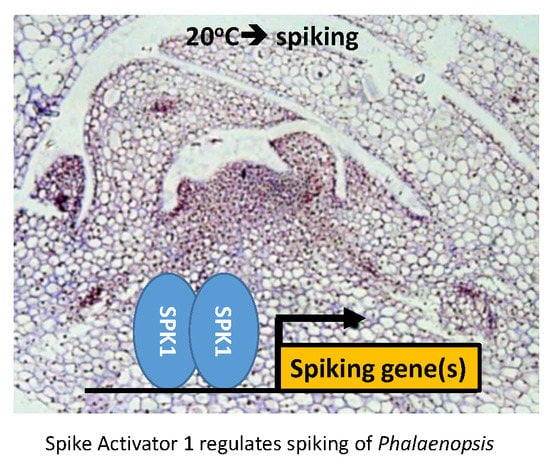
Spike Activator 1, Encoding a bHLH, Mediates Axillary Bud Development and Spike Initiation in Phalaenopsis aphrodite
Abstract
:1. Introduction
2. Results
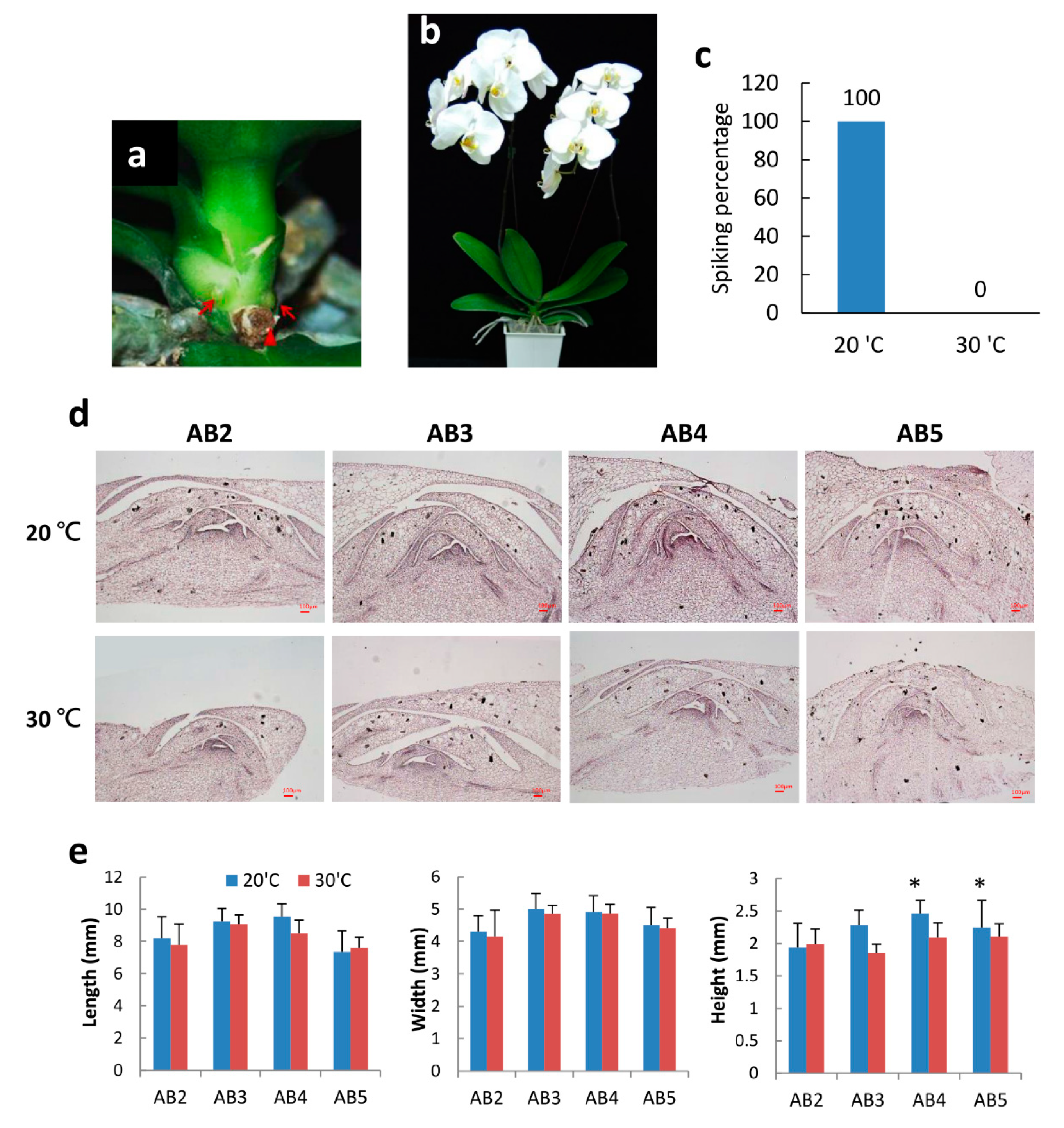
2.1. Morphology of Axillary Bud Development after Spike Induction of Phal. Orchid
2.2. Identification of a bHLH Gene Responding to Spiking
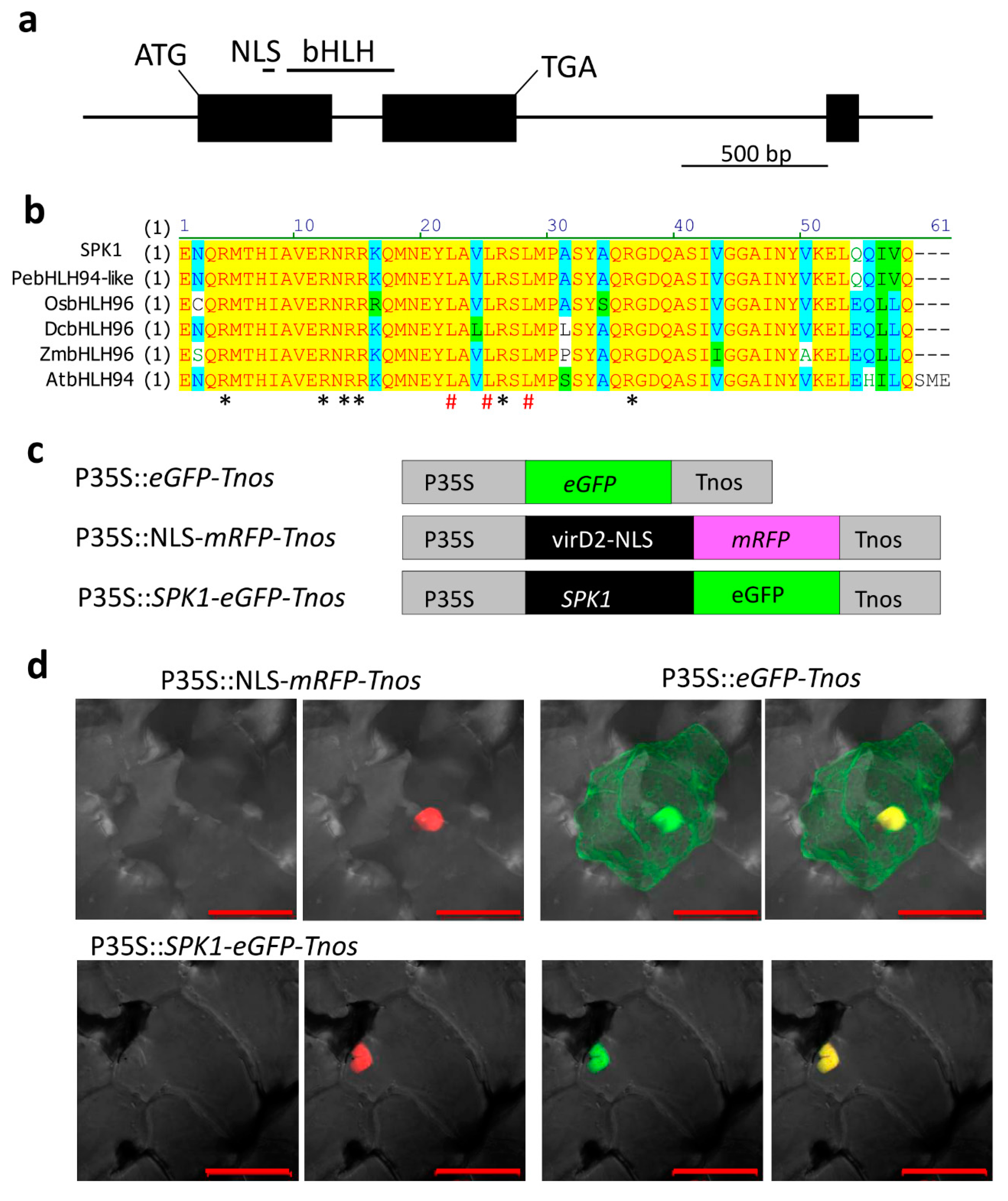
2.3. Isolation of SPK1 Full-Length cDNA and Its Protein Subcellular Localization in the Nucleus
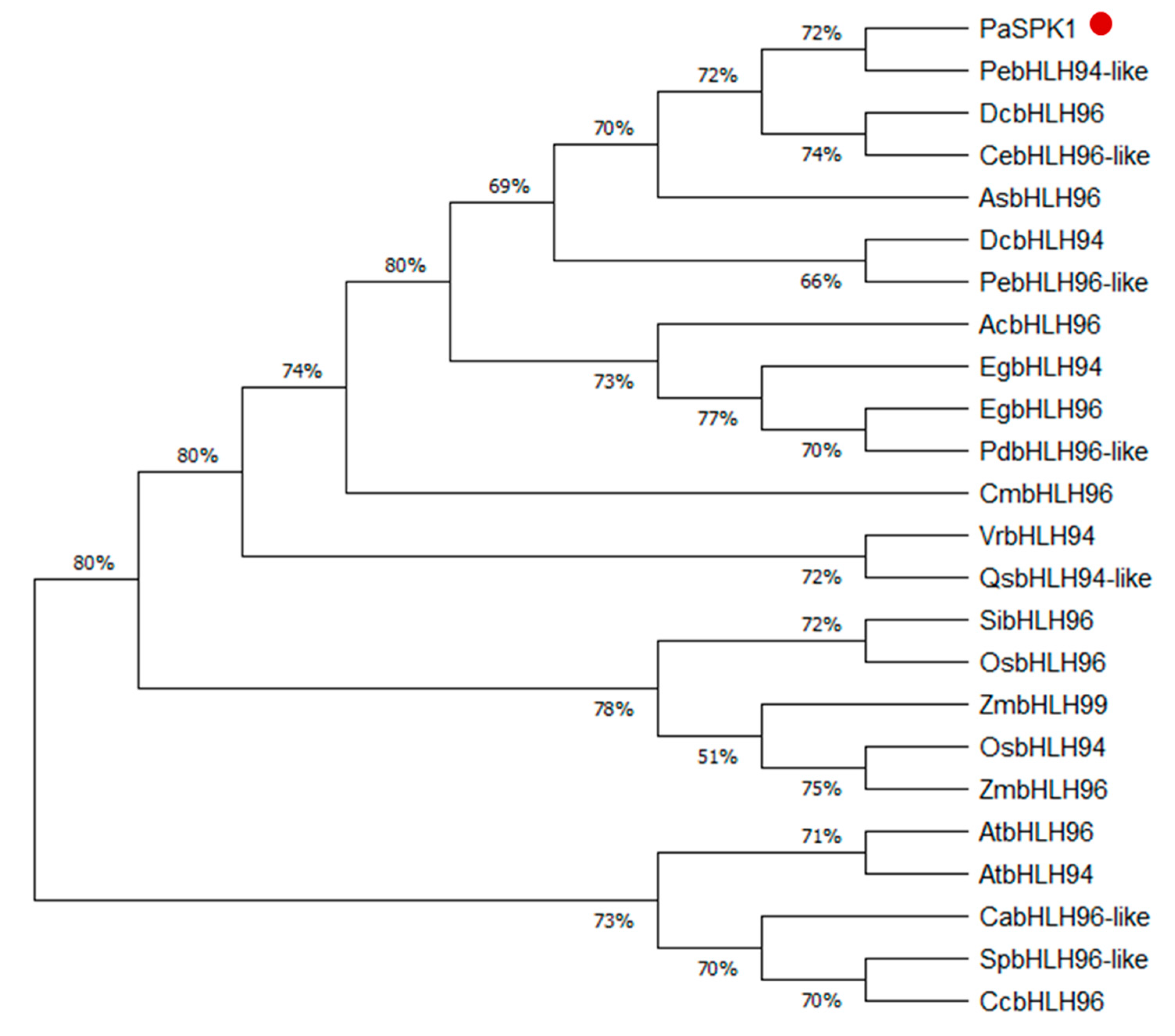
2.4. Phylogenetic Tree
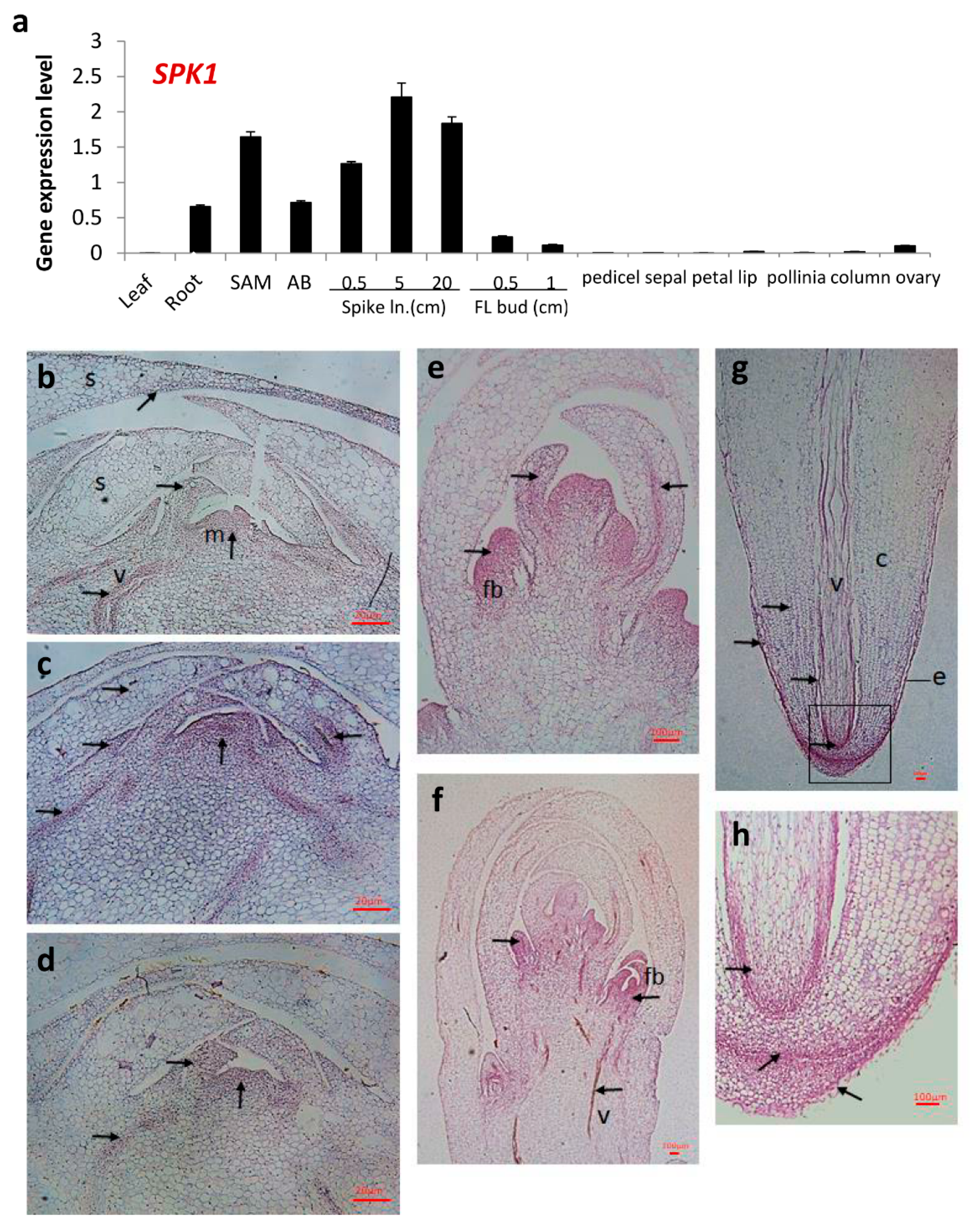
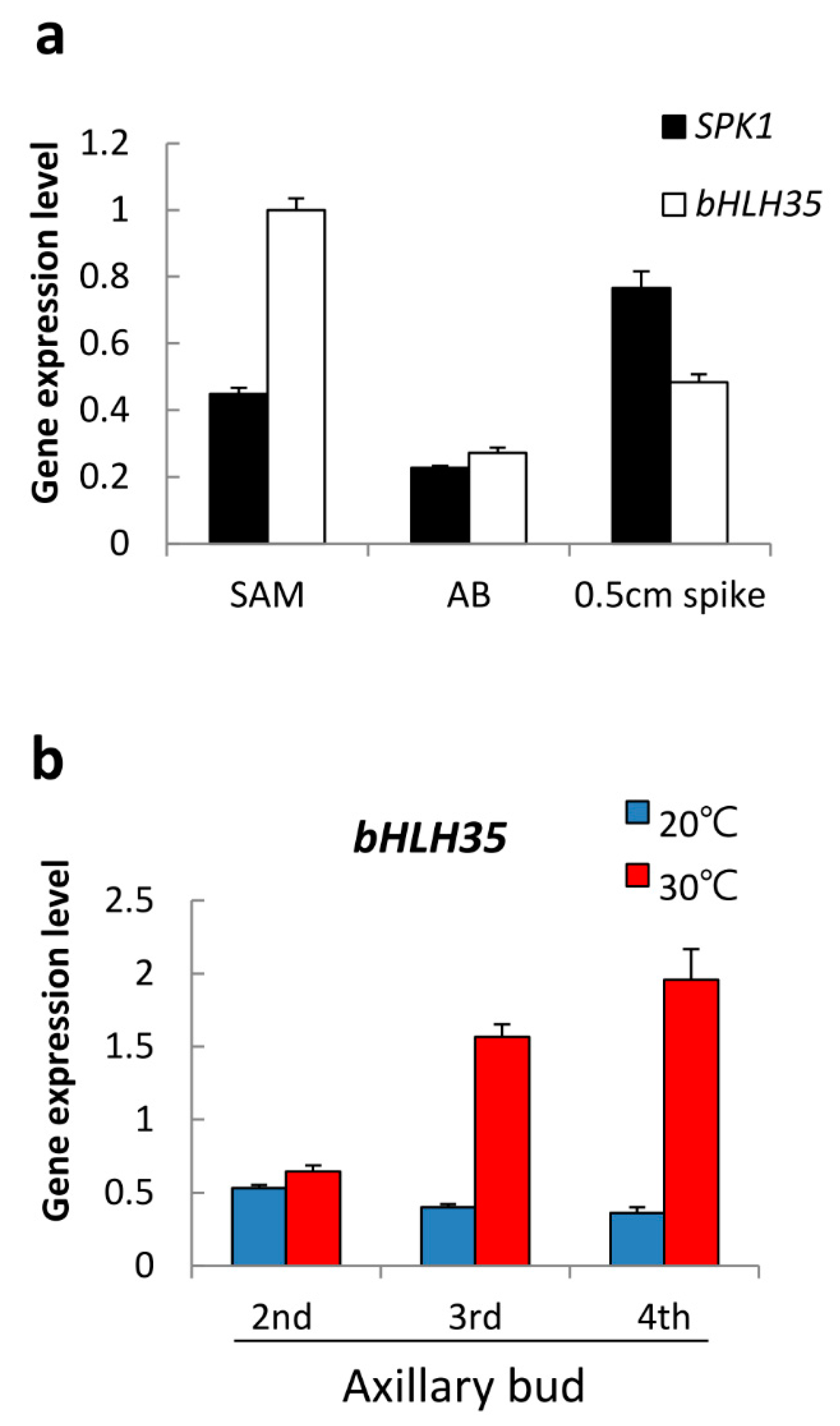
2.5. Gene Expression Patterns of SPK1
2.6. bHLH35 Was Upregulated by High Temperature
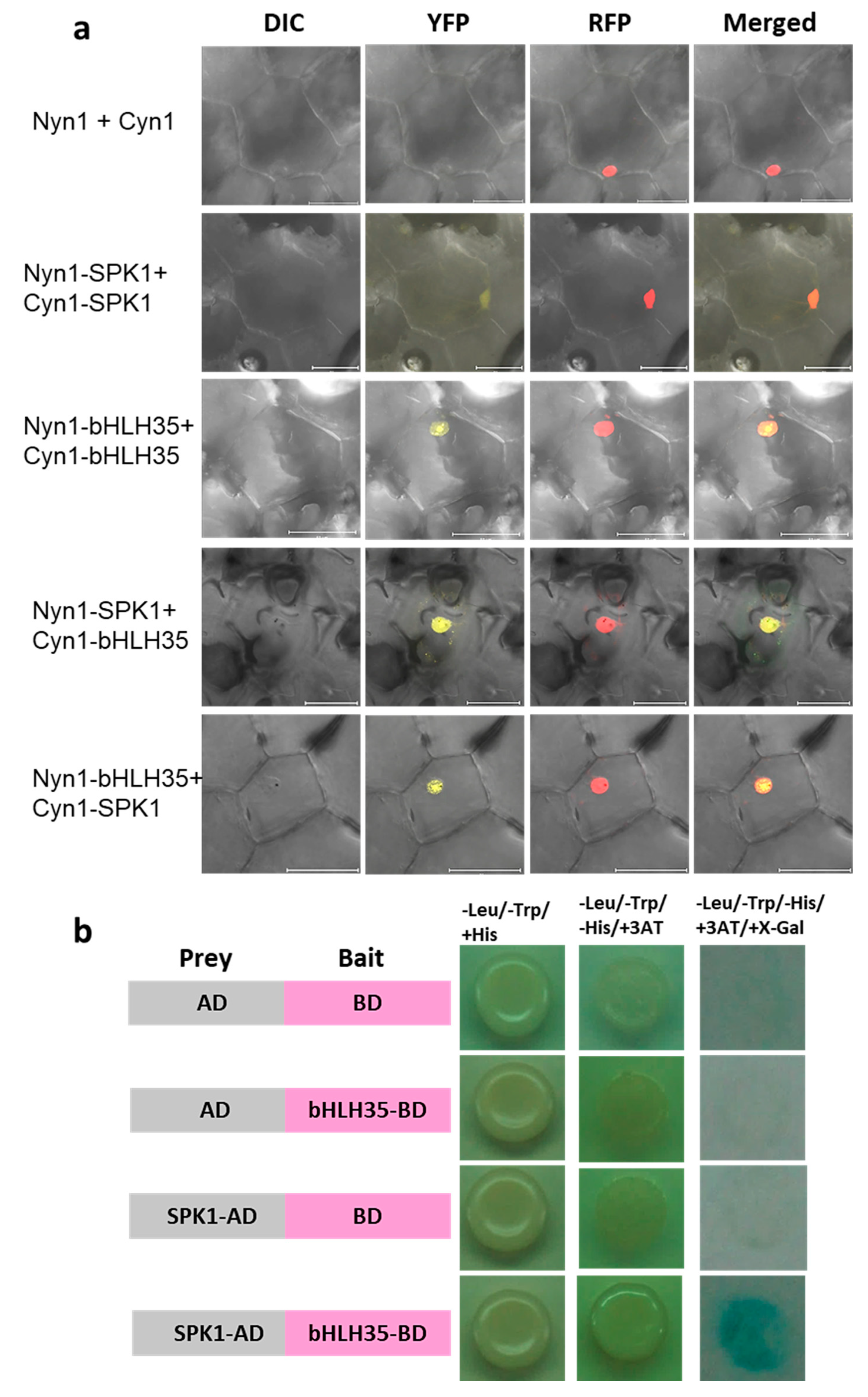
2.7. SPK1 and bHLH35 Form a Heterodimer
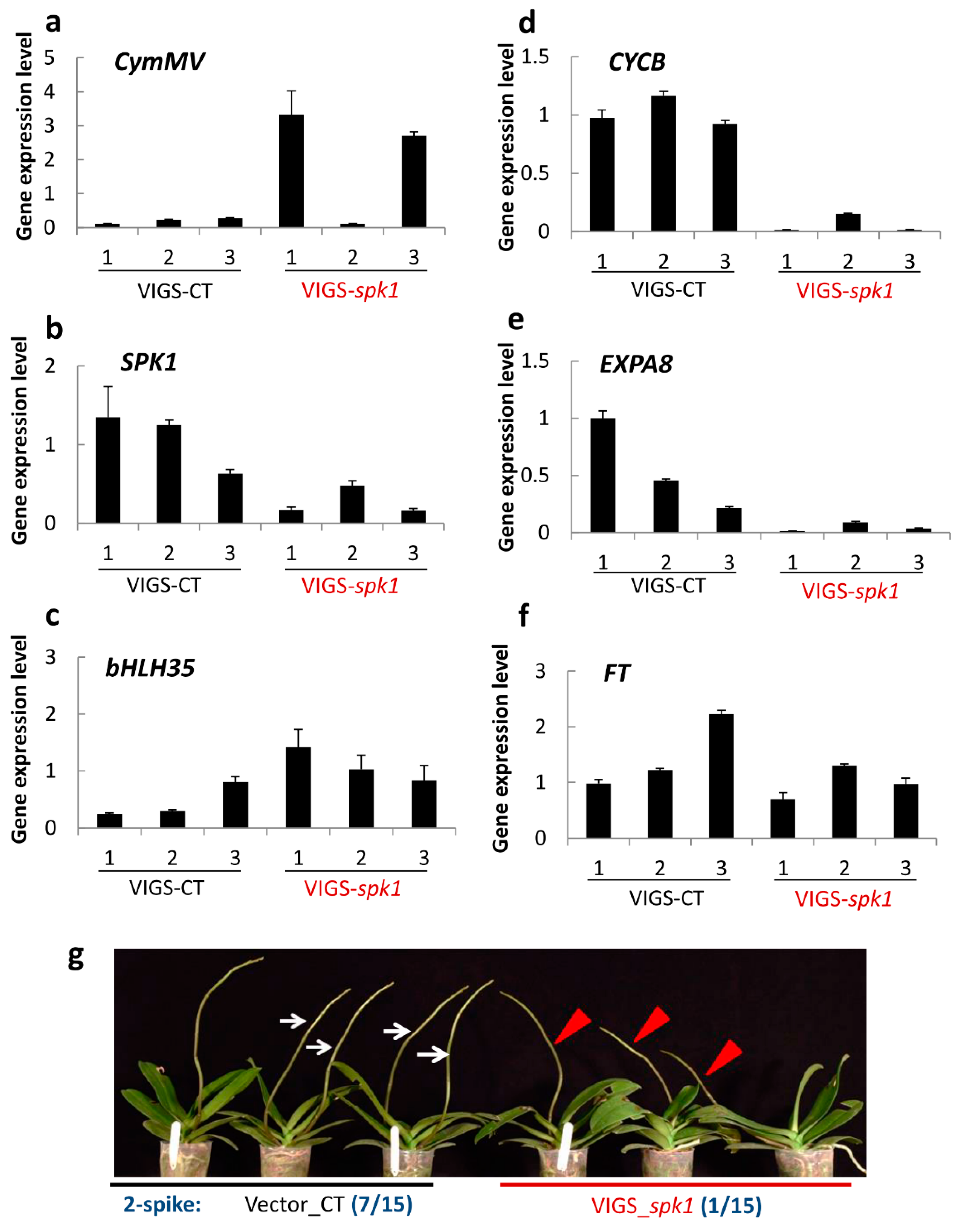
2.8. Verification of SPK1 Function
3. Discussion
3.1. Physiology of Spiking in Phalaenopsis Orchid
3.2. Identification of a Novel Orchid Spiking Activator, SPK1
3.3. SPK1 Controls Meristematic Cell Proliferation
3.4. SPK1 Controls Orchid Spiking
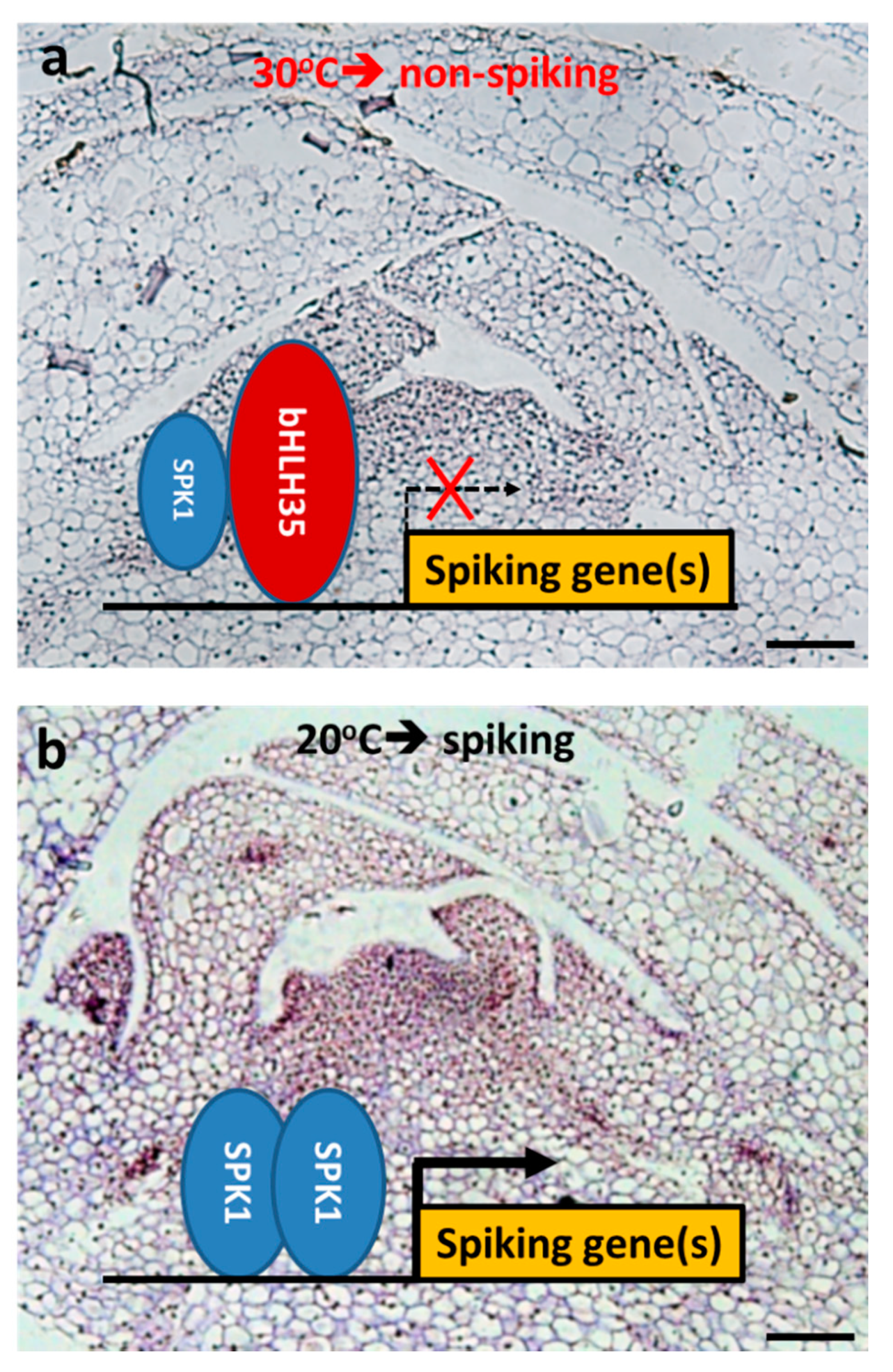
3.5. Coordination among bHLH TFs for Spiking
3.6. Proposed Model for SPK1 Mediates Spiking of Phal. Orchid
3.7. Research Prospects for Control Spiking in Orchid
4. Materials and Methods
4.1. Plant Materials and Growth Conditions
4.2. Cloning of the SPK1 Gene from Phal. aphrodite
4.3. Phylogenetic Analysis
4.4. Gene Expression Analysis
4.5. Tissue Section
4.6. In Situ Hybridization (ISH)
4.7. Bimolecular Florescence Complementation Assay
4.8. Yeast-Two Hybrid (Y2H)
4.9. Virus-Induced Gene Silencing (VIGS)
4.10. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| AB | Axillary bud |
| BiFC | Bimolecular florescence complementation |
| FT | FLOWERING LOCUS T |
| Phal. | Phalaenopsis aphrodite |
| SPK1 | Spike Activator 1 |
| RT-qPCR | Quantitative real-time polymerase chain reaction |
| VIGS | Virus-induced gene silencing |
| Y2H | Yeast two hybrid |
References
- Lee, N.; Lin, G.M. Effect of temperature on growth and flowering of Phalaenopsis white hybrid. J. Chin. Soc. Hort. Sci. 1984, 30, 223–231. [Google Scholar]
- Blanchard, M.G.; Runkle, E.S. Temperature during the day, but not during the night, controls flowering of Phalaenopsis orchids J. Exp. Bot. 2006, 57, 4043–4049. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.H.; Tseng, Y.C.; Liu, Y.C.; Chuo, C.M.; Chen, P.T.; Tseng, K.M.; Yeh, Y.C.; Ger, M.J.; Wang, H.L. Cool-night temperature induces spike emergence and affects photosynthetic efficiency and metabolizable carbohydrate and organic acid pools in Phalaenopsis aphrodite. Plant Cell Rep. 2008, 27, 1667–1675. [Google Scholar] [CrossRef] [PubMed]
- Newton, L.A.; Runkle, E.S. High-temperature inhibition of flowering of Phalaenopsis and Doritaenopsis orchids. Hortscience 2009, 44, 1271–1276. [Google Scholar] [CrossRef]
- Liu, Y.C.; Liu, C.H.; Lin, Y.C.; Lu, C.H.; Chen, W.H.; Wang, H.L. Effect of low irradiance on the photosynthetic performance and spiking of Phalaenopsis. Photosynthetica 2016, 54, 259–266. [Google Scholar] [CrossRef]
- Sakanishi, Y.; Imanishi, H.; Ishida, G. Effect of temperature on growth and flowering of Phalaenopsis amabilis. Bull. Uni. Osaka Prefect. Series B: Agri. Biol. 1980, 32, 1–9. [Google Scholar]
- Abe, M.; Kobayashi, Y.; Yamamoto, S.; Daimon, Y.; Yamaguchi, A.; Ikeda, Y.; Ichinoki, H.; Notaguchi, M.; Goto, K.; Araki, T. FD, a bZIP protein mediating signals from the floral pathway integrator FT at the shoot apex. Science 2005, 309, 1052–1056. [Google Scholar] [CrossRef]
- Jang, S.; Choi, S.C.; Li, H.Y.; An, G.; Schmelzer, E. Functional characterization of Phalaenopsis aphrodite flowering genes PaFT1 and PaFD. PLoS ONE 2015, 10, e0134987. [Google Scholar] [CrossRef]
- Corbesier, L.; Vincent, C.; Jang, S.; Fornara, F.; Fan, Q.; Searle, I.; Giakountis, A.; Farrona, S.; Gissot, L.; Turnbull, C.; et al. FT protein movement contributes to long-distance signaling in floral induction of Arabidopsis. Science 2007, 316, 1030–1033. [Google Scholar] [CrossRef]
- Lin, H.Y.; Chen, J.C.; Wei, M.J.; Lien, Y.C.; Li, H.H.; Ko, S.S.; Liu, Z.H.; Fang, S.C. Genome-wide annotation, expression profiling, and protein interaction studies of the core cell-cycle genes in Phalaenopsis aphrodite. Plant Mol. Biol. 2014, 84, 203–226. [Google Scholar] [CrossRef]
- Inze, D.; De Veylder, L. Cell cycle regulation in plant development. Annu. Rev. Genet. 2006, 40, 77–105. [Google Scholar] [CrossRef] [PubMed]
- Chen, T.K.; Yang, H.T.; Fang, S.C.; Lien, Y.C.; Yang, T.T.; Ko, S.S. Hybrid-Cut: An improved sectioning method for recalcitrant plant tissue samples. J. Vis. Exp. 2016, 117, e54754. [Google Scholar] [CrossRef] [PubMed]
- Vergara, R.; Noriega, X.; Parada, F.; Dantas, D.; Perez, F.J. Relationship between endodormancy, FLOWERING LOCUS T and cell cycle genes in Vitis vinifera. Planta 2015. [Google Scholar] [CrossRef] [PubMed]
- Cosgrove, D.J. Plant expansins: Diversity and interactions with plant cell walls. Curr. Opin. Plant Biol. 2015, 25, 162–172. [Google Scholar] [CrossRef]
- Cosgrove, D.J. New genes and new biological roles for expansins. Curr. Opin. Plant Biol. 2000, 3, 73–78. [Google Scholar] [CrossRef]
- Cosgrove, D.J. Loosening of plant cell walls by expansins. Nature 2000, 407, 321–326. [Google Scholar] [CrossRef]
- Feller, A.; Machemer, K.; Braun, E.L.; Grotewold, E. Evolutionary and comparative analysis of MYB and bHLH plant transcription factors. Plant J. 2011, 66, 94–116. [Google Scholar] [CrossRef]
- Liu, H.; Yu, X.; Li, K.; Klejnot, J.; Yang, H.; Lisiero, D.; Lin, C. Photoexcited CRY2 interacts with CIB1 to regulate transcription and floral initiation in Arabidopsis. Science 2008, 322, 1535–1539. [Google Scholar] [CrossRef]
- Fursova, O.V.; Pogorelko, G.V.; Tarasov, V.A. Identification of ICE2, a gene involved in cold acclimation which determines freezing tolerance in Arabidopsis thaliana. Gene 2009, 429, 98–103. [Google Scholar] [CrossRef]
- Chinnusamy, V.; Ohta, M.; Kanrar, S.; Lee, B.H.; Hong, X.; Agarwal, M.; Zhu, J.K. ICE1: A regulator of cold-induced transcriptome and freezing tolerance in Arabidopsis. Genes Dev. 2003, 17, 1043–1054. [Google Scholar] [CrossRef]
- Lee, S.; Lee, S.; Yang, K.Y.; Kim, Y.M.; Park, S.Y.; Kim, S.Y.; Soh, M.S. Overexpression of PRE1 and its homologous genes activates Gibberellin-dependent responses in Arabidopsis thaliana. Plant Cell Physiol. 2006, 47, 591–600. [Google Scholar] [CrossRef] [PubMed]
- Ni, M.; Tepperman, J.M.; Quail, P.H. PIF3, a phytochrome-interacting factor necessary for normal photoinduced signal transduction, is a novel basic helix-loop-helix protein. Cell 1998, 95, 657–667. [Google Scholar] [CrossRef]
- Bernhardt, C.; Lee, M.M.; Gonzalez, A.; Zhang, F.; Lloyd, A.; Schiefelbein, J. The bHLH genes GLABRA3 (GL3) and ENHANCER OF GLABRA3 (EGL3) specify epidermal cell fate in the Arabidopsis root. Development 2003, 130, 6431–6439. [Google Scholar] [CrossRef] [PubMed]
- Friedrichsen, D.M.; Nemhauser, J.; Muramitsu, T.; Maloof, J.N.; Alonso, J.; Ecker, J.R.; Furuya, M.; Chory, J. Three redundant brassinosteroid early response genes encode putative bHLH transcription factors required for normal growth. Genetics 2002, 162, 1445–1456. [Google Scholar] [PubMed]
- Ohashi-Ito, K.; Bergmann, D.C. Arabidopsis FAMA controls the final proliferation/differentiation switch during stomatal development. Plant Cell 2006, 18, 2493–2505. [Google Scholar] [CrossRef]
- Heisler, M.G.; Atkinson, A.; Bylstra, Y.H.; Walsh, R.; Smyth, D.R. SPATULA, a gene that controls development of carpel margin tissues in Arabidopsis, encodes a bHLH protein. Development 2001, 128, 1089–1098. [Google Scholar]
- Oikawa, T.; Kyozuka, J. Two-step regulation of LAX PANICLE1 protein accumulation in axillary meristem formation in rice. Plant Cell 2009, 21, 1095–1108. [Google Scholar] [CrossRef]
- Ohashi-Ito, K.; Saegusa, M.; Iwamoto, K.; Oda, Y.; Katayama, H.; Kojima, M.; Sakakibara, H.; Fukuda, H. A bHLH complex activates vascular cell division via cytokinin action in root apical meristem. Curr. Biol. 2014, 24, 2053–2058. [Google Scholar] [CrossRef]
- Su, C.L.; Chao, Y.T.; Yen, S.H.; Chen, C.Y.; Chen, W.C.; Chang, Y.C.; Shih, M.C. Orchidstra: An integrated orchid functional genomics database. Plant Cell Physiol. 2013, 54, e11. [Google Scholar] [CrossRef]
- Lu, H.C.; Chen, H.H.; Tsai, W.C.; Chen, W.H.; Su, H.J.; Chang, D.C.; Yeh, H.H. Strategies for functional validation of genes involved in reproductive stages of orchids. Plant Physiol. 2007, 143, 558–569. [Google Scholar] [CrossRef]
- Sheldon, C.C.; Finnegan, E.J.; Rouse, D.T.; Tadege, M.; Bagnall, D.J.; Helliwell, C.A.; Peacock, W.J.; Dennis, E.S. The control of flowering by vernalization. Curr. Opin. Plant Biol. 2000, 3, 418–422. [Google Scholar] [CrossRef]
- Haywood, V.; Yu, T.S.; Huang, N.C.; Lucas, W.J. Phloem long-distance trafficking of GIBBERELLIC ACID-INSENSITIVE RNA regulates leaf development. Plant. J. 2005, 42, 49–68. [Google Scholar] [CrossRef] [PubMed]
- Hanano, S.; Goto, K. Arabidopsis TERMINAL FLOWER1 is involved in the regulation of flowering time and inflorescence development through transcriptional repression. Plant. Cell 2011, 23, 3172–3184. [Google Scholar] [CrossRef] [PubMed]
- Saitou, N.; Nei, M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Zuckerkandl, E.; Pauling, L. Evolutionary Divergence and Convergence in Proteins; Academic Press: New York, NY, USA, 1965. [Google Scholar]
- Lin, Y.J.; Chen, Y.C.; Tseng, K.C.; Chang, W.C.; Ko, S.S. Phototropins Mediate Chloroplast Movement in Phalaenopsis aphrodite (Moth Orchid). Plant. Cell Physiol. 2019, 60, 2243–2254. [Google Scholar] [CrossRef]
- Hsu, C.T.; Liao, D.C.; Wu, F.H.; Liu, N.T.; Shen, S.C.; Chou, S.J.; Tung, S.Y.; Yang, C.H.; Chan, M.T.; Lin, C.S. Integration of molecular biology tools for identifying promoters and genes abundantly expressed in flowers of Oncidium Gower Ramsey. BMC Plant. Biol. 2011, 11, 60. [Google Scholar] [CrossRef]
- Ko, S.S.; Li, M.J.; Ku, M.S.-B.; Ho, Y.C.; Lin, Y.J.; Chuang, M.H.; Hsing, H.X.; Lien, Y.C.; Yang, H.T.; Chang, H.C.; et al. The bHLH142 transcription factor coordinates with TDR1 to modulate the expression of EAT1 and regulate pollen development in rice. Plant. Cell 2014, 26, 2486–2504. [Google Scholar] [CrossRef]

© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lin, Y.-J.; Li, M.-J.; Hsing, H.-C.; Chen, T.-K.; Yang, T.-T.; Ko, S.-S. Spike Activator 1, Encoding a bHLH, Mediates Axillary Bud Development and Spike Initiation in Phalaenopsis aphrodite. Int. J. Mol. Sci. 2019, 20, 5406. https://doi.org/10.3390/ijms20215406
Lin Y-J, Li M-J, Hsing H-C, Chen T-K, Yang T-T, Ko S-S. Spike Activator 1, Encoding a bHLH, Mediates Axillary Bud Development and Spike Initiation in Phalaenopsis aphrodite. International Journal of Molecular Sciences. 2019; 20(21):5406. https://doi.org/10.3390/ijms20215406
Chicago/Turabian StyleLin, Yi-Jyun, Min-Jeng Li, Hung-Chien Hsing, Tien-Kuan Chen, Ting-Ting Yang, and Swee-Suak Ko. 2019. "Spike Activator 1, Encoding a bHLH, Mediates Axillary Bud Development and Spike Initiation in Phalaenopsis aphrodite" International Journal of Molecular Sciences 20, no. 21: 5406. https://doi.org/10.3390/ijms20215406
APA StyleLin, Y.-J., Li, M.-J., Hsing, H.-C., Chen, T.-K., Yang, T.-T., & Ko, S.-S. (2019). Spike Activator 1, Encoding a bHLH, Mediates Axillary Bud Development and Spike Initiation in Phalaenopsis aphrodite. International Journal of Molecular Sciences, 20(21), 5406. https://doi.org/10.3390/ijms20215406