Determination of Ligand Profiles for Pseudomonas aeruginosa Solute Binding Proteins
Abstract
1. Introduction
2. Results and Discussion
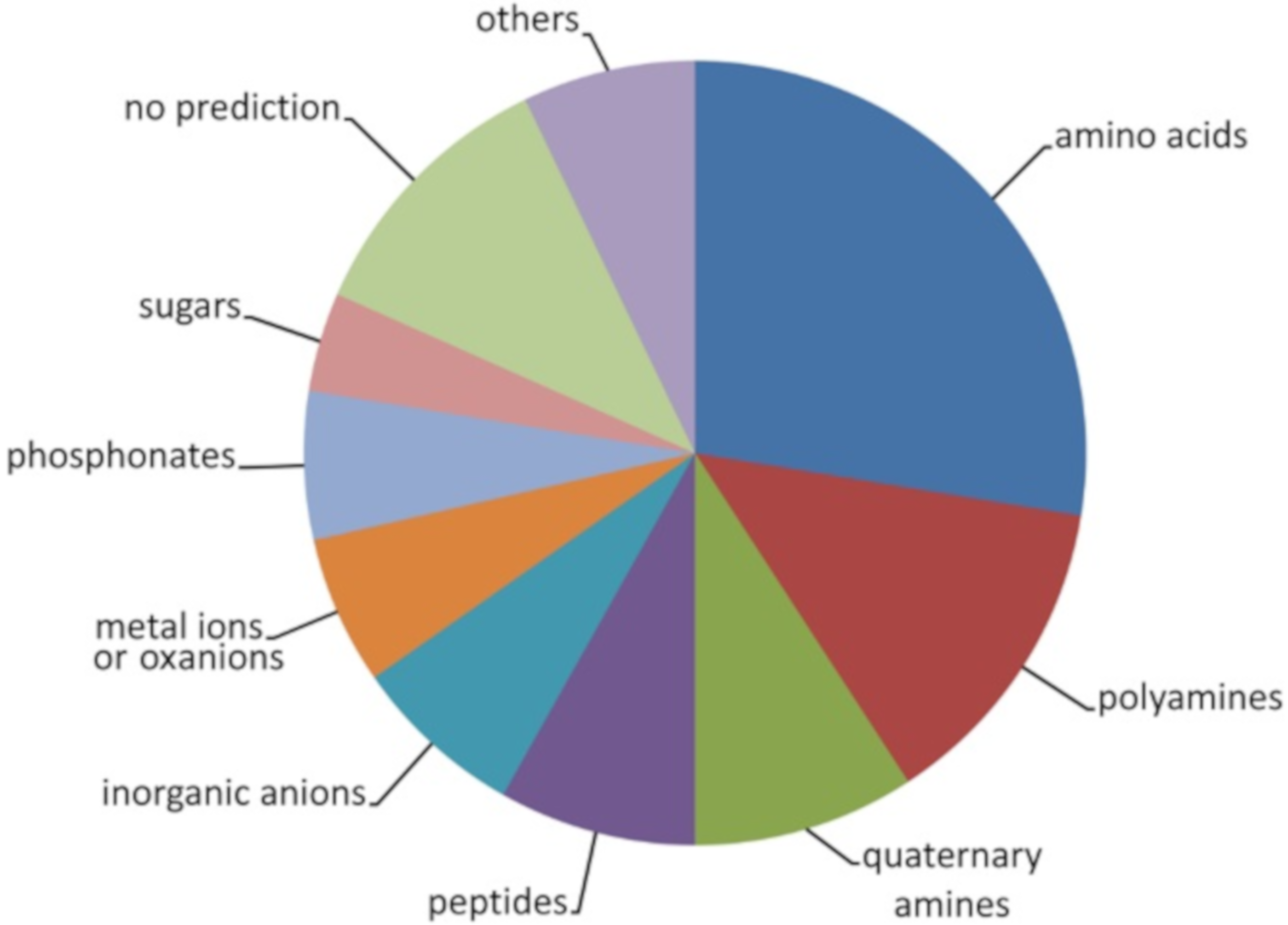
2.1. The Solute Binding Protein Repertoire of P. aeruginosa PAO1
2.2. Study of Ligand Binding to Selected SBPs
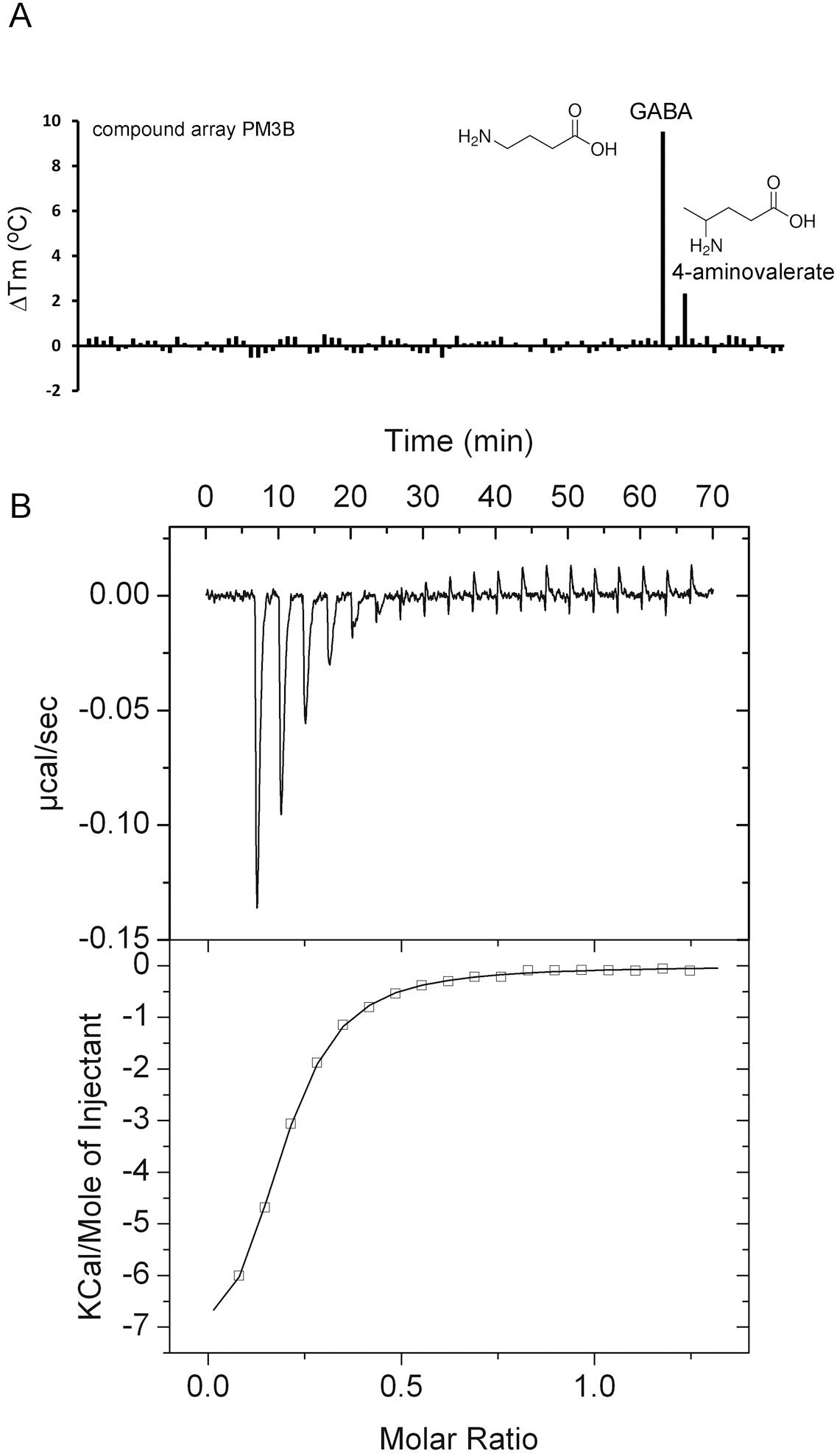
2.3. Proteins Predicted to Bind Polyamines
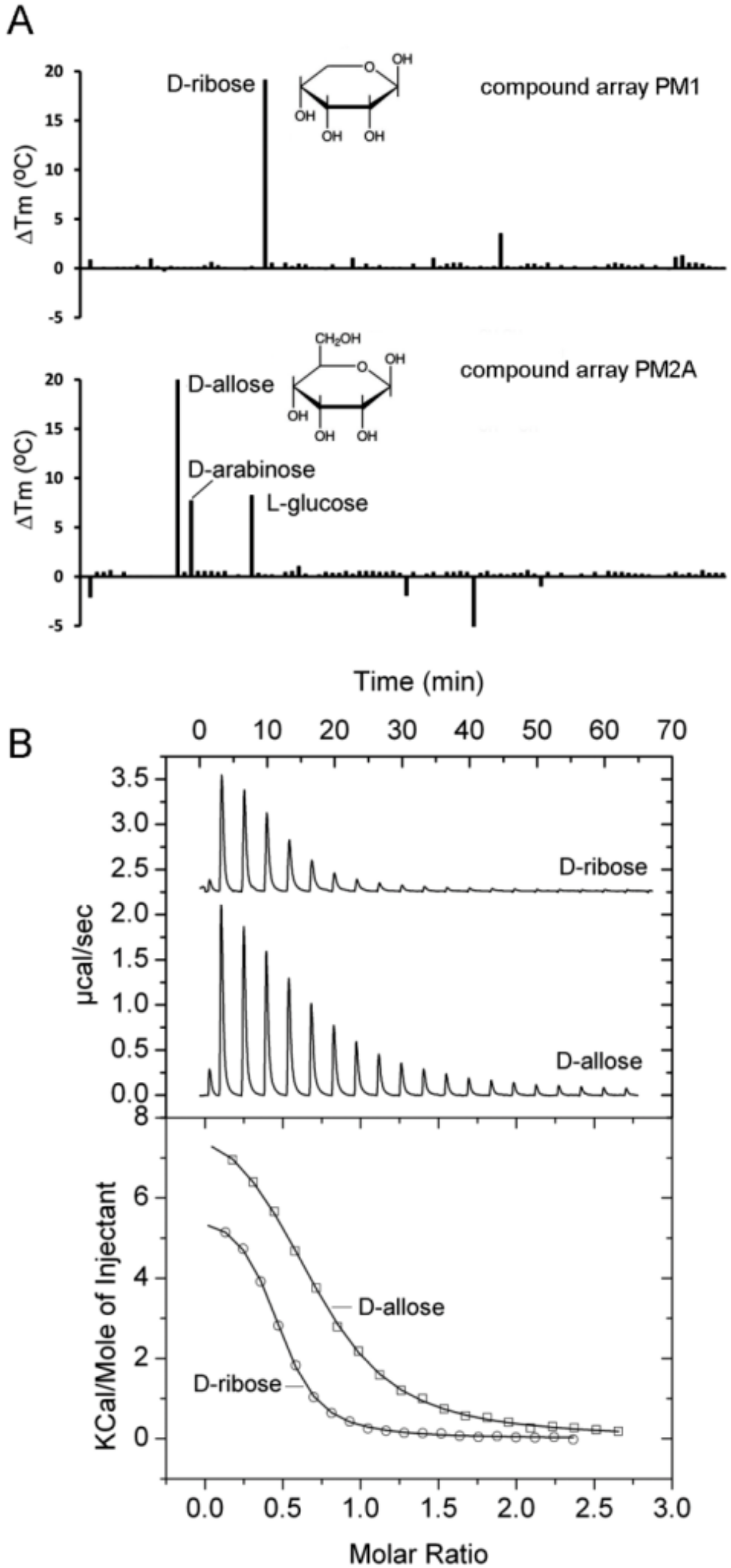
2.4. Proteins Predicted to Bind Sugars
2.5. Proteins Predicted to Bind Amino Acids
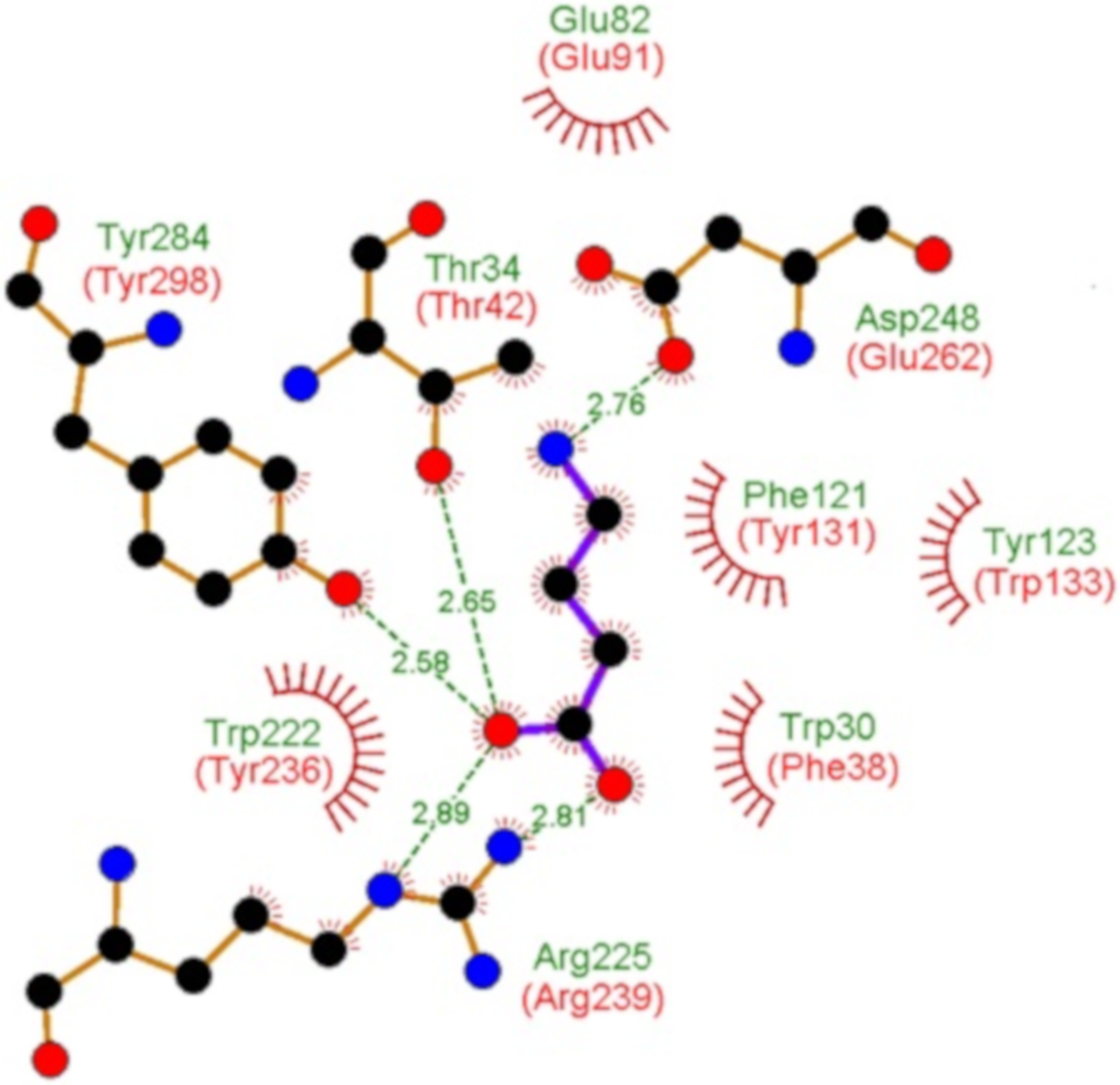
2.6. Proteins Predicted to Bind Peptides
2.7. Proteins Predicted to Bind Sulphate/Thiosulphate
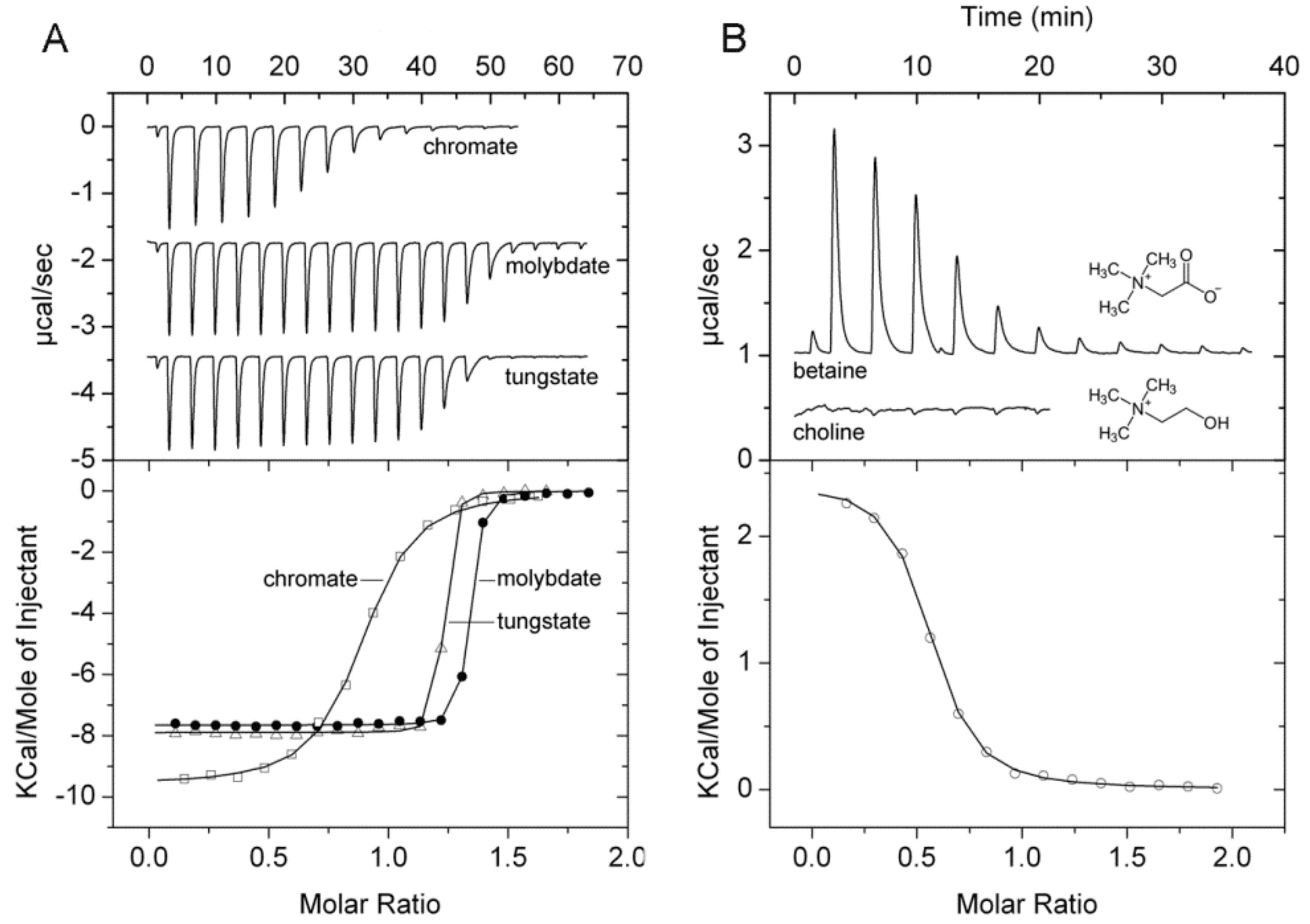
2.8. Proteins Predicted to Bind Metal Ions and Oxanions
2.9. Protein Predicted to Bind Glycine-Betaine
2.10. Chemotaxis to Ligands Recognized by SBPs
3. Materials and Methods
3.1. Establishment of the Solute Binding Protein Repertoire
3.2. Cloning, Expression, and Purification of Solute Binding Proteins
3.3. Differential Scanning Fluorimetry-Based Thermal Shift Assays
3.4. Isothermal Titration Calorimetry
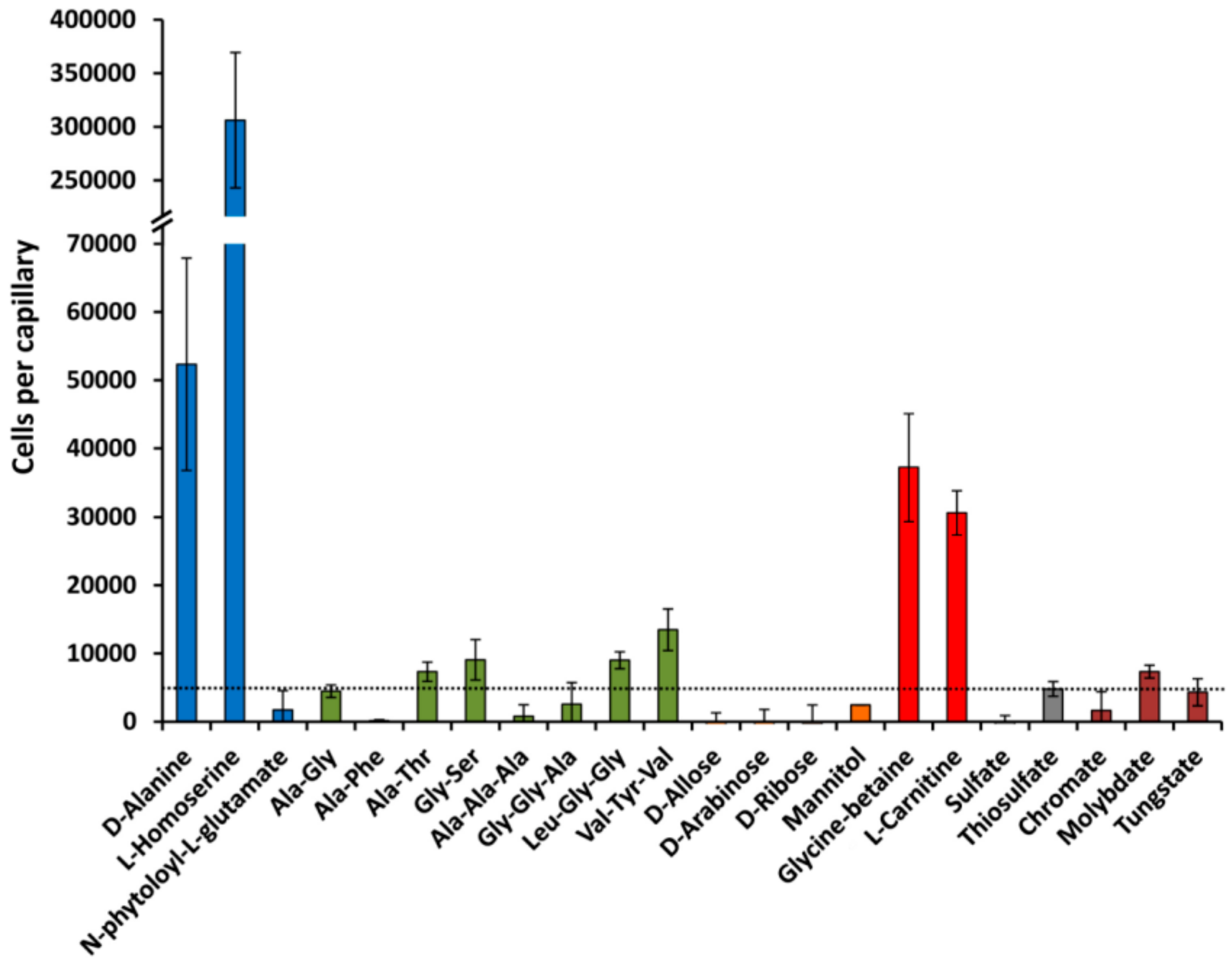
3.5. Quantitative Capillary Chemotaxis Assays
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Wuichet, K.; Zhulin, I.B. Origins and diversification of a complex signal transduction system in prokaryotes. Sci. Signal. 2010, 3, ra50. [Google Scholar] [CrossRef]
- Parkinson, J.S.; Hazelbauer, G.L.; Falke, J.J. Signaling and sensory adaptation in Escherichia coli chemoreceptors: 2015 update. Trends Microbiol. 2015, 23, 257–266. [Google Scholar] [CrossRef] [PubMed]
- Bi, S.; Sourjik, V. Stimulus sensing and signal processing in bacterial chemotaxis. Curr. Opin. Microbiol. 2018, 45, 22–29. [Google Scholar] [CrossRef] [PubMed]
- Hickman, J.W.; Tifrea, D.F.; Harwood, C.S. A chemosensory system that regulates biofilm formation through modulation of cyclic diguanylate levels. Proc. Natl. Acad. Sci. USA 2005, 102, 14422–14427. [Google Scholar] [CrossRef] [PubMed]
- Whitchurch, C.B.; Leech, A.J.; Young, M.D.; Kennedy, D.; Sargent, J.L.; Bertrand, J.J.; Semmler, A.B.; Mellick, A.S.; Martin, P.R.; Alm, R.A.; et al. Characterization of a complex chemosensory signal transduction system which controls twitching motility in Pseudomonas aeruginosa. Mol. Microbiol. 2004, 52, 873–893. [Google Scholar] [CrossRef]
- Hazelbauer, G.L.; Falke, J.J.; Parkinson, J.S. Bacterial chemoreceptors: High-performance signaling in networked arrays. Trends Biochem. Sci. 2008, 33, 9–19. [Google Scholar] [CrossRef]
- Manson, M.D.; Blank, V.; Brade, G.; Higgins, C.F. Peptide chemotaxis in E. coli involves the Tap signal transducer and the dipeptide permease. Nature 1986, 321, 253–256. [Google Scholar] [CrossRef]
- Hegde, M.; Englert, D.L.; Schrock, S.; Cohn, W.B.; Vogt, C.; Wood, T.K.; Manson, M.D.; Jayaraman, A. Chemotaxis to the quorum-sensing signal AI-2 requires the Tsr chemoreceptor and the periplasmic LsrB AI-2-binding protein. J. Bacteriol. 2011, 193, 768–773. [Google Scholar] [CrossRef]
- Springer, M.S.; Goy, M.F.; Adler, J. Sensory transduction in Escherichia coli: Two complementary pathways of information processing that involve methylated proteins. Proc. Natl. Acad. Sci. USA 1977, 74, 3312–3316. [Google Scholar] [CrossRef]
- Kondoh, H.; Ball, C.B.; Adler, J. Identification of a methyl-accepting chemotaxis protein for the ribose and galactose chemoreceptors of Escherichia coli. Proc. Natl. Acad. Sci. USA 1979, 76, 260–264. [Google Scholar] [CrossRef]
- Alexandre, G.; Greer-Phillips, S.; Zhulin, I.B. Ecological role of energy taxis in microorganisms. FEMS Microbiol. Rev. 2004, 28, 113–126. [Google Scholar] [CrossRef] [PubMed]
- Scharf, B.E.; Hynes, M.F.; Alexandre, G.M. Chemotaxis signaling systems in model beneficial plant-bacteria associations. Plant Mol. Biol. 2016, 90, 549–559. [Google Scholar] [CrossRef] [PubMed]
- Ortega, A.; Zhulin, I.B.; Krell, T. Sensory repertoire of bacterial chemoreceptors. Microbiol. Mol. Biol. Rev. 2017, 81, e00033-17. [Google Scholar] [CrossRef] [PubMed]
- Reyes-Darias, J.A.; Garcia, V.; Rico-Jimenez, M.; Corral-Lugo, A.; Lesouhaitier, O.; Juarez-Hernandez, D.; Yang, Y.; Bi, S.; Feuilloley, M.; Munoz-Rojas, J.; et al. Specific gamma-aminobutyrate chemotaxis in pseudomonads with different lifestyle. Mol. Microbiol. 2015, 97, 488–501. [Google Scholar] [CrossRef] [PubMed]
- Taguchi, K.; Fukutomi, H.; Kuroda, A.; Kato, J.; Ohtake, H. Genetic identification of chemotactic transducers for amino acids in Pseudomonas aeruginosa. Microbiology 1997, 143(Pt. 10), 3223–3229. [Google Scholar] [CrossRef] [PubMed]
- Corral-Lugo, A.; Matilla, M.A.; Martin-Mora, D.; Silva Jimenez, H.; Mesa Torres, N.; Kato, J.; Hida, A.; Oku, S.; Conejero-Muriel, M.; Gavira, J.A.; et al. High-affinity chemotaxis to histamine mediated by the tlpq chemoreceptor of the human pathogen Pseudomonas aeruginosa. MBio 2018, 9, e01894-18. [Google Scholar] [CrossRef]
- Martin-Mora, D.; Ortega, A.; Matilla, M.A.; Martinez-Rodriguez, S.; Gavira, J.A.; Krell, T. The molecular mechanism of nitrate chemotaxis via direct ligand binding to the pilj domain of McpN. MBio 2019, 10, e02334–e02418. [Google Scholar] [CrossRef]
- Martin-Mora, D.; Ortega, A.; Reyes-Darias, J.A.; García, V.; López-Farfán, D.; Matilla, M.A.; Krell, T. Identification of a Chemoreceptor in Pseudomonas aeruginosa that specifically mediates Chemotaxis towards alpha-Ketoglutarate. Front. Microbiol. 2016, 7, 1937. [Google Scholar] [CrossRef]
- Rico-Jimenez, M.; Reyes-Darias, J.A.; Ortega, A.; Diez Pena, A.I.; Morel, B.; Krell, T. Two different mechanisms mediate chemotaxis to inorganic phosphate in Pseudomonas aeruginosa. Sci. Rep. 2016, 6, 28967. [Google Scholar] [CrossRef]
- Wu, H.; Kato, J.; Kuroda, A.; Ikeda, T.; Takiguchi, N.; Ohtake, H. Identification and characterization of two chemotactic transducers for inorganic phosphate in Pseudomonas aeruginosa. J. Bacteriol. 2000, 182, 3400–3404. [Google Scholar] [CrossRef]
- Anderson, J.K.; Huang, J.Y.; Wreden, C.; Sweeney, E.G.; Goers, J.; Remington, S.J.; Guillemin, K. Chemorepulsion from the Quorum Signal Autoinducer-2 Promotes Helicobacter pylori Biofilm Dispersal. MBio 2015, 6, e00379. [Google Scholar] [CrossRef] [PubMed]
- Glekas, G.D.; Mulhern, B.J.; Kroc, A.; Duelfer, K.A.; Lei, V.; Rao, C.V.; Ordal, G.W. The Bacillus subtilis chemoreceptor McpC senses multiple ligands using two discrete mechanisms. J. Biol. Chem. 2012, 287, 39412–39418. [Google Scholar] [CrossRef] [PubMed]
- Machuca, M.A.; Liu, Y.C.; Beckham, S.A.; Gunzburg, M.J.; Roujeinikova, A. The crystal structure of the tandem-PAS sensing domain of Campylobacter jejuni chemoreceptor Tlp1 suggests indirect mechanism of ligand recognition. J. Struct. Biol. 2016, 194, 205–213. [Google Scholar] [CrossRef] [PubMed]
- Scheepers, G.H.; Lycklama, A.N.J.A.; Poolman, B. An updated structural classification of substrate-binding proteins. FEBS Lett. 2016, 590, 4393–4401. [Google Scholar] [CrossRef] [PubMed]
- Dwyer, M.A.; Hellinga, H.W. Periplasmic binding proteins: A versatile superfamily for protein engineering. Curr. Opin. Struct. Biol. 2004, 14, 495–504. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Ghimire-Rijal, S.; Lucas, S.L.; Stanley, C.B.; Wright, E.; Agarwal, P.K.; Myles, D.A.; Cuneo, M.J. Periplasmic binding protein dimer has a second allosteric event tied to ligand binding. Biochemistry 2017, 56, 5328–5337. [Google Scholar] [CrossRef] [PubMed]
- Song, L.; Zhang, Y.; Chen, W.; Gu, T.; Zhang, S.Y.; Ji, Q. Mechanistic insights into staphylopine-mediated metal acquisition. Proc. Natl. Acad. Sci. USA 2018, 115, 3942–3947. [Google Scholar] [CrossRef]
- Lamarche, M.G.; Wanner, B.L.; Crepin, S.; Harel, J. The phosphate regulon and bacterial virulence: A regulatory network connecting phosphate homeostasis and pathogenesis. FEMS Microbiol. Rev. 2008, 32, 461–473. [Google Scholar] [CrossRef]
- Hsieh, Y.J.; Wanner, B.L. Global regulation by the seven-component Pi signaling system. Curr. Opin. Microbiol. 2010, 13, 198–203. [Google Scholar] [CrossRef]
- Zaborina, O.; Holbrook, C.; Chen, Y.; Long, J.; Zaborin, A.; Morozova, I.; Fernandez, H.; Wang, Y.; Turner, J.R.; Alverdy, J.C. Structure-function aspects of PstS in multi-drug-resistant Pseudomonas aeruginosa. PLoS Pathog. 2008, 4, e43. [Google Scholar] [CrossRef]
- Madhusudhan, K.T.; McLaughlin, R.; Komori, N.; Matsumoto, H. Identification of a major protein upon phosphate starvation of Pseudomonas aeruginosa PAO1. J. Basic Microbiol. 2003, 43, 36–46. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Wang, C.; Yang, G.; Sun, Z.; Guo, H.; Shao, K.; Gu, Y.; Jiang, W.; Zhang, P. Molecular mechanism of environmental d-xylose perception by a XylFII-LytS complex in bacteria. Proc. Natl. Acad. Sci. USA 2017, 114, 8235–8240. [Google Scholar] [CrossRef] [PubMed]
- Moore, J.O.; Hendrickson, W.A. An asymmetry-to-symmetry switch in signal transmission by the histidine kinase receptor for TMAO. Structure 2012, 20, 729–741. [Google Scholar] [CrossRef] [PubMed]
- Neiditch, M.B.; Federle, M.J.; Pompeani, A.J.; Kelly, R.C.; Swem, D.L.; Jeffrey, P.D.; Bassler, B.L.; Hughson, F.M. Ligand-induced asymmetry in histidine sensor kinase complex regulates quorum sensing. Cell 2006, 126, 1095–1108. [Google Scholar] [CrossRef] [PubMed]
- Elbourne, L.D.; Tetu, S.G.; Hassan, K.A.; Paulsen, I.T. TransportDB 2.0: A database for exploring membrane transporters in sequenced genomes from all domains of life. Nucleic Acids Res. 2017, 45, D320–D324. [Google Scholar] [CrossRef] [PubMed]
- Johnson, D.A.; Tetu, S.G.; Phillippy, K.; Chen, J.; Ren, Q.; Paulsen, I.T. High-throughput phenotypic characterization of Pseudomonas aeruginosa membrane transport genes. PLoS Genet. 2008, 4, e1000211. [Google Scholar] [CrossRef] [PubMed]
- El-Gebali, S.; Mistry, J.; Bateman, A.; Eddy, S.R.; Luciani, A.; Potter, S.C.; Qureshi, M.; Richardson, L.J.; Salazar, G.A.; Smart, A.; et al. The Pfam protein families database in 2019. Nucleic Acids Res. 2019, 47, D427–D432. [Google Scholar] [CrossRef]
- Mitchell, A.L.; Attwood, T.K.; Babbitt, P.C.; Blum, M.; Bork, P.; Bridge, A.; Brown, S.D.; Chang, H.Y.; El-Gebali, S.; Fraser, M.I.; et al. InterPro in 2019: Improving coverage, classification and access to protein sequence annotations. Nucleic Acids Res. 2019, 47, D351–D360. [Google Scholar] [CrossRef]
- Stover, C.K.; Pham, X.Q.; Erwin, A.L.; Mizoguchi, S.D.; Warrener, P.; Hickey, M.J.; Brinkman, F.S.; Hufnagle, W.O.; Kowalik, D.J.; Lagrou, M.; et al. Complete genome sequence of Pseudomonas aeruginosa PAO1, an opportunistic pathogen. Nature 2000, 406, 959–964. [Google Scholar] [CrossRef]
- UniProt, C. UniProt: A worldwide hub of protein knowledge. Nucleic Acids Res. 2019, 47, D506–D515. [Google Scholar]
- Rico-Jimenez, M.; Munoz-Martinez, F.; Garcia-Fontana, C.; Fernandez, M.; Morel, B.; Ortega, A.; Ramos, J.L.; Krell, T. Paralogous chemoreceptors mediate chemotaxis towards protein amino acids and the non-protein amino acid gamma-aminobutyrate (GABA). Mol. Microbiol. 2013, 88, 1230–1243. [Google Scholar] [CrossRef] [PubMed]
- Ekiert, D.C.; Bhabha, G.; Isom, G.L.; Greenan, G.; Ovchinnikov, S.; Henderson, I.R.; Cox, J.S.; Vale, R.D. Architectures of lipid transport systems for the bacterial outer membrane. Cell 2017, 169, 273–285.e17. [Google Scholar] [CrossRef] [PubMed]
- Martin-Mora, D.; Fernandez, M.; Velando, F.; Ortega, A.; Gavira, J.A.; Matilla, M.A.; Krell, T. Functional annotation of bacterial signal transduction systems: Progress and challenges. Int. J. Mol. Sci. 2018, 19, 3755. [Google Scholar] [CrossRef] [PubMed]
- Wu, R.; Sanishvili, R.; Belitsky, B.R.; Juncosa, J.I.; Le, H.V.; Lehrer, H.J.; Farley, M.; Silverman, R.B.; Petsko, G.A.; Ringe, D.; et al. PLP and GABA trigger GabR-mediated transcription regulation in Bacillus subtilis via external aldimine formation. Proc. Natl. Acad. Sci. USA 2017, 114, 3891–3896. [Google Scholar] [CrossRef]
- Spurny, R.; Ramerstorfer, J.; Price, K.; Brams, M.; Ernst, M.; Nury, H.; Verheij, M.; Legrand, P.; Bertrand, D.; Bertrand, S.; et al. Pentameric ligand-gated ion channel ELIC is activated by GABA and modulated by benzodiazepines. Proc. Natl. Acad. Sci. USA 2012, 109, E3028–E3034. [Google Scholar] [CrossRef]
- Planamente, S.; Mondy, S.; Hommais, F.; Vigouroux, A.; Morera, S.; Faure, D. Structural basis for selective GABA binding in bacterial pathogens. Mol. Microbiol. 2012, 86, 1085–1099. [Google Scholar] [CrossRef]
- Guthrie, G.D.; Nicholson-Guthrie, C.S.; Leary, H.L., Jr. A bacterial high-affinity GABA binding protein: Isolation and characterization. Biochem. Biophys. Res. Commun. 2000, 268, 65–68. [Google Scholar] [CrossRef]
- Reimer, A.; Yagur-Kroll, S.; Belkin, S.; Roy, S.; Van der Meer, J.R. Escherichia coli ribose binding protein based bioreporters revisited. Sci. Rep. 2014, 4, 5626. [Google Scholar] [CrossRef]
- Iida, A.; Harayama, S.; Iino, T.; Hazelbauer, G.L. Molecular cloning and characterization of genes required for ribose transport and utilization in Escherichia coli K-12. J. Bacteriol. 1984, 158, 674–682. [Google Scholar]
- Chaudhuri, B.N.; Ko, J.; Park, C.; Jones, T.A.; Mowbray, S.L. Structure of D-allose binding protein from Escherichia coli bound to D-allose at 1.8 A resolution. J. Mol. Biol. 1999, 286, 1519–1531. [Google Scholar] [CrossRef]
- Mowbray, S.L.; Cole, L.B. 1.7 Å X-ray structure of the periplasmic ribose receptor from Escherichia coli. J. Mol. Biol. 1992, 225, 155–175. [Google Scholar] [CrossRef]
- Hare, N.J.; Solis, N.; Harmer, C.; Marzook, N.B.; Rose, B.; Harbour, C.; Crossett, B.; Manos, J.; Cordwell, S.J. Proteomic profiling of Pseudomonas aeruginosa AES-1R, PAO1 and PA14 reveals potential virulence determinants associated with a transmissible cystic fibrosis-associated strain. BMC Microbiol. 2012, 12, 16. [Google Scholar] [CrossRef] [PubMed]
- Nishijyo, T.; Park, S.M.; Lu, C.D.; Itoh, Y.; Abdelal, A.T. Molecular characterization and regulation of an operon encoding a system for transport of arginine and ornithine and the ArgR regulatory protein in Pseudomonas aeruginosa. J. Bacteriol. 1998, 180, 5559–5566. [Google Scholar] [PubMed]
- Wissenbach, U.; Six, S.; Bongaerts, J.; Ternes, D.; Steinwachs, S.; Unden, G. A third periplasmic transport system for L-arginine in Escherichia coli: Molecular characterization of the artPIQMJ genes, arginine binding and transport. Mol. Microbiol. 1995, 17, 675–686. [Google Scholar] [CrossRef] [PubMed]
- Stamp, A.L.; Owen, P.; El Omari, K.; Lockyer, M.; Lamb, H.K.; Charles, I.G.; Hawkins, A.R.; Stammers, D.K. Crystallographic and microcalorimetric analyses reveal the structural basis for high arginine specificity in the Salmonella enterica serovar Typhimurium periplasmic binding protein STM4351. Proteins 2011, 79, 2352–2357. [Google Scholar] [CrossRef]
- Oh, B.H.; Ames, G.F.; Kim, S.H. Structural basis for multiple ligand specificity of the periplasmic lysine-, arginine-, ornithine-binding protein. J. Biol. Chem. 1994, 269, 26323–26330. [Google Scholar]
- Nikaido, K.; Ames, G.F. Purification and characterization of the periplasmic lysine-, arginine-, ornithine-binding protein (LAO) from Salmonella typhimurium. J. Biol. Chem. 1992, 267, 20706–20712. [Google Scholar]
- Hoshino, T.; Kose-Terai, K.; Sato, K. Solubilization and reconstitution of the Pseudomonas aeruginosa high affinity branched-chain amino acid transport system. J. Biol. Chem. 1992, 267, 21313–21318. [Google Scholar]
- Singh, B.; Rohm, K.H. A new subfamily of bacterial glutamate/aspartate receptors. Biol. Chem. 2008, 389, 33–36. [Google Scholar] [CrossRef]
- Singh, B.; Rohm, K.H. Characterization of a Pseudomonas putida ABC transporter (AatJMQP) required for acidic amino acid uptake: Biochemical properties and regulation by the Aau two-component system. Microbiology 2008, 154, 797–809. [Google Scholar] [CrossRef]
- Letoffe, S.; Delepelaire, P.; Wandersman, C. The housekeeping dipeptide permease is the Escherichia coli heme transporter and functions with two optional peptide binding proteins. Proc. Natl. Acad. Sci. USA 2006, 103, 12891–12896. [Google Scholar] [CrossRef] [PubMed]
- Pletzer, D.; Lafon, C.; Braun, Y.; Kohler, T.; Page, M.G.; Mourez, M.; Weingart, H. High-throughput screening of dipeptide utilization mediated by the ABC transporter DppBCDF and its substrate-binding proteins DppA1-A5 in Pseudomonas aeruginosa. PLoS ONE 2014, 9, e111311. [Google Scholar] [CrossRef] [PubMed]
- Nickitenko, A.V.; Trakhanov, S.; Quiocho, F.A. 2 A resolution structure of DppA, a periplasmic dipeptide transport/chemosensory receptor. Biochemistry 1995, 34, 16585–16595. [Google Scholar] [CrossRef] [PubMed]
- Guyer, C.A.; Morgan, D.G.; Staros, J.V. Binding specificity of the periplasmic oligopeptide-binding protein from Escherichia coli. J. Bacteriol. 1986, 168, 775–779. [Google Scholar] [CrossRef]
- Berntsson, R.P.; Doeven, M.K.; Fusetti, F.; Duurkens, R.H.; Sengupta, D.; Marrink, S.J.; Thunnissen, A.M.; Poolman, B.; Slotboom, D.J. The structural basis for peptide selection by the transport receptor OppA. EMBO J. 2009, 28, 1332–1340. [Google Scholar] [CrossRef] [PubMed]
- Wright, B.W.; Kamath, K.S.; Krisp, C.; Molloy, M.P. Proteome profiling of Pseudomonas aeruginosa PAO1 identifies novel responders to copper stress. BMC Microbiol. 2019, 19, 69. [Google Scholar] [CrossRef]
- Hryniewicz, M.; Sirko, A.; Palucha, A.; Bock, A.; Hulanicka, D. Sulfate and thiosulfate transport in Escherichia coli K-12: Identification of a gene encoding a novel protein involved in thiosulfate binding. J. Bacteriol. 1990, 172, 3358–3366. [Google Scholar] [CrossRef]
- Sirko, A.; Zatyka, M.; Sadowy, E.; Hulanicka, D. Sulfate and thiosulfate transport in Escherichia coli K-12: Evidence for a functional overlapping of sulfate- and thiosulfate-binding proteins. J. Bacteriol. 1995, 177, 4134–4136. [Google Scholar] [CrossRef]
- Murphy, T.F.; Kirkham, C.; Johnson, A.; Brauer, A.L.; Koszelak-Rosenblum, M.; Malkowski, M.G. Sulfate-binding protein, CysP, is a candidate vaccine antigen of Moraxella catarrhalis. Vaccine 2016, 34, 3855–3861. [Google Scholar] [CrossRef]
- Pereira, C.T.; Roesler, C.; Faria, J.N.; Fessel, M.R.; Balan, A. Sulfate-Binding Protein (Sbp) from Xanthomonas citri: Structure and Functional Insights. Mol. Plant. Microbe Interact. 2017, 30, 578–588. [Google Scholar] [CrossRef]
- He, J.J.; Quiocho, F.A. Dominant role of local dipoles in stabilizing uncompensated charges on a sulfate sequestered in a periplasmic active transport protein. Protein Sci. 1993, 2, 1643–1647. [Google Scholar] [CrossRef] [PubMed]
- Jacobson, B.L.; Quiocho, F.A. Sulfate-binding protein dislikes protonated oxyacids. A molecular explanation. J. Mol. Biol. 1988, 204, 783–787. [Google Scholar] [CrossRef]
- Dudev, T.; Lim, C. Oxyanion selectivity in sulfate and molybdate transport proteins: An ab initio/CDM study. J. Am. Chem. Soc. 2004, 126, 10296–10305. [Google Scholar] [CrossRef] [PubMed]
- Pederick, V.G.; Eijkelkamp, B.A.; Ween, M.P.; Begg, S.L.; Paton, J.C.; McDevitt, C.A. Acquisition and role of molybdate in Pseudomonas aeruginosa. Appl. Environ. Microbiol. 2014, 80, 6843–6852. [Google Scholar] [CrossRef]
- Tirado-Lee, L.; Lee, A.; Rees, D.C.; Pinkett, H.W. Classification of a Haemophilus influenzae ABC transporter HI1470/71 through its cognate molybdate periplasmic binding protein, MolA. Structure 2011, 19, 1701–1710. [Google Scholar] [CrossRef]
- Rech, S.; Wolin, C.; Gunsalus, R.P. Properties of the periplasmic ModA molybdate-binding protein of Escherichia coli. J. Biol. Chem. 1996, 271, 2557–2562. [Google Scholar] [CrossRef]
- Vigonsky, E.; Ovcharenko, E.; Lewinson, O. Two molybdate/tungstate ABC transporters that interact very differently with their substrate binding proteins. Proc. Natl. Acad. Sci. USA 2013, 110, 5440–5445. [Google Scholar] [CrossRef]
- Otrelo-Cardoso, A.R.; Nair, R.R.; Correia, M.A.; Rivas, M.G.; Santos-Silva, T. TupA: A tungstate binding protein in the periplasm of Desulfovibrio alaskensis G20. Int. J. Mol. Sci. 2014, 15, 11783–11798. [Google Scholar] [CrossRef]
- Bevers, L.E.; Hagedoorn, P.L.; Krijger, G.C.; Hagen, W.R. Tungsten transport protein A (WtpA) in Pyrococcus furiosus: The first member of a new class of tungstate and molybdate transporters. J. Bacteriol. 2006, 188, 6498–6505. [Google Scholar] [CrossRef]
- Karpus, J.; Bosscher, M.; Ajiboye, I.; Zhang, L.; He, C. Chromate Binding and Removal by the Molybdate-Binding Protein ModA. ChemBioChem 2017, 18, 633–637. [Google Scholar] [CrossRef]
- Fernandez, M.; Ortega, A.; Rico-Jimenez, M.; Martin-Mora, D.; Daddaoua, A.; Matilla, M.A.; Krell, T. High-throughput screening to identify chemoreceptor ligands. Methods Mol. Biol. 2018, 1729, 291–301. [Google Scholar] [PubMed]
- Abril, M.A.; Michan, C.; Timmis, K.N.; Ramos, J.L. Regulator and enzyme specificities of the TOL plasmid-encoded upper pathway for degradation of aromatic hydrocarbons and expansion of the substrate range of the pathway. J. Bacteriol. 1989, 171, 6782–6790. [Google Scholar] [CrossRef] [PubMed]
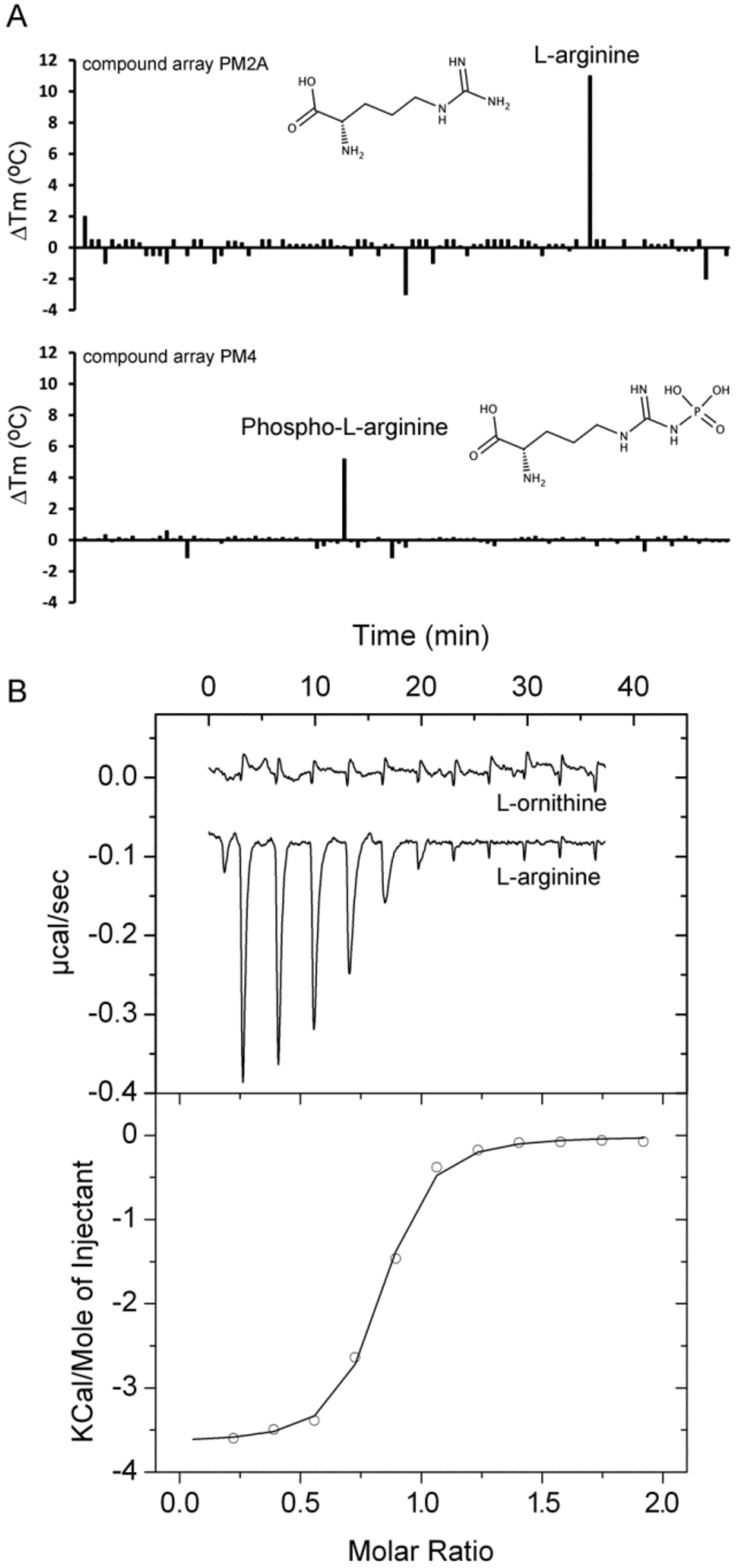
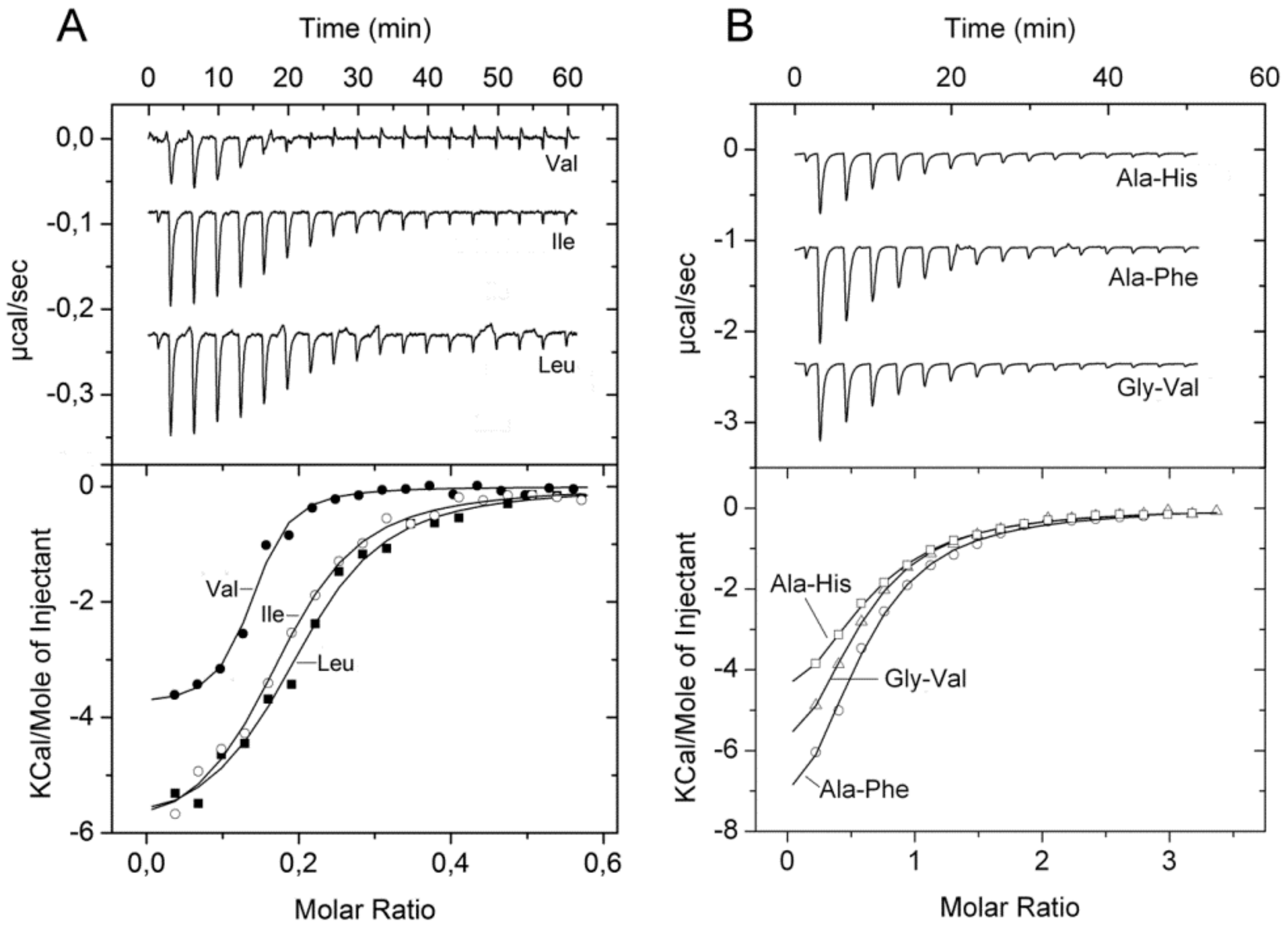
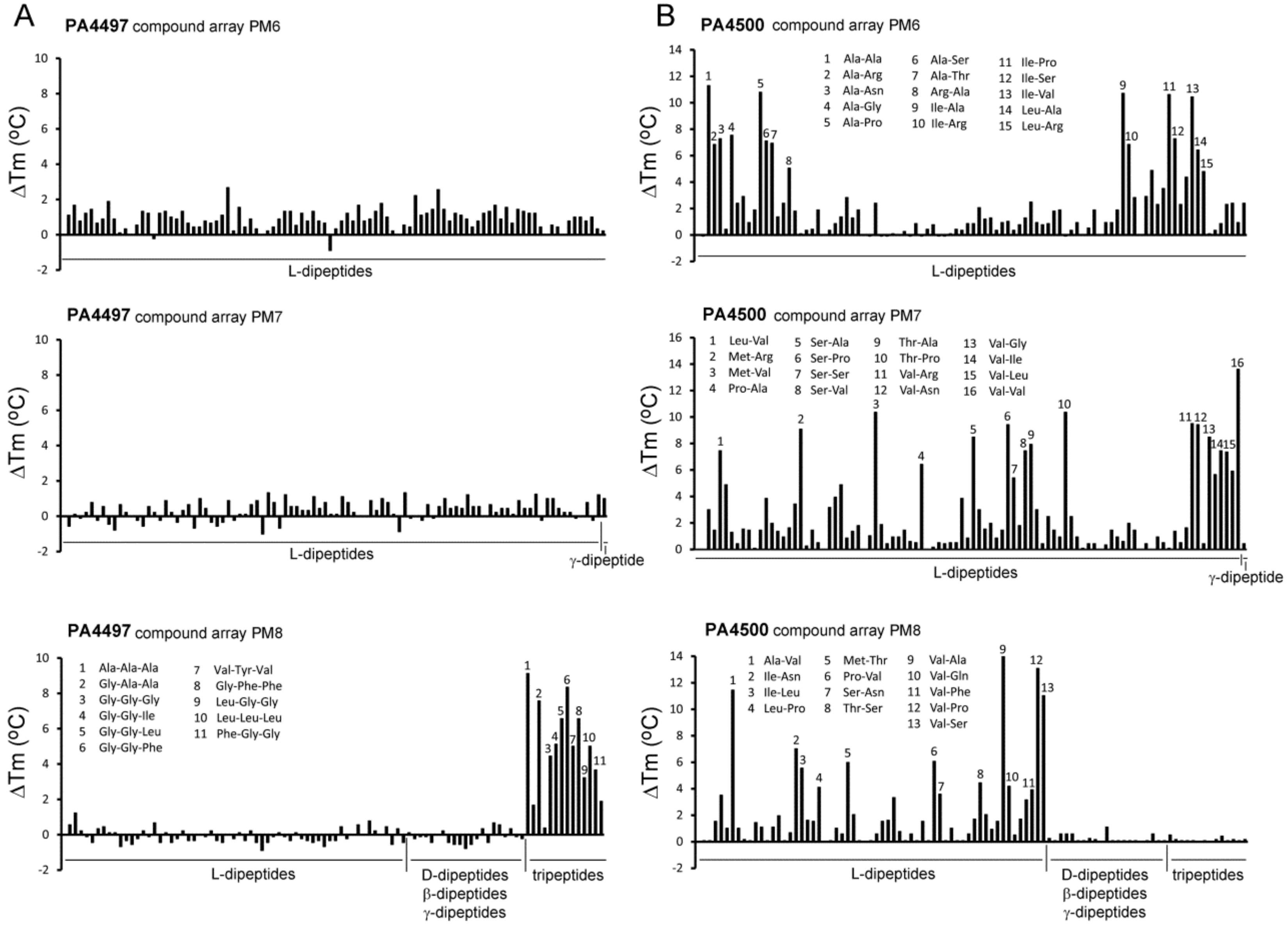
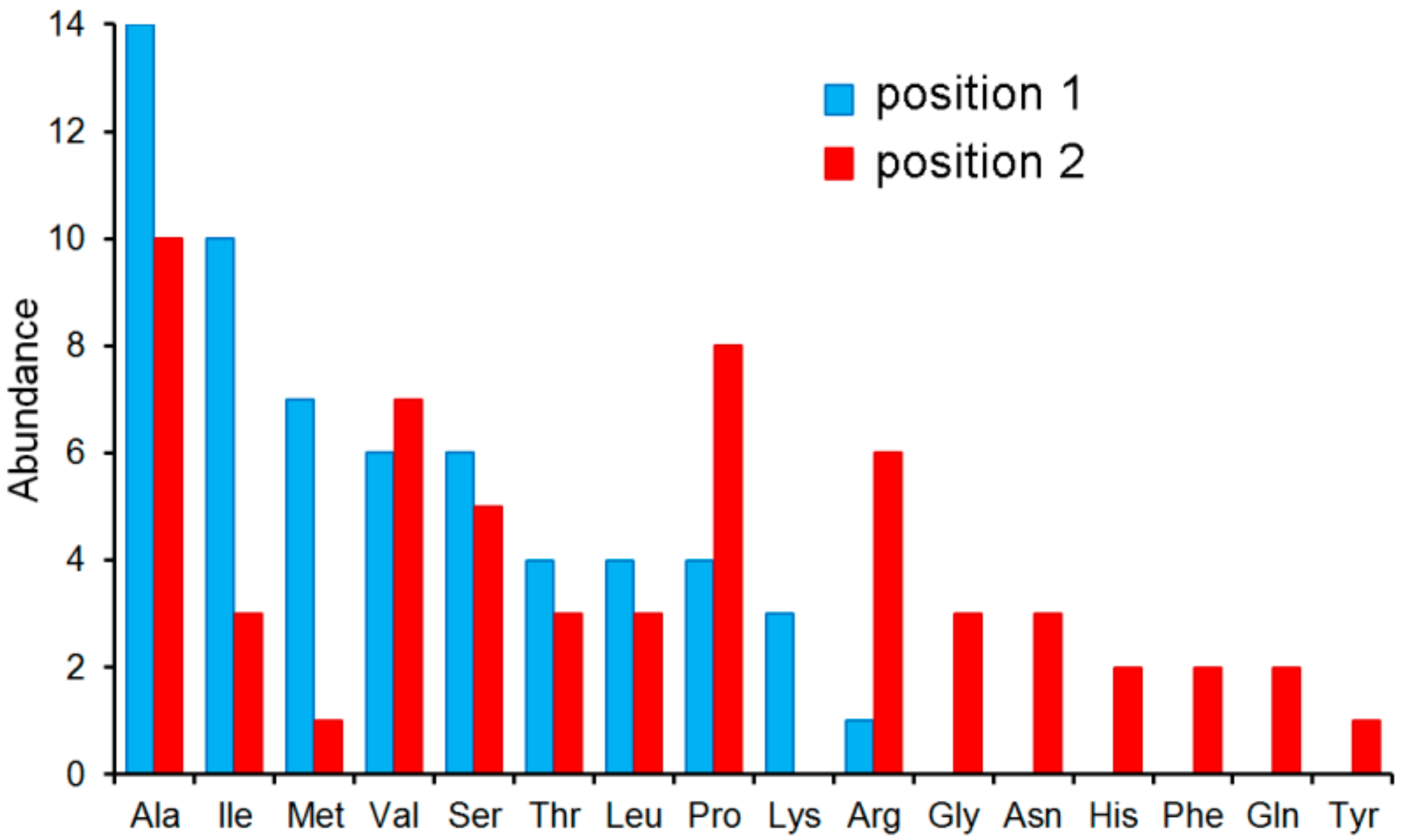
| ORF (Gene Name) | Ligand | Tm DSF (°C) | KD by ITC (µM) | Chemotaxis 1 | No Binding Observed by ITC |
|---|---|---|---|---|---|
| PA0222 | GABA 4-aminovalerate | 9.6 2.2 | 0.29 ± 0.05 No binding | Yes [14] n. d. | Butyrate, spermidine, histamine |
| PA0283 (sbp) | Sulphate Thiosulphate | 12.2 2.1 | No binding No binding | No Yes | |
| PA0888 (aotJ) | L-Arg Phospho-L-arginine | 11 5.8 | 0.12 ± 0.02 n. d. | Yes [15,41] n. d. | L-ornithine, D-Arg, L-canavanine |
| PA1074 (braC) | L-Ala L-Ile L-Leu L-Val L-Thr L-Met L-Homoserine D,L-α-Amino-N-butyrate α-amino-N-valerate | 4.0 12.1 10.1 10.2 3.0 3.5 5.0 3.8 4.2 | 0.21 ± 0.03 0.28 ± 0.05 0.29 ± 0.06 0.07 ± 0.01 0.48 ± 0.1 No binding 0.72 ± 0.1 n. d. n. d. | Yes [15,41] Yes [15,41] Yes [15,41] Yes [15,41] Yes [15,41] Yes [15,41] Yes n. d. n. d. | L-Met, D-Ala |
| PA1342 (aatJ) | L-Asp L-Glu N-phthaloyl-L-glutamate | 10.9 13.1 11.5 | 20.4 ± 5 1.4 ± 0.1 n. d. | Yes [15] Yes [15] No | L-Ala, D-Glu, L-Arg, L-Cys, L-Gln, L-Asn |
| PA1493 (cysP) | Thiosulphate | 7.4 | 0.29 ± 0.2 | No | Sulphate, sulfite, molybdate, phosphate, selenite |
| PA1863 (modA) | Chromate Molybdate Tungstate | 9.9 2,3 12.0 2,3 n. d. 2 | 0.44 ± 0.02 0.01 ± 0.001 Ultratight 4 | No Yes No | Vanadate, manganate |
| PA1946 (rspB) | D-Ribose D-Allose D-Arabinose | 21.7 22.0 7.5 | 2.1 ± 0.1 6.6 ± 0.1 No binding | No No No | |
| PA2204 | No compound increased the Tm by more than 1.5 degrees. | ||||
| PA2338 | Mannitol | 4.1 | 0.83 ± 0.2 | No | Maltose |
| PA2592 | Putrescine Agmatine Histamine | 4.4 5.3 3.9 | 31 ± 4 15 ± 2 No binding | Yes [16] Yes [16] Yes [16] | Spermidine |
| PA3610 (potD) | Putrescine Cadaverine | 13 n. d.2 | 4.8 ± 0.4 65 ± 9 | Yes [16] Yes [16] | Spermidine, histamine, homoserine, L-Asn, butyrate |
| PA3889 (opuCC) | Glycine-betaine L-carnitine | 3.2 2.8 | 3.0 ± 0.4 No binding | Yes Yes | Choline, L-Pro |
| PA4497 (dppA2) | Ala-Ala-Ala Gly-Gly-Ala Gly-Gly-Gly Gly-Gly-Ile Gly-Gly-Leu Phe-Gly-Gly Val-Tyr-Val Gly-Phe-Phe Leu-Gly-Gly Leu-Leu-Leu Gly-Gly-Phe | 9.1 7.6 4.4 5.1 6.6 8.3 5.0 6.6 3.2 5.0 3.7 | Insufficient protein to do ITC | No No n. d. n. d. n. d. n. d. Yes n. d. Yes n. d. n. d. | |
| PA4500 (dppA3) | 59 dipeptides increased the Tm by more than 3 °C. (see Table S2 and Figure 5B and Figure 6) Ala-Ala Ala-Thr Ala-His Ala-Phe Gly-Val Gly-Ser Ala-Gly | 11.3 7.0 3.0 2.0 2.0 n. d. n. d. | 0.21 ± 0.02 0.98 ± 0.1 7.4 ± 0.4 6.1 ± 0.6 6.3 ± 0.4 n. d. n. d. | n. d. Yes n. d. No n. d. Yes No | L-Ala, Ala-Ala-Ala, Glu-Glu |
| PA4687 (hitA) | No compound increased the Tm by more than 1.5 degrees. | Iron(III) citrate, FeCl3, Fe(SO4)3 | |||
| PA4913 | L-Ala L-Pro L-Ser L-Thr L-Val D-Ala Gly α-aminobutyrate | 12.7 7.6 5.3 3.2 4.1 6.8 4.5 8.1 | Insufficient protein for ITC | Yes [15,41] Yes [15,41] Yes [15,41] Yes [15,41] Yes [15,41] Yes Yes [15,41] n. d. |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fernández, M.; Rico-Jiménez, M.; Ortega, Á.; Daddaoua, A.; García García, A.I.; Martín-Mora, D.; Mesa Torres, N.; Tajuelo, A.; Matilla, M.A.; Krell, T. Determination of Ligand Profiles for Pseudomonas aeruginosa Solute Binding Proteins. Int. J. Mol. Sci. 2019, 20, 5156. https://doi.org/10.3390/ijms20205156
Fernández M, Rico-Jiménez M, Ortega Á, Daddaoua A, García García AI, Martín-Mora D, Mesa Torres N, Tajuelo A, Matilla MA, Krell T. Determination of Ligand Profiles for Pseudomonas aeruginosa Solute Binding Proteins. International Journal of Molecular Sciences. 2019; 20(20):5156. https://doi.org/10.3390/ijms20205156
Chicago/Turabian StyleFernández, Matilde, Miriam Rico-Jiménez, Álvaro Ortega, Abdelali Daddaoua, Ana Isabel García García, David Martín-Mora, Noel Mesa Torres, Ana Tajuelo, Miguel A. Matilla, and Tino Krell. 2019. "Determination of Ligand Profiles for Pseudomonas aeruginosa Solute Binding Proteins" International Journal of Molecular Sciences 20, no. 20: 5156. https://doi.org/10.3390/ijms20205156
APA StyleFernández, M., Rico-Jiménez, M., Ortega, Á., Daddaoua, A., García García, A. I., Martín-Mora, D., Mesa Torres, N., Tajuelo, A., Matilla, M. A., & Krell, T. (2019). Determination of Ligand Profiles for Pseudomonas aeruginosa Solute Binding Proteins. International Journal of Molecular Sciences, 20(20), 5156. https://doi.org/10.3390/ijms20205156







