Pharmacological Enhancement of Regeneration-Dependent Regulatory T Cell Recruitment in Zebrafish
Abstract
:1. Introduction
2. Results
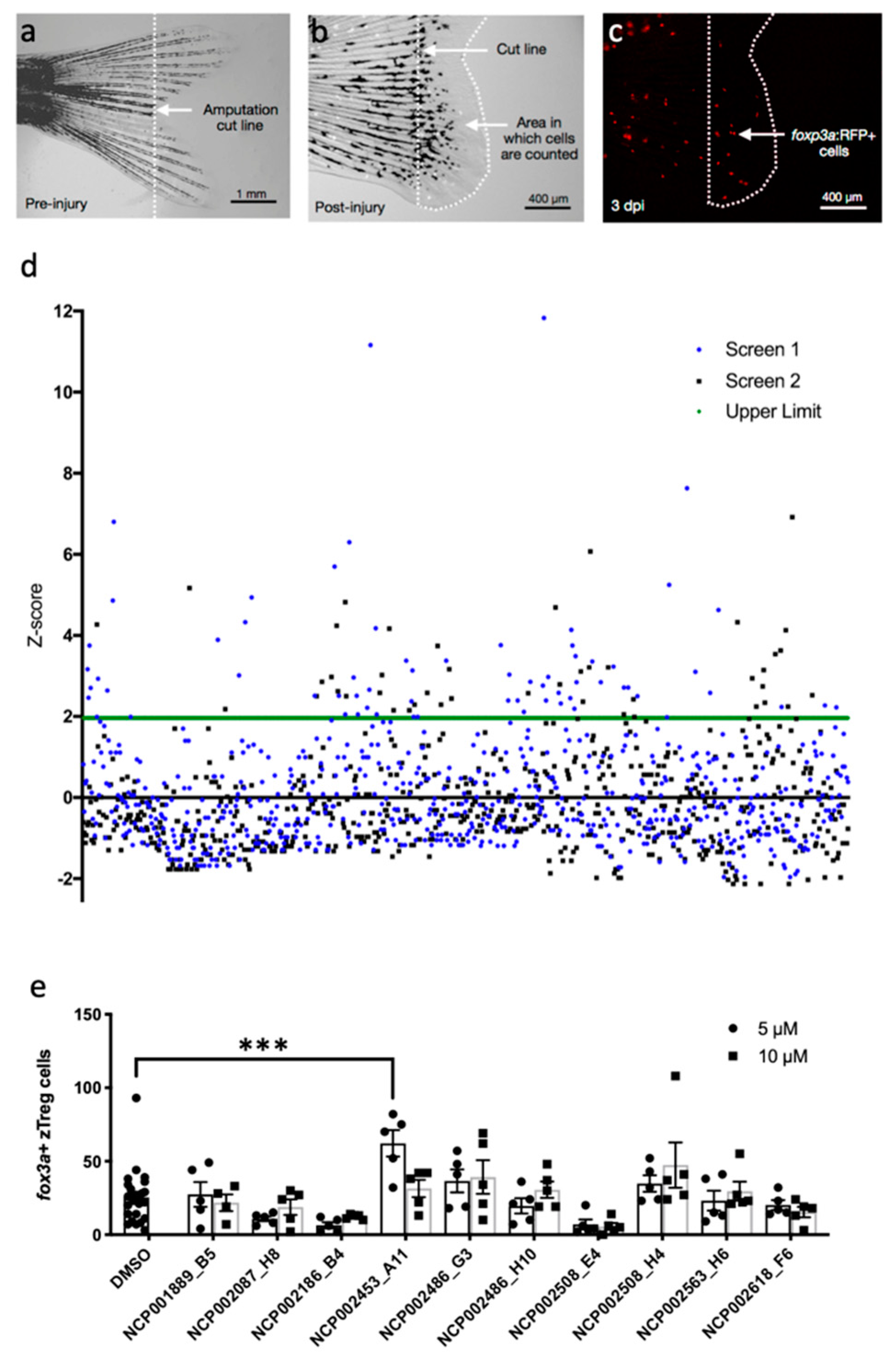
2.1. Chemical Screen for Modulators of zTreg Recruitment to Regenerating Tail Tissue
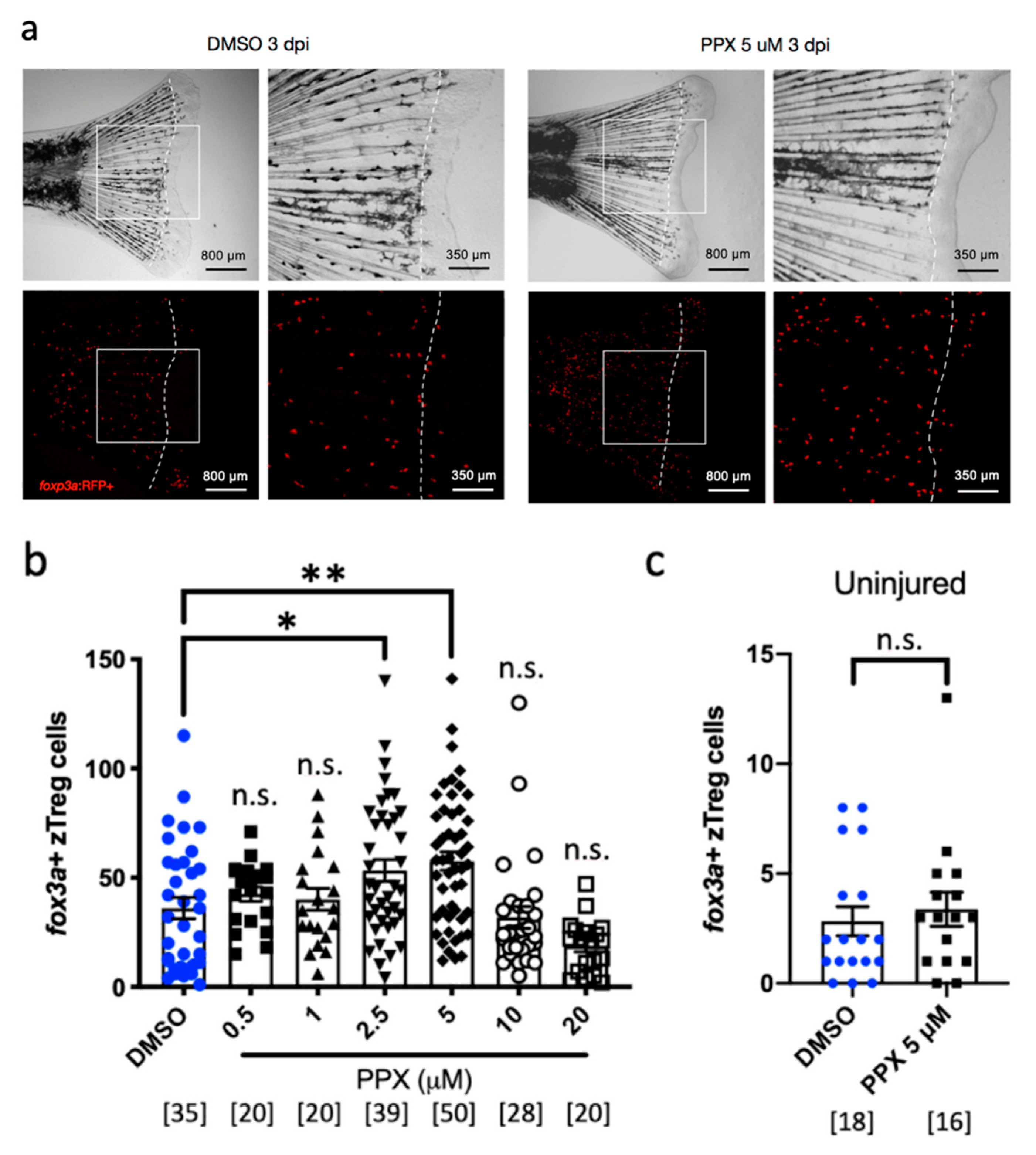
2.2. Pramipexole Stimulates zTreg Recruitment in an Injury-Dependent Manner
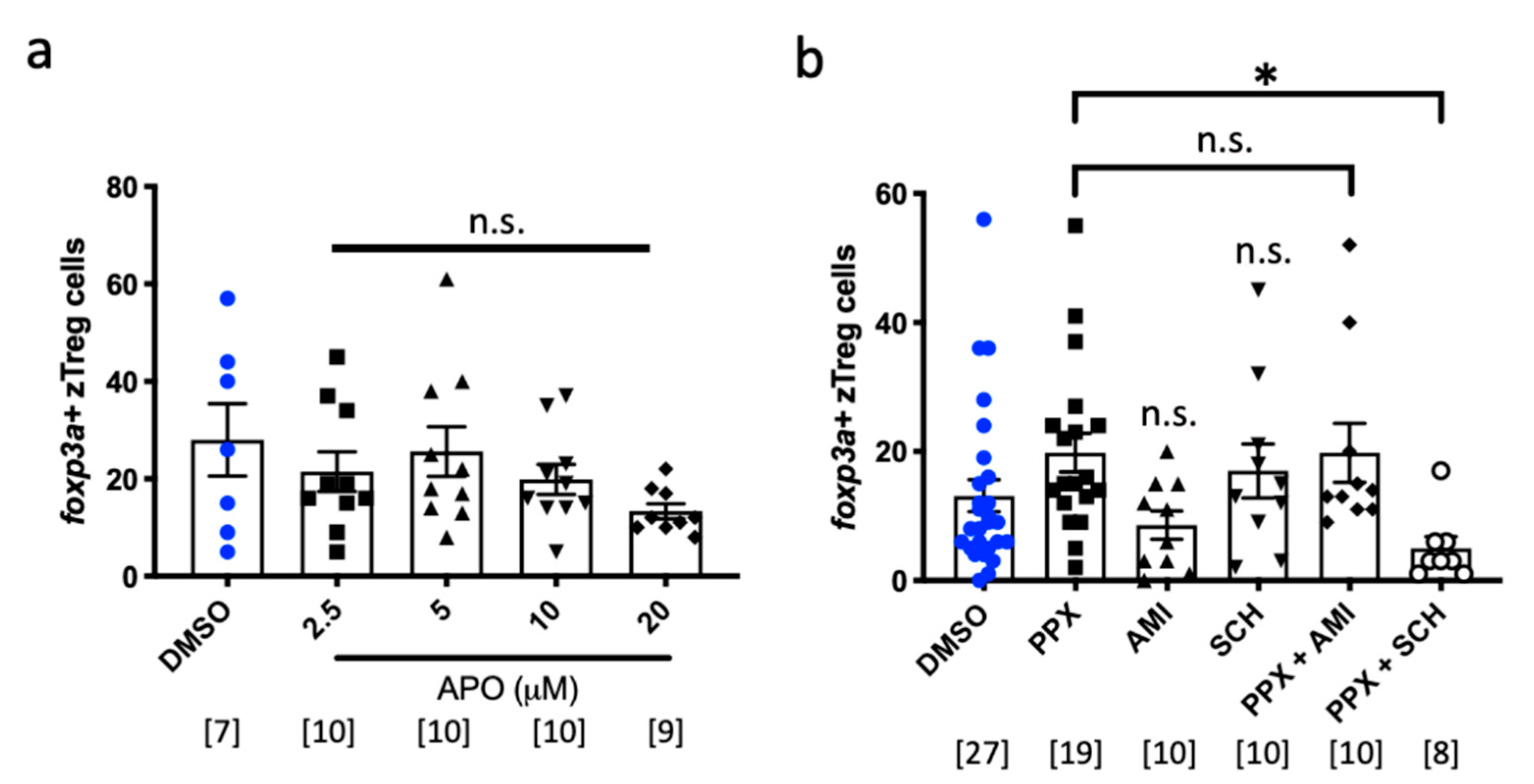
2.3. Pramipexole Stimulates zTreg Recruitment via Dopaminergic Pathways
3. Discussion
4. Materials and Methods
4.1. Zebrafish Maintenance and Breeding
4.2. Zebrafish Transgenic Lines
4.3. Juvenile Fin Amputation
4.4. Chemical Screen and Drug Treatments
4.5. zTreg Quantification
4.6. Statistical Analysis
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| AMI | amisulpride |
| APO | apomorphine |
| BAC | Bacterial Artificial Chromosome |
| DMSO | dimethyl sulfoxide |
| dpi | days post-injury |
| EGFP | Enhanced GFP |
| FOXP3 | Forkhead Box P3 |
| PD | Parkinson’s Disease |
| PPX | pramipexole |
| RFP | Red Fluorescent Protein |
| SCH | SCH-23390 |
| Treg | regulatory T cell |
| wpf | weeks post-fertilization |
| zTreg | zebrafish regulatory T cell |
Appendix A
| Drug Type 1 | Drug Class | Identity |
|---|---|---|
| Immune | Glucocorticoid | CPD000058331 |
| suppressants | Glucocorticoid | CPD000058335 |
| Glucocorticoid | Amcinonide | |
| Glucocorticoid | Beclomethasone | |
| Glucocorticoid | Clobetasol proprionate | |
| Glucocorticoid | Fluorometholone | |
| Glucocorticoid | Fluticasone proprionate | |
| Glucocorticoid | Halometasone monohydrate | |
| Glucocorticoid | Westcort | |
| Glucocorticoid | Beclomethasone dipropionate | |
| Glucocorticoid | Prednisolone acetate | |
| Glucocorticoid | (R)-Budesonide | |
| Glucocorticoid | Loteprednol Etabonate | |
| IL-2 immunosuppressant | Tacrolimus | |
| CNS agents | Phenothiazine | Thioridazine hydrochloride |
| Phenothiazine | Chlorpromazine hydrochloride | |
| Tricyclic antidepressant | Amoxapine | |
| Tricyclic antidepressant | Maprotilline HCl | |
| Tricyclic antidepressant | Desipramine hydrochloride | |
| Selective serotonin reuptake inhibitor | Paroxetine | |
| Serotonin receptor antagonist | Tropisetron hydrochloride | |
| Cyclopyrrolone | Eszopiclone | |
| Benzodiazepine | Chlordiazepoxide | |
| Opioid antagonist | Naloxone hydrochloride | |
| Nicotine receptor agonist | Nicotine | |
| Anaesthetic | Procaine hydrochloride | |
| Antimicrobial | Antibiotic | Doxycycline |
| Antibiotic | Sulfisoxazole | |
| Antiparasitic | Thiabendazole | |
| Antiparasitic | Mebendazole | |
| Antifungal | Griseofulvin | |
| Antiviral | Ganciclovir | |
| Diuretic | Diuretic | Bendrofluazide |
| Carbonic anhydrase inhibitor | Methazolamide | |
| Sodium channel blocker | Triamterene | |
| Hypoglycemic | Enzyme inhibitor | Etomoxir |
| Potassium channel blocker | Tolbutamide | |
| Antineoplastic | Antineoplastic | 6-Azauridine |
| Antineoplastic | Mitoxantrone | |
| Other | PDE5 inhibitor | Tadalafil |
| Adrenoreceptor agonist | Brimonidine | |
| Uricosuric | Sulfinpyrazone | |
| Anticoagulant | Phylloquinone | |
| Antiarrhythmic | Pronestyl |
References
- Goss, R.J. Principles of Regeneration; Academic Press: Cambridge, MA, USA, 1969; p. 287. [Google Scholar]
- Stocum, D.L. Regenerative Biology and Medicine; Academic Press: Cambridge, MA, USA, 2012; p. 15. [Google Scholar]
- Aurora, A.B.; Olson, E.N. Immune modulation of stem cells and regeneration. Cell Stem Cell 2014, 15, 14–25. [Google Scholar] [CrossRef]
- Hui, S.P.; Sheng, D.Z.; Sugimoto, K.; Gonzalez-Rajal, A.; Nakagawa, S.; Hesselson, D.; Kikuchi, K. Zebrafish regulatory T cells mediate organ-specific regenerative programs. Dev. Cell 2017, 43, 659–672.e5. [Google Scholar] [CrossRef]
- Sharma, A.; Rudra, D. Emerging Functions of Regulatory T Cells in Tissue Homeostasis. Front Immunol. 2018, 9, 883. [Google Scholar] [CrossRef]
- Tanaka, E.M. Regeneration: if they can do it, why cant we? Cell 2003, 113, 559–562. [Google Scholar] [CrossRef]
- Garza-Garcia, A.A.; Driscoll, P.C.; Brockes, J.P. Evidence for the local evolution of mechanisms underlying limb regeneration in salamanders. Integr. Comp. Biol. 2010, 50, 528–535. [Google Scholar] [CrossRef]
- Stewart, S.; Tsun, Z.Y.; Izpisua Belmonte, J.C. A histone demethylase is necessary for regeneration in zebrafish. Proc. Natl. Acad. Sci. USA 2009, 106, 19889–19894. [Google Scholar] [CrossRef] [Green Version]
- Yakushiji, N.; Suzuki, M.; Satoh, A.; Sagai, T.; Shiroishi, T.; Kobayashi, H.; Sasaki, H.; Ide, H.; Tamura, K. Correlation between Shh expression and DNA methylation status of the limb-specific Shh enhancer region during limb regeneration in amphibians. Dev. Biol. 2007, 312, 171–182. [Google Scholar] [CrossRef]
- Sanchez Alvarado, A.; Tsonis, P.A. Bridging the regeneration gap: genetic insights from diverse animal models. Nat. Rev. Genet. 2006, 7, 873–884. [Google Scholar] [CrossRef]
- Drenckhahn, J.D.; Schwarz, Q.P.; Gray, S.; Laskowski, A.; Kiriazis, H.; Ming, Z.; Harvey, R.P.; Du, X.J.; Thorburn, D.R.; Cox, T.C. Compensatory growth of healthy cardiac cells in the presence of diseased cells restores tissue homeostasis during heart development. Dev. Cell 2008, 15, 521–533. [Google Scholar] [CrossRef]
- Porrello, E.R.; Mahmoud, A.I.; Simpson, E.; Hill, J.A.; Richardson, J.A.; Olson, E.N.; Sadek, H.A. Transient regenerative potential of the neonatal mouse heart. Science 2011, 331, 1078–1080. [Google Scholar] [CrossRef]
- Illingworth, C.M. Trapped fingers and amputated finger tips in children. J. Pediatr. Surg. 1974, 9, 853–858. [Google Scholar] [CrossRef]
- Douglas, B.S. Conservative management of guillotine amputation of the finger in children. Aust. Paediatr. J. 1972, 8, 86–89. [Google Scholar] [CrossRef] [PubMed]
- Poss, K.D. Advances in understanding tissue regenerative capacity and mechanisms in animals. Nat. Rev. Genet. 2010, 11, 710. [Google Scholar] [CrossRef]
- Howe, K.; Clark, M.D.; Torroja, C.F.; Torrance, J.; Berthelot, C.; Muffato, M.; Collins, J.E.; Humphray, S.; McLaren, K.; Matthews, L.; et al. The zebrafish reference genome sequence and its relationship to the human genome. Nature 2013, 496, 498–503. [Google Scholar] [CrossRef] [Green Version]
- MacRae, C.A.; Peterson, R.T. Zebrafish as tools for drug discovery. Nat. Rev. Drug Discov. 2015, 14, 721. [Google Scholar] [CrossRef]
- Arpaia, N.; Clark, M.D.; Torroja, C.F.; Torrance, J.; Berthelot, C.; Muffato, M.; Collins, J.E.; Humphray, S.; McLaren, K.; Matthews, L.; et al. A distinct function of regulatory T cells in tissue protection. Cell 2015, 162, 1078–1089. [Google Scholar] [CrossRef]
- Brusko, T.; Atkinson, M. Treg in type 1 diabetes. Cell Biochem. Biophys. 2007, 48, 165–175. [Google Scholar] [CrossRef]
- Burzyn, D.; Kuswanto, W.; Kolodin, D.; Shadrach, J.L.; Cerletti, M.; Jang, Y.; Sefik, E.; Tan, T.G.; Wagers, A.J.; Benoist, C.; et al. A special population of regulatory T cells potentiates muscle repair. Cell 2013, 155, 1282–1295. [Google Scholar] [CrossRef]
- Sugimoto, K.; Hui, S.P.; Sheng, D.Z.; Nakayama, M.; Kikuchi, K. Zebrafish FOXP3 is required for the maintenance of immune tolerance. Dev. Comp. Immunol. 2017, 73, 156–162. [Google Scholar] [CrossRef]
- Kvernmo, T.; Hartter, S.; Burger, E. A review of the receptor-binding and pharmacokinetic properties of dopamine agonists. Clin. Ther. 2006, 28, 1065–1078. [Google Scholar] [CrossRef]
- Millan, M.J.; Maiofiss, L.; Cussac, D.; Audinot, V.; Boutin, J.A.; Newman-Tancredi, A. Differential actions of antiparkinson agents at multiple classes of monoaminergic receptor. I. A multivariate analysis of the binding profiles of 14 drugs at 21 native and cloned human receptor subtypes. J. Pharmacol. Exp. Ther. 2002, 303, 791–804. [Google Scholar] [CrossRef] [PubMed]
- Piercey, M.F. Pharmacology of pramipexole, a dopamine D3-preferring agonist useful in treating Parkinson’s disease. Clin. Neuropharmacol. 1998, 21, 141–151. [Google Scholar] [PubMed]
- Wang, Y.; Yu, X.; Zhang, P.; Ma, Y.; Wang, L.; Xu, H.; Sui, D. Neuroprotective effects of pramipexole transdermal patch in the MPTP-induced mouse model of Parkinson’s disease. J. Pharmacol. Sci. 2018, 138, 31–37. [Google Scholar] [CrossRef] [PubMed]
- Sethy, V.H.; Wu, H.; Oostveen, J.A.; Hall, E.D. Neuroprotective effects of the dopamine agonists pramipexole and bromocriptine in 3-acetylpyridine-treated rats. Brain Res. 1997, 754, 181–186. [Google Scholar] [CrossRef]
- Lachowicz, J.E.; Sibley, D.R. Molecular characteristics of mammalian dopamine receptors. Pharmacol. Toxicol. 1997, 81, 105–113. [Google Scholar] [CrossRef] [PubMed]
- Klee, E.W.; Schneider, H.; Clark, K.J.; Cousin, M.A.; Ebbert, J.O.; Hooten, W.M.; Karpyak, V.M.; Warner, D.O.; Ekker, S.C. Zebrafish: A model for the study of addiction genetics. Hum. Genet. 2012, 131, 977–1008. [Google Scholar] [CrossRef] [PubMed]
- Li, P.; Shah, S.; Huang, L.; Carr, A.L.; Gao, Y.; Thisse, C.; Thisse, B.; Li, L. Cloning and spatial and temporal expression of the zebrafish dopamine D1 receptor. Dev. Dyn. 2007, 236, 1339–1346. [Google Scholar] [CrossRef]
- Cosentino, M.; Fietta, A.M.; Ferrari, M.; Rasini, E.; Bombelli, R.; Carcano, E.; Saporiti, F.; Meloni, F.; Marino, F.; Lecchini, S. Human CD4+CD25+ regulatory T cells selectively express tyrosine hydroxylase and contain endogenous catecholamines subserving an autocrine/paracrine inhibitory functional loop. Blood 2007, 109, 632–642. [Google Scholar] [CrossRef]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zwi, S.F.; Choron, C.; Zheng, D.; Nguyen, D.; Zhang, Y.; Roshal, C.; Kikuchi, K.; Hesselson, D. Pharmacological Enhancement of Regeneration-Dependent Regulatory T Cell Recruitment in Zebrafish. Int. J. Mol. Sci. 2019, 20, 5189. https://doi.org/10.3390/ijms20205189
Zwi SF, Choron C, Zheng D, Nguyen D, Zhang Y, Roshal C, Kikuchi K, Hesselson D. Pharmacological Enhancement of Regeneration-Dependent Regulatory T Cell Recruitment in Zebrafish. International Journal of Molecular Sciences. 2019; 20(20):5189. https://doi.org/10.3390/ijms20205189
Chicago/Turabian StyleZwi, Stephanie F., Clarisse Choron, Dawei Zheng, David Nguyen, Yuxi Zhang, Camilla Roshal, Kazu Kikuchi, and Daniel Hesselson. 2019. "Pharmacological Enhancement of Regeneration-Dependent Regulatory T Cell Recruitment in Zebrafish" International Journal of Molecular Sciences 20, no. 20: 5189. https://doi.org/10.3390/ijms20205189





