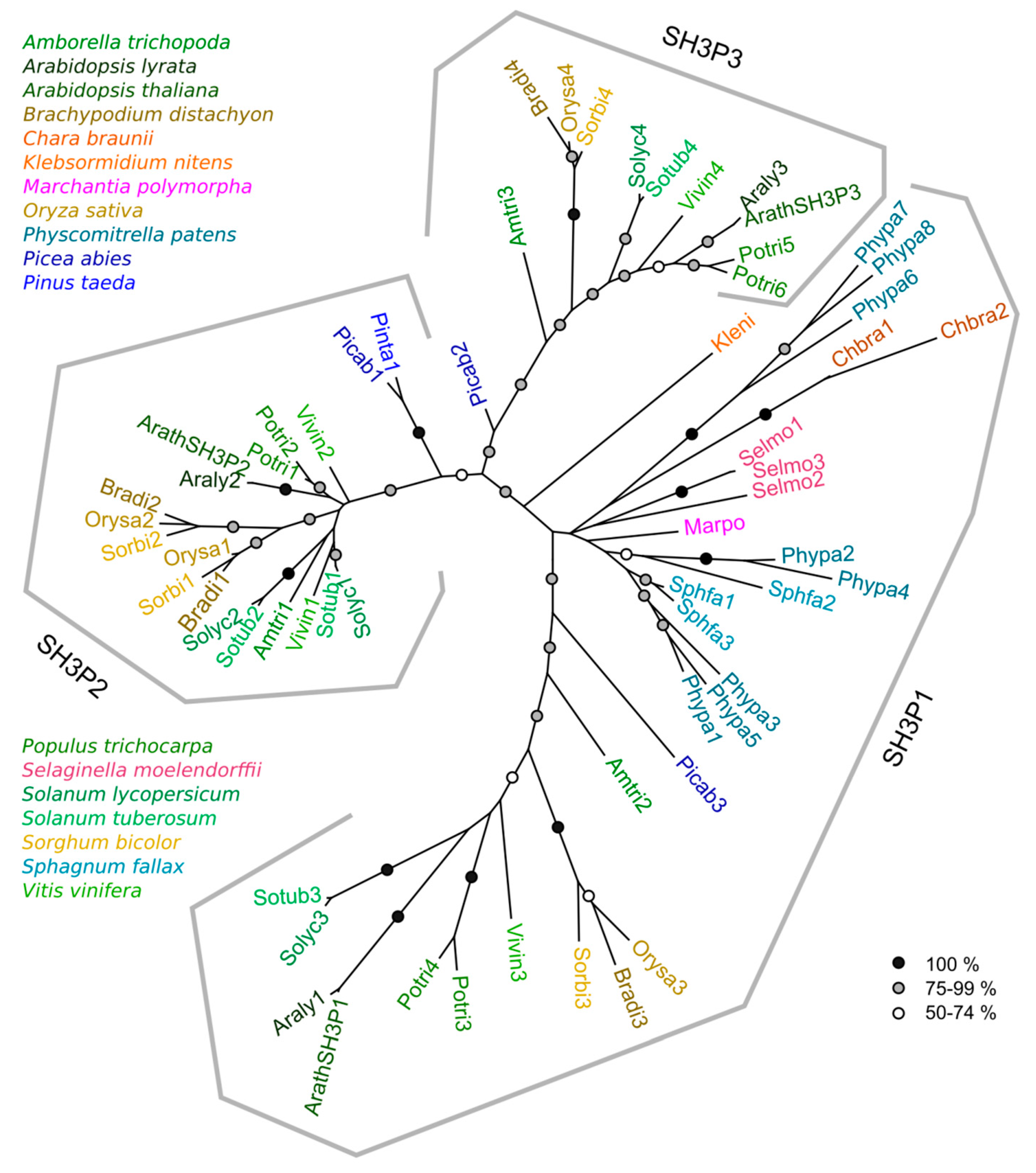
2.2. Phylogeny of Plant SH3P Proteins
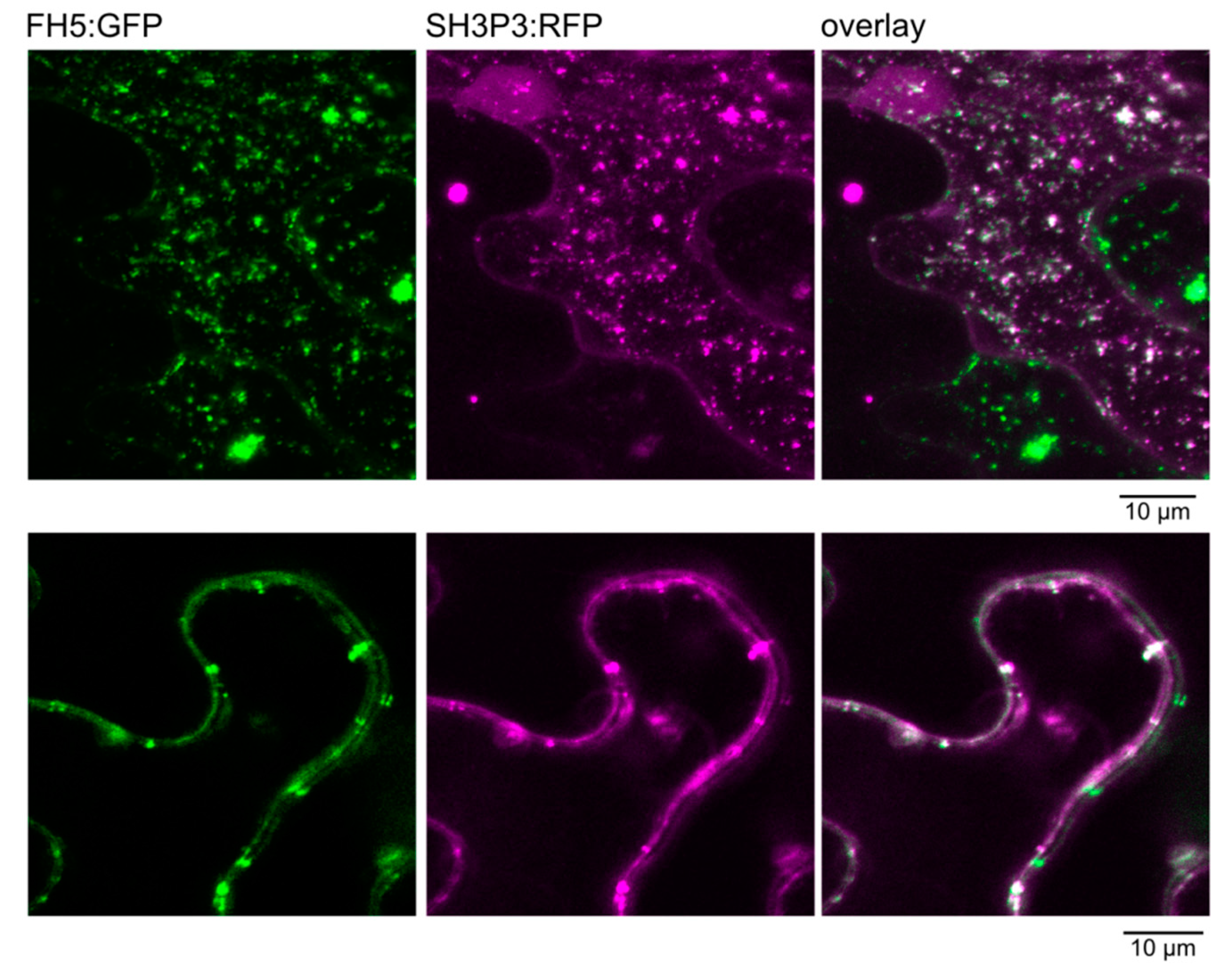
2.3. Differential Binding of Arabidopsis SH3P2 and SH3P3 to the Cytokinetic Formin AtFH5 and to DRP1A
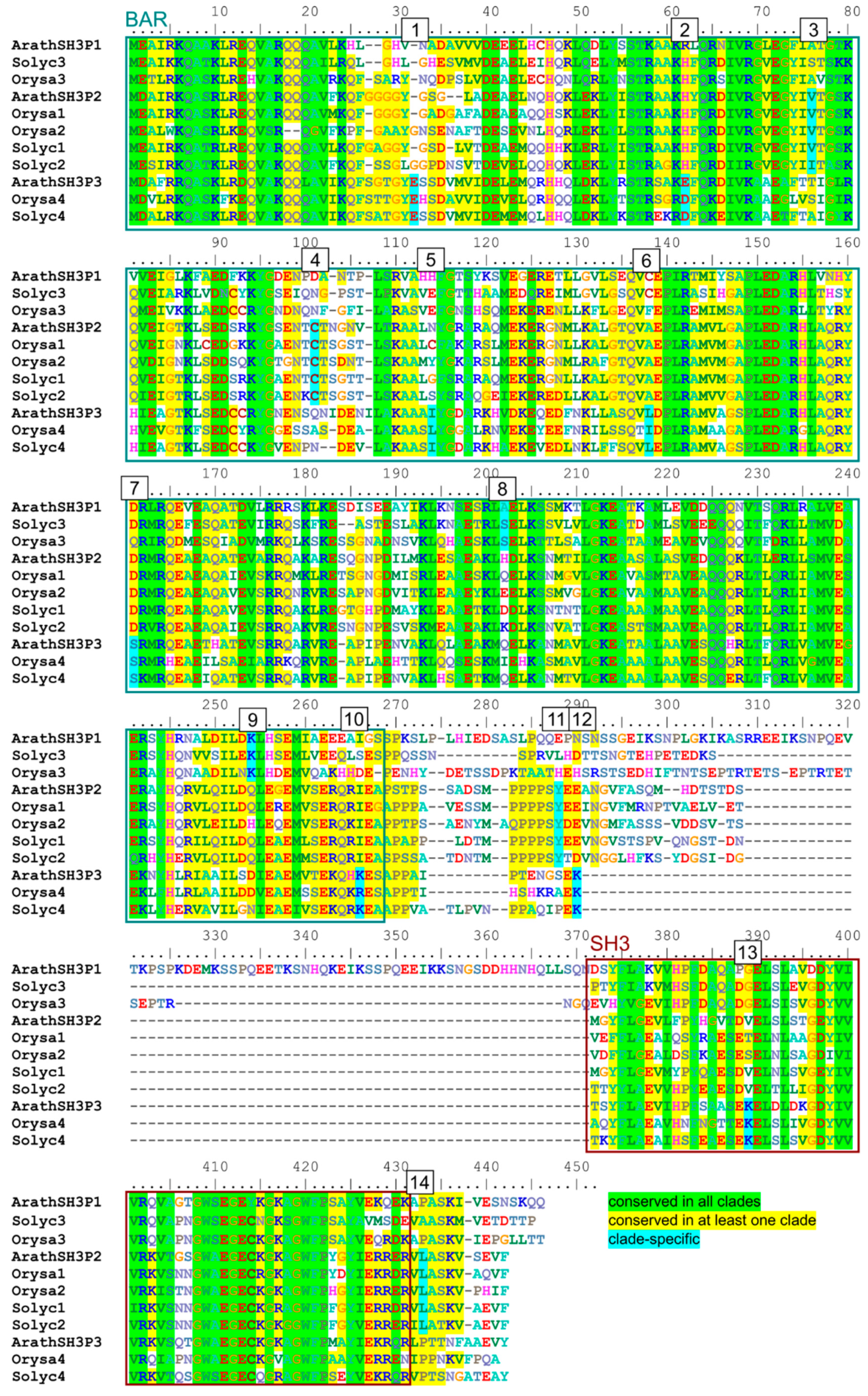
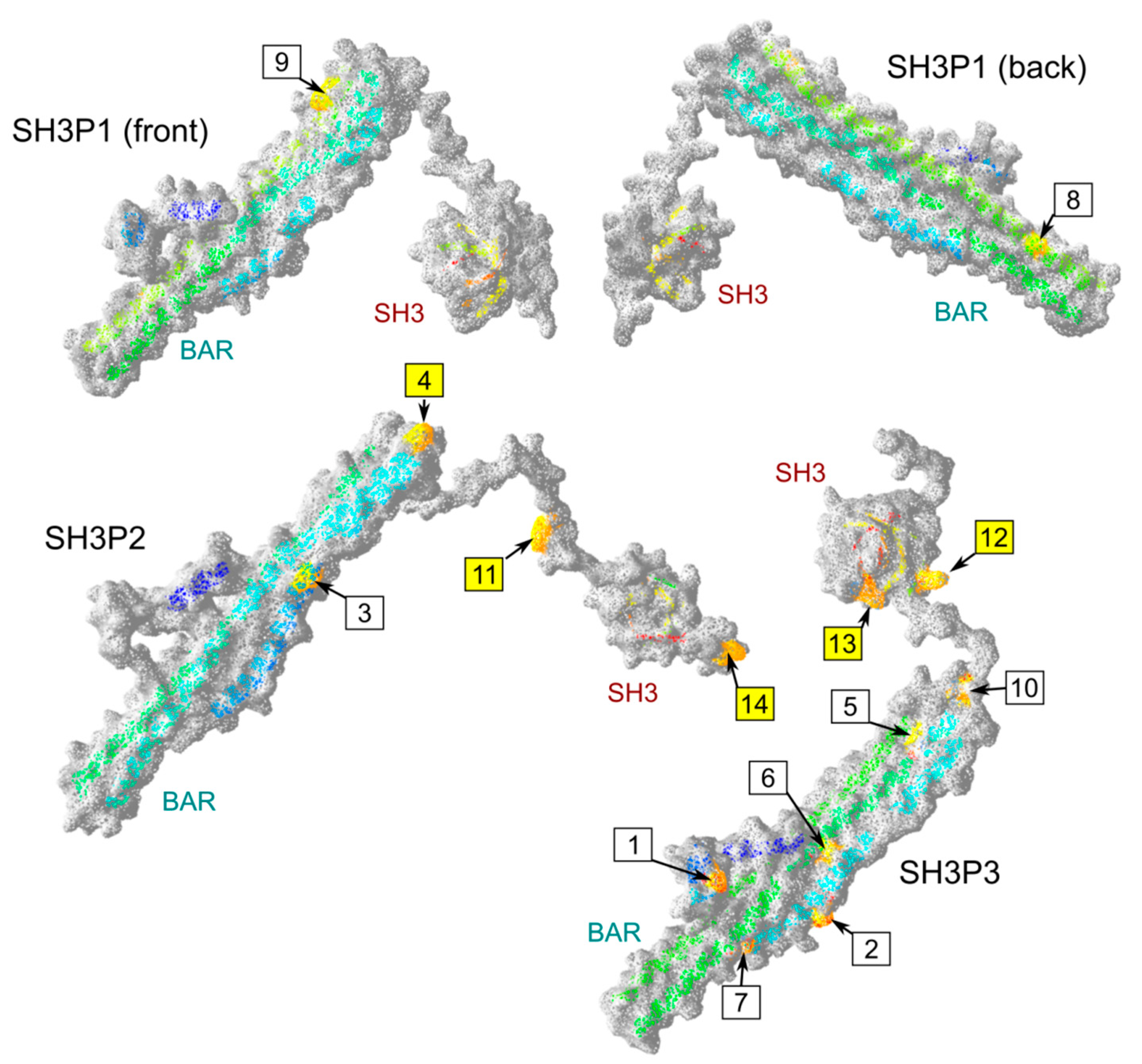
2.4. Possible Determinants of Interaction Specificity Among Angiosperm SH3P Proteins
3. Discussion
4. Materials and Methods
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| AA | amino acid |
| AD | activation domain |
| BAR | Bin/amphiphysin/Rvs |
| DBD | DNA binding domain |
| DRP1A | dynamin-related protein 1A |
| FH1 | formin homology domain 1 |
| FH2 | formin homology domain 2 |
| FH5 | Arabidopsis formin homolog 5 |
| GFP | green fluorescent protein |
| RFP | red fluorescent protein |
| SH3 | Sarc homology 3 |
| SH3P | SH3 and BAR domain-containing protein |
References
- Panteris, E. Cortical actin filaments at the division site of mitotic plant cells: A reconsideration of the actin-depleted zone. New Phytol. 2008, 179, 334–341. [Google Scholar] [CrossRef] [PubMed]
- Rasmussen, C.G.; Wright, A.J.; Müller, S. The role of the cytoskeleton and associated proteins in determination of the plant cell division plane. Plant J. 2013, 75, 258–269. [Google Scholar] [CrossRef] [PubMed]
- Müller, S. Plant cell division—Defining and finding the sweet spot for cell plate insertion. Curr. Opin. Cell Biol. 2019, 60, 9–18. [Google Scholar] [CrossRef] [PubMed]
- Fujimoto, M.; Arimura, S.; Nakazono, M.; Tsutsumi, N. Arabidopsis dynamin-related protein DRP2B is co-localized with DRP1A on the leading edge of the forming cell plate. Plant Cell Rep. 2008, 27, 1581–1586. [Google Scholar] [CrossRef] [PubMed]
- Bednarek, S.Y.; Backues, S.K. Plant dynamin-related protein families DRP1 and DRP2 in plant development. Biochem. Soc. Trans. 2010, 38, 797–806. [Google Scholar] [CrossRef] [PubMed]
- Smertenko, A. Phragmoplast expansion: The four-stroke engine that powers plant cytokinesis. Curr. Opin. Plant Biol. 2018, 46, 130–137. [Google Scholar] [CrossRef] [PubMed]
- Buschmann, H.; Zachgo, S. The evolution of cell division: From streptophyte algae to land plants. Trends Plant Sci. 2016, 21, 872–883. [Google Scholar] [CrossRef] [PubMed]
- Cross, F.R.; Umen, J.G. The Chlamydomonas cell cycle. Plant J. 2015, 82, 370–392. [Google Scholar] [CrossRef] [PubMed]
- Smertenko, A.; Assaad, F.; Baluška, F.; Bezanilla, M.; Buschmann, H.; Drakakaki, G.; Hauser, M.T.; Janson, M.; Mineyuki, Y.; Moore, I.; et al. Plant cytokinesis: Terminology for structures and processes. Trends Cell Biol. 2017, 27, 885–894. [Google Scholar] [CrossRef] [PubMed]
- Ahn, G.; Kim, H.; Kim, D.H.; Hanh, H.; Yoon, Y.; Singaram, I.; Wijesinghe, K.J.; Johnson, K.A.; Zhuang, X.; Liang, Z.; et al. SH3 domain-containing protein 2 plays a crucial role at the step of membrane tubulation during cell plate formation. Plant Cell 2017, 29, 1388–1405. [Google Scholar] [CrossRef] [PubMed]
- Peter, B.J.; Kent, H.M.; Mills, I.G.; Vallis, Y.; Butler, P.J.G.; Evans, P.R.; McMahon, H.T. BAR domains as sensors of membrane curvature: The amphiphysin BAR structure. Science 2004, 303, 495–499. [Google Scholar] [CrossRef] [PubMed]
- Dawson, J.C.; Legg, J.A.; Machesky, L.M. Bar domain proteins: A role in tubulation, scission and actin assembly in clathrin-mediated endocytosis. Trends Cell Biol. 2006, 16, 493–498. [Google Scholar] [CrossRef] [PubMed]
- Musacchio, A.; Noble, M.; Pauptit, R.; Wierenga, R.; Saraste, M. Crystal structure of a Src-homology 3 (SH3) domain. Nature 1992, 359, 851–855. [Google Scholar] [CrossRef] [PubMed]
- Shah, N.H.; Amacher, J.F.; Nocka, L.M.; Kuriyan, J. The Src module: An ancient scaffold in the evolution of cytoplasmic tyrosine kinases. Crit. Rev. Biochem. Mol. Biol. 2018, 53, 535–563. [Google Scholar] [CrossRef] [PubMed]
- Carman, P.J.; Dominguez, R. BAR domain proteins-a linkage between cellular membranes, signaling pathways, and the actin cytoskeleton. Biophys. Rev. 2018, 10, 1587–1604. [Google Scholar] [CrossRef] [PubMed]
- Aspenström, P.; Richnau, N.; Johansson, A.S. The diaphanous-related formin DAAM1 collaborates with the Rho GTPases RhoA and Cdc42, CIP4 and Src in regulating cell morphogenesis and actin dynamics. Exp. Cell Res. 2006, 312, 2180–2194. [Google Scholar] [CrossRef] [PubMed]
- Huett, A.; Ng, A.; Cao, Z.; Kuballa, P.; Komatsu, M.; Daly, M.J.; Podolsky, D.K.; Xavier, R.J. A novel hybrid yeast-human network analysis reveals an essential role for FNBP1L in antibacterial autophagy. J. Immunol. 2009, 182, 4917–4930. [Google Scholar] [CrossRef] [PubMed]
- Wakita, Y.; Kakimoto, T.; Katoh, H.; Negishi, M. The F-BAR protein Rapostlin regulates dendritic spine formation in hippocampal neurons. J. Biol. Chem. 2011, 286, 32672–32683. [Google Scholar] [CrossRef] [PubMed]
- Suetsugu, S.; Gautreau, A. synergistic BAR-NPF interactions in actin-driven membrane remodeling. Trends Cell Biol. 2012, 22, 141–150. [Google Scholar] [CrossRef] [PubMed]
- Cvrčková, F. Formins and membranes: Anchoring cortical actin to the cell wall and beyond. Front. Plant Sci. 2013, 4, 436. [Google Scholar] [CrossRef] [PubMed]
- Lam, B.C.; Sage, T.L.; Bianchi, F.; Blumwald, E. Role of SH3 domain-containing proteins in clathrin-mediated vesicle trafficking in Arabidopsis. Plant Cell 2001, 13, 2499–2512. [Google Scholar] [CrossRef] [PubMed]
- Kolb, C.; Nagel, M.K.; Kalinowska, K.; Hagmann, J.; Ichikawa, M.; Anzenberger, F.; Alkofer, A.; Sato, M.H.; Braun, P.; Isono, E. FYVE1 is essential for vacuole biogenesis and intracellular trafficking in Arabidopsis. Plant Physiol. 2015, 167, 1361–1373. [Google Scholar] [CrossRef] [PubMed]
- Lam, B.C.; Sage, T.L.; Bianchi, F.; Blumwald, E. Regulation of ADL6 activity by its associated molecular network. Plant J. 2002, 31, 565–576. [Google Scholar] [CrossRef] [PubMed]
- Zhuang, X.; Wang, H.; Lam, S.K.; Gao, C.; Wang, X.; Cai, Y.; Jiang, L. A BAR-domain protein, SH3P2, which binds to phosphatidylinositol 3-phosphate and ATG8, regulates autophagosome formation in Arabidopsis. Plant Cell 2013, 25, 4596–4615. [Google Scholar] [CrossRef] [PubMed]
- Zhuang, X.; Jiang, L. Autophagosome biogenesis in plants: Roles of SH3P2. Autophagy 2014, 10, 704–705. [Google Scholar] [CrossRef] [PubMed]
- Gao, C.; Zhuang, X.; Cui, Y.; Fu, X.; He, Y.; Zhao, Q.; Zeng, Y.; Shen, J.; Luo, M.; Jiang, L. Dual roles of an Arabidopsis ESCRT component FREE1 in regulating vacuolar protein transport and autophagic degradation. Proc. Natl. Acad. Sci. USA 2015, 112, 1886–1891. [Google Scholar] [CrossRef] [PubMed]
- Nagel, M.K.; Kalinowska, K.; Vogel, K.; Reynolds, G.D.; Wu, Z.; Anzenberger, F.; Ichikawa, M.; Tsutsumi, C.; Sato, M.H.; Kuster, B.; et al. Arabidopsis SH3P2 is an ubiquitin-binding protein that functions together with ESCRT-I and the deubiquitylating enzyme AMSH3. Proc. Natl. Acad. Sci. USA 2017, 114, 7197–7204. [Google Scholar] [CrossRef] [PubMed]
- Johnson, A.; Vert, G. Unraveling K63 polyubiquitination networks by sensor-based proteomics. Plant Physiol. 2016, 171, 1808–1820. [Google Scholar] [CrossRef] [PubMed]
- Winter, D.; Vinegar, B.; Nahal, H.; Ammar, R.; Wilson, G.V.; Provart, N.J. An “Electronic Fluorescent Pictograph” browser for exploring and analyzing large-scale biological data sets. PLoS ONE 2007, 2, e718. [Google Scholar] [CrossRef] [PubMed]
- Cvrčková, F.; Novotný, M.; Pícková, D.; Žárský, V. Formin homology 2 domains occur in multiple contexts in angiosperms. BMC Genom. 2004, 5, 44. [Google Scholar] [CrossRef] [PubMed]
- Ingouff, M.; Fitz Gerald, J.N.; Guérin, C.; Robert, H.; Sørensen, M.B.; Van Damme, D.; Geelen, D.; Blanchoin, L.; Berger, F. Plant formin AtFH5 is an evolutionarily conserved actin nucleator involved in cytokinesis. Nat. Cell Biol. 2005, 7, 374–380. [Google Scholar] [CrossRef] [PubMed]
- Hori, K.; Maruyama, F.; Fujisawa, T.; Togashi, T.; Yamamoto, N.; Seo, M.; Sato, S.; Yamada, T.; Mori, H.; Tajima, N.; et al. Klebsormidium flaccidum genome reveals primary factors for plant terrestrial adaptation. Nat. Commun. 2014, 5, 3978. [Google Scholar] [CrossRef] [PubMed]
- Vitkup, D.; Sander, C.; Church, G.M. The amino-acid mutational spectrum of human genetic disease. Genome Biol. 2003, 4, R72. [Google Scholar] [CrossRef] [PubMed]
- Kelley, L.A.; Mezulis, S.; Yates, C.M.; Wass, M.N.; Sternberg, M.J. The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 2015, 10, 845–858. [Google Scholar] [CrossRef] [PubMed]
- Waterhouse, A.; Bertoni, M.; Bienert, S.; Studer, G.; Tauriello, G.; Gumienny, R.; Heer, F.T.; de Beer, T.A.P.; Rempfer, C.; Bordoli, L.; et al. SWISS-MODEL: Homology modelling of protein structures and complexes. Nucleic Acids Res. 2018, 46, 296–303. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.; Wang, S.; Bu, D.; Xu, J. Protein threading using residue co-variation and deep learning. Bioinformatics 2018, 34, 263–273. [Google Scholar] [CrossRef] [PubMed]
- Adamowski, M.; Narasimhan, M.; Kania, U.; Glanc, M.; De Jaeger, G.; Friml, J. A functional study of AUXILIN-LIKE1 and 2, two putative clathrin uncoating factors in Arabidopsis. Plant Cell 2018, 30, 700–716. [Google Scholar] [CrossRef] [PubMed]
- Xin, X.; Gfeller, D.; Cheng, J.; Tonikian, R.; Sun, L.; Guo, A.; Lopez, L.; Pavlenco, A.; Akintobi, A.; Zhang, Y.; et al. SH3 interactome conserves general function over specific form. Mol. Syst. Biol. 2013, 9, 652. [Google Scholar] [CrossRef] [PubMed]
- Yoon, Y.; Zhang, X.; Cho, W. Phosphatidylinositol 4,5-bisphosphate (PtdIns (4,5) P2) specifically induces membrane penetration and deformation by Bin/Amphiphysin/Rvs (BAR) domains. J. Biol. Chem. 2012, 287, 34078–34090. [Google Scholar] [CrossRef] [PubMed]
- Neumann, S.; Schmid, S.L. Dual role of BAR domain-containing proteins in regulating vesicle release catalyzed by the GTPase, dynamin-2. J. Biol. Chem. 2013, 288, 25119–25128. [Google Scholar] [CrossRef] [PubMed]
- Daumke, O.; Roux, A.; Haucke, V. BAR domain scaffolds in dynamin-mediated membrane fission. Cell 2014, 156, 882–892. [Google Scholar] [CrossRef] [PubMed]
- Pollard, T.D. Nine unanswered questions about cytokinesis. J. Cell Biol. 2017, 216, 3007–3016. [Google Scholar] [CrossRef] [PubMed]
- Roberts-Galbraith, R.H.; Chen, J.S.; Wang, J.; Gould, K.L. The SH3 domains of two PCH family members cooperate in assembly of the Schizosaccharomyces pombe contractile ring. J. Cell Biol. 2009, 184, 113–127. [Google Scholar] [CrossRef] [PubMed]
- Ren, L.; Willet, A.H.; Roberts-Galbraith, R.H.; McDonald, N.A.; Feoktistova, A.; Chen, J.S.; Huang, H.; Guillen, R.; Boone, C.; Sidhu, S.S.; et al. The Cdc15 and Imp2 SH3 domains cooperatively scaffold a network of proteins that redundantly ensure efficient cell division in fission yeast. Mol. Biol. Cell. 2015, 26, 256–269. [Google Scholar] [CrossRef] [PubMed]
- Oh, Y.; Schreiter, J.; Nishihama, R.; Wloka, C.; Bi, E. Targeting and functional mechanisms of the cytokinesis-related F-BAR protein Hof1 during the cell cycle. Mol. Biol. Cell. 2013, 24, 1305–1320. [Google Scholar] [CrossRef] [PubMed]
- Spencer, S.; Dowbenko, D.; Cheng, J.; Li, W.; Brush, J.; Utzig, S.; Simanis, V.; Lasky, L.A. PSTPIP: A tyrosine phosphorylated cleavage furrow-associated protein that is a substrate for a PEST tyrosine phosphatase. J. Cell Biol. 1997, 138, 845–860. [Google Scholar] [CrossRef] [PubMed]
- Arakaki, Y.; Fujiwara, T.; Kawai-Toyooka, H.; Kawafune, K.; Featherston, J.; Durand, P.M.; Miyagishima, S.Y.; Nozaki, H. Evolution of cytokinesis-related protein localization during the emergence of multicellularity in volvocine green algae. BMC Evol. Biol. 2017, 17, 243. [Google Scholar] [CrossRef] [PubMed]
- Pertl-Obermeyer, H.; Lackner, P.; Schulze, W.X.; Hoepflinger, M.C.; Hoeftberger, M.; Foissner, I.; Obermeyer, G. Dissecting the subcellular membrane proteome reveals enrichment of H+ (co-)transporters and vesicle trafficking proteins in acidic zones of Chara internodal cells. PLoS ONE 2018, 13, e0201480. [Google Scholar] [CrossRef] [PubMed]
- Erffelinck, M.L.; Ribeiro, B.; Perassolo, M.; Pauwels, L.; Pollier, J.; Storme, V.; Goossens, A. A user-friendly platform for yeast two-hybrid library screening using next generation sequencing. PLoS ONE 2018, 13, e0201270. [Google Scholar] [CrossRef] [PubMed]
- Fàbregas, N.; Li, N.; Boeren, S.; Nash, T.E.; Goshe, M.B.; Clouse, S.D.; de Vries, S.; Caño-Delgado, A.I. The Brassinosteroid Insensitive1-like3 signalosome complex regulates Arabidopsis root development. Plant Cell 2013, 25, 3377–3388. [Google Scholar] [CrossRef] [PubMed]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Goodstein, D.M.; Shu, S.; Howson, R.; Neupane, R.; Hayes, R.D.; Fazo, J.; Mitros, T.; Dirks, W.; Hellsten, U.; Putnam, N.; et al. Phytozome: A comparative platform for green plant genomics. Nucleic Acids Res. 2012, 40, 1178–1186. [Google Scholar] [CrossRef] [PubMed]
- Sundell, D.; Mannapperuma, C.; Netotea, S.; Delhomme, N.; Lin, Y.C.; Sjödin, A.; Van de Peer, Y.; Jansson, S.; Hvidsten, T.R.; Street, N.R. The plant genome integrative explorer resource: PlantGenIE.org. New Phytol. 2015, 208, 1149–1156. [Google Scholar] [CrossRef] [PubMed]
- Rawat, A.; Brejšková, L.; Hála, M.; Cvrčková, F.; Žárský, V. The Physcomitrella patens exocyst subunit EXO70.3d has distinct roles in growth and development, and is essential for completion of the moss life cycle. New Phytol. 2017, 216, 438–454. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Rosero, A.; Oulehlová, D.; Stillerová, L.; Schiebertová, P.; Grunt, M.; Žárský, V.; Cvrčková, F. Arabidopsis FH1 formin affects cotyledon pavement cell shape by modulating cytoskeleton dynamics. Plant Cell Physiol. 2016, 57, 488–504. [Google Scholar] [CrossRef] [PubMed]
- Cvrčková, F.; Oulehlová, D.; Žárský, V. Formins: Linking cytoskeleton and endomembranes in plant cells. Int. J. Mol. Sci. 2015, 16, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Lu, Q.; Tang, X.; Tian, G.; Wang, F.; Liu, K.; Nguyen, V.; Kohalmi, S.E.; Keller, W.A.; Tsang, E.W.; Harada, J.J.; et al. Arabidopsis homolog of the yeast TREX-2 mRNA export complex: Components and anchoring nucleoporin. Plant J. 2010, 61, 259–270. [Google Scholar] [CrossRef] [PubMed]
- Konopka, C.A.; Bednarek, S.Y. Comparison of the dynamics and functional redundancy of the Arabidopsis dynamin-related isoforms DRP1A and DRP1C during plant development. Plant Physiol. 2008, 147, 1590–1602. [Google Scholar] [CrossRef] [PubMed]
- Karimi, M.; Bleys, A.; Vanderhaeghen, R.; Hilson, P. Building blocks for plant gene assembly. Plant Physiol. 2007, 145, 1183–1191. [Google Scholar] [CrossRef] [PubMed]
- Pečenková, T.; Pleskot, R.; Žárský, V. Subcellular localization of Arabidopsis Pathogenesis-related 1 (PR1) protein. Int. J. Mol. Sci. 2017, 18, 825. [Google Scholar] [CrossRef] [PubMed]
- Oulehlová, D.; Kollárová, E.; Cifrová, P.; Pejchar, P.; Žárský, V.; Cvrčková, F. Arabidopsis class I formin FH1 relocates between membrane compartments during root cell ontogeny and associates with plasmodesmata. Plant Cell Physiol. 2019, 60, 1855–1870. [Google Scholar] [CrossRef] [PubMed]
- Schindelin, J.; Rueden, C.T.; Hiner, M.C.; Eliceiri, K.W. The ImageJ ecosystem: An open platform for biomedical image analysis. Mol. Reprod. Dev. 2015, 82, 518–529. [Google Scholar] [CrossRef] [PubMed]
- Johansson, M.U.; Zoete, V.; Michielin, O.; Guex, N. Defining and searching for structural motifs using DeepView/Swiss-PdbViewer. BMC Bioinform. 2012, 13, 173. [Google Scholar] [CrossRef] [PubMed]

| Group | Species | Total SH3Ps | SH3P1 | SH3P2 | SH3P3 |
|---|---|---|---|---|---|
| Chlorophyta | Chlamydomonas reinhardtii | none | - | - | - |
| Volvox carteri | none | - | - | - | |
| Charophyta | Chara braunii | 2 | 2 | none | none |
| Klebsormidium nitens | 1 | 1 | none | none | |
| Liverworts | Marchantia polymorpha | 1 | 1 | none | none |
| Mosses | Physcomitrella patens | 8 | 8 | none | none |
| Sphagnum fallax | 5 | 5 | none | none | |
| Lycophyta | Selaginella moelendorffii | 3 | 3 | none | none |
| Gymnosperms | Picea abies | 3 | 1 | 1 | 1 |
| Pinus taeda1 | 3 | ? | 1 | 1? | |
| Angiosperms/Dicots | Amborella trichopoda | 3 | 1 | 1 | 1 |
| Angiosperms/Dicots/Rosidae | Arabidopsis thaliana | 3 | 1 | 1 | 1 |
| Arabidopsis lyrata | 3 | 1 | 1 | 1 | |
| Populus trichocarpa | 6 | 2 | 2 | 2 | |
| Vitis vinifera | 4 | 1 | 2 | 1 | |
| Angiosperms/Dicots/Asteridae | Solanum lycopersicum | 4 | 1 | 2 | 1 |
| Solanum tuberosum | 4 | 1 | 2 | 1 | |
| Angiosperms/Monocots | Brachypodium distachyon | 4 | 1 | 2 | 1 |
| Oryza sativa japonica | 4 | 1 | 2 | 1 | |
| Sorghum bicolor | 4 | 1 | 2 | 1 |
| Site | Clade | Domain | Position 1 | Conserved AA | AA in Other Clades | Exposed |
|---|---|---|---|---|---|---|
| 1 | SH3P3 | BAR | 32 | E | G or gap | + |
| 2 | SH3P3 | BAR | 62 | D or E | H or R | + 2 |
| 3 | SH3P2 | BAR | 73 | V or I | small hydrophilic (STA) | +/− 2 |
| 4 | SH3P2 | BAR | 98 | C | various | + + |
| 5 | SH3P3 | BAR | 114 | I or L | various | +/− 2 |
| 6 | SH3P3 | BAR | 138 | L or I | various | +/− 2 |
| 7 | SH3P3 | BAR | 161 | S | acid or amide (DEQ) | +/−2 |
| 8 | SH3P1 | BAR | 197 | small hydrophilic (SAT) | various charged | +/−2 |
| 9 | SH3P1 | BAR | 248 | K | various (mostly QDE) | + 2 |
| 10 | SH3P3 | BAR | 265 | K or R | various (mostly I) | +/− |
| 11 | SH3P2 | linker | 281 | Y | various | + + |
| 12 | SH3P3 | linker | 280 | K | various (mostly NDE) | + + |
| 13 | SH3P3 | SH3 | 298 | K | various | + + |
| 14 | SH3P2 | C-terminal | 360 | L | small hydrophilic (PAS) | + + |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Baquero Forero, A.; Cvrčková, F. SH3Ps—Evolution and Diversity of a Family of Proteins Engaged in Plant Cytokinesis. Int. J. Mol. Sci. 2019, 20, 5623. https://doi.org/10.3390/ijms20225623
Baquero Forero A, Cvrčková F. SH3Ps—Evolution and Diversity of a Family of Proteins Engaged in Plant Cytokinesis. International Journal of Molecular Sciences. 2019; 20(22):5623. https://doi.org/10.3390/ijms20225623
Chicago/Turabian StyleBaquero Forero, Anežka, and Fatima Cvrčková. 2019. "SH3Ps—Evolution and Diversity of a Family of Proteins Engaged in Plant Cytokinesis" International Journal of Molecular Sciences 20, no. 22: 5623. https://doi.org/10.3390/ijms20225623
APA StyleBaquero Forero, A., & Cvrčková, F. (2019). SH3Ps—Evolution and Diversity of a Family of Proteins Engaged in Plant Cytokinesis. International Journal of Molecular Sciences, 20(22), 5623. https://doi.org/10.3390/ijms20225623





