Pyridine-Containing Macrocycles Display MMP-2/9 Inhibitory Activity and Distinct Effects on Migration and Invasion of 2D and 3D Breast Cancer Models
Abstract
:1. Introduction
2. Results
2.1. Viable Spheroids were Obtained from MDA-MB-231 Cells
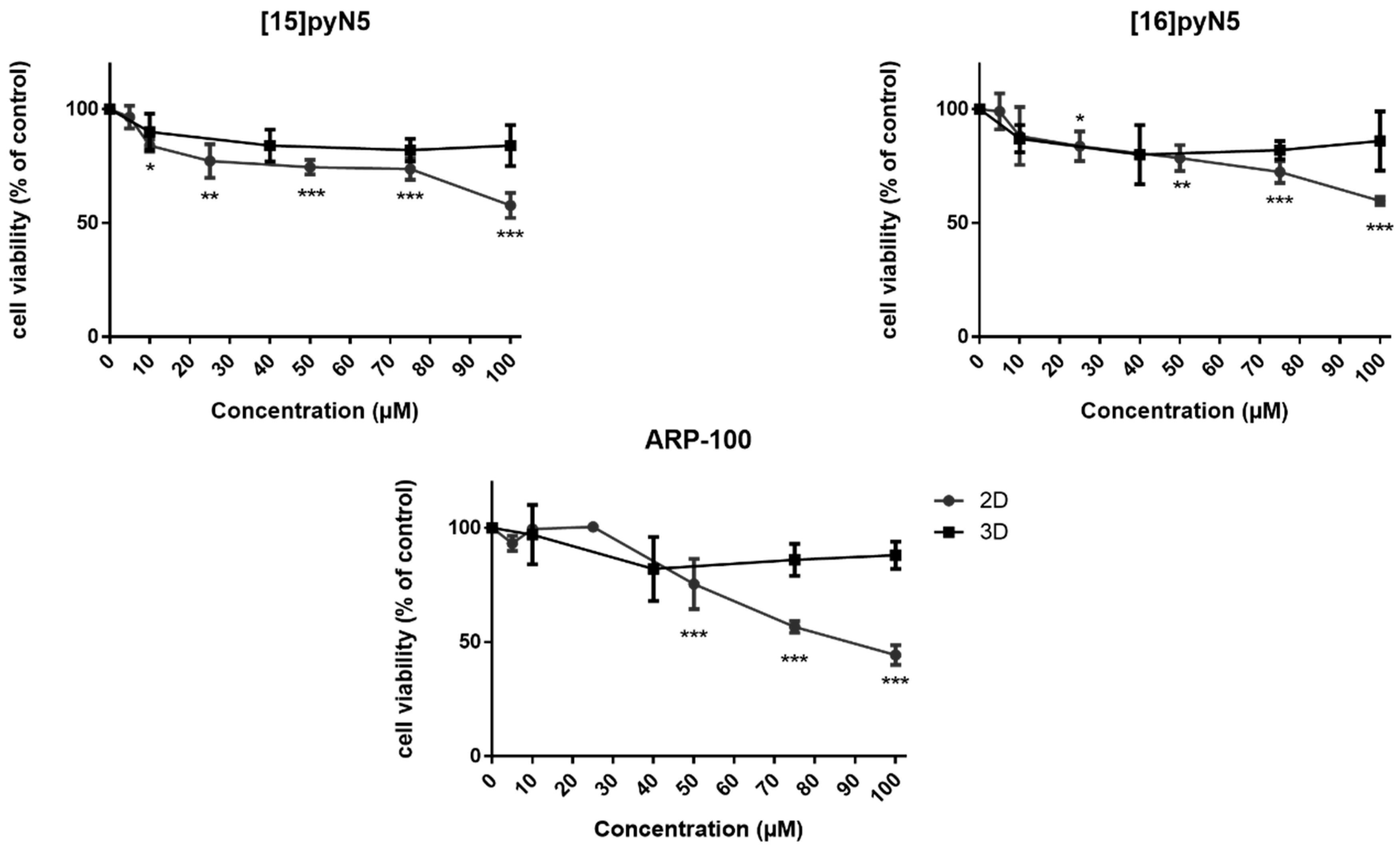
2.2. [15]pyN5, [16]pyN5 and ARP-100 Present a Differential Cytotoxic Profile in 2D and 3D Cultures of MDA-MB-231
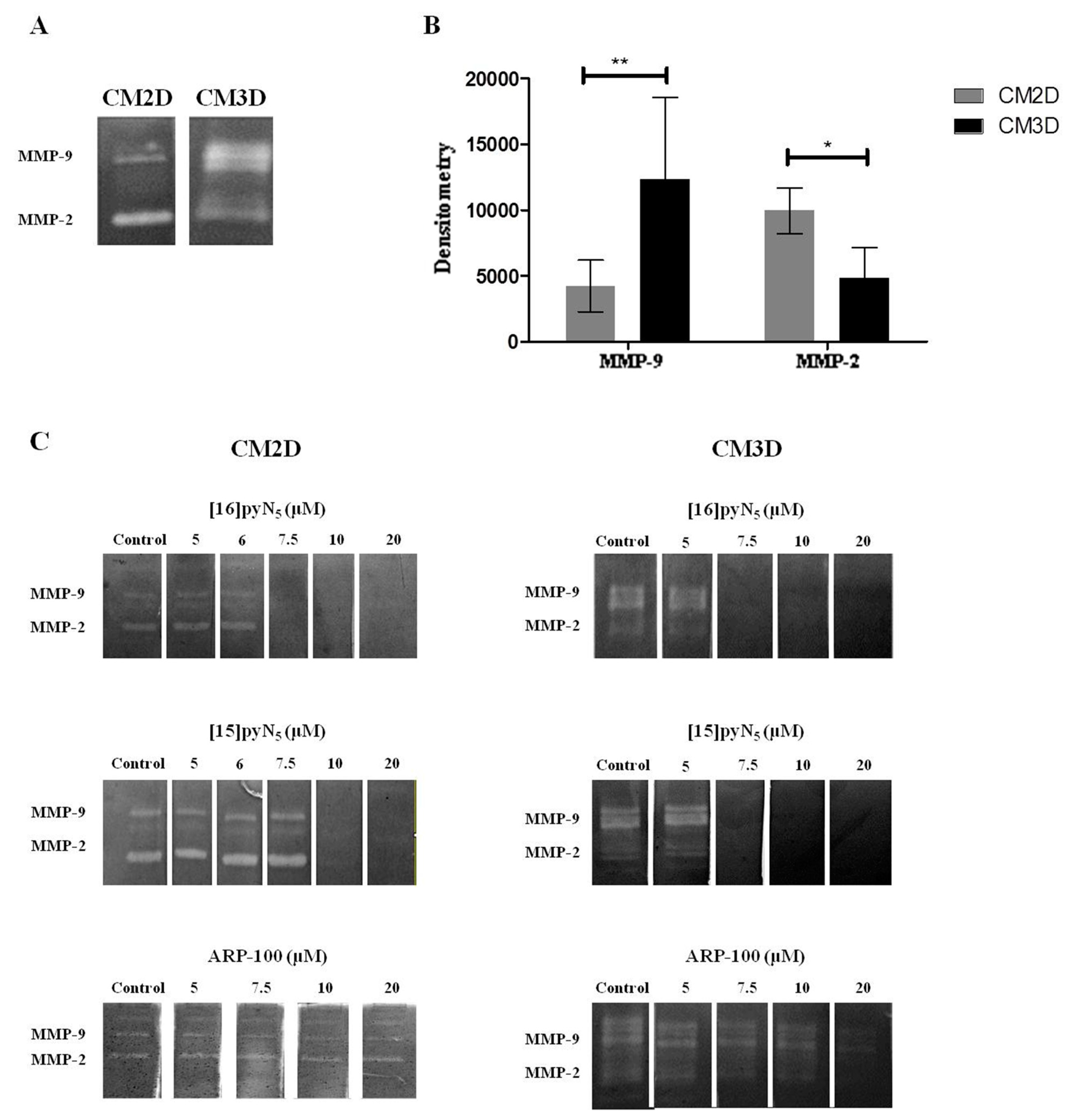
2.3. MDA-MB-231 Reveal a Culture-Dependent MMP-2/9 Secretion Profile
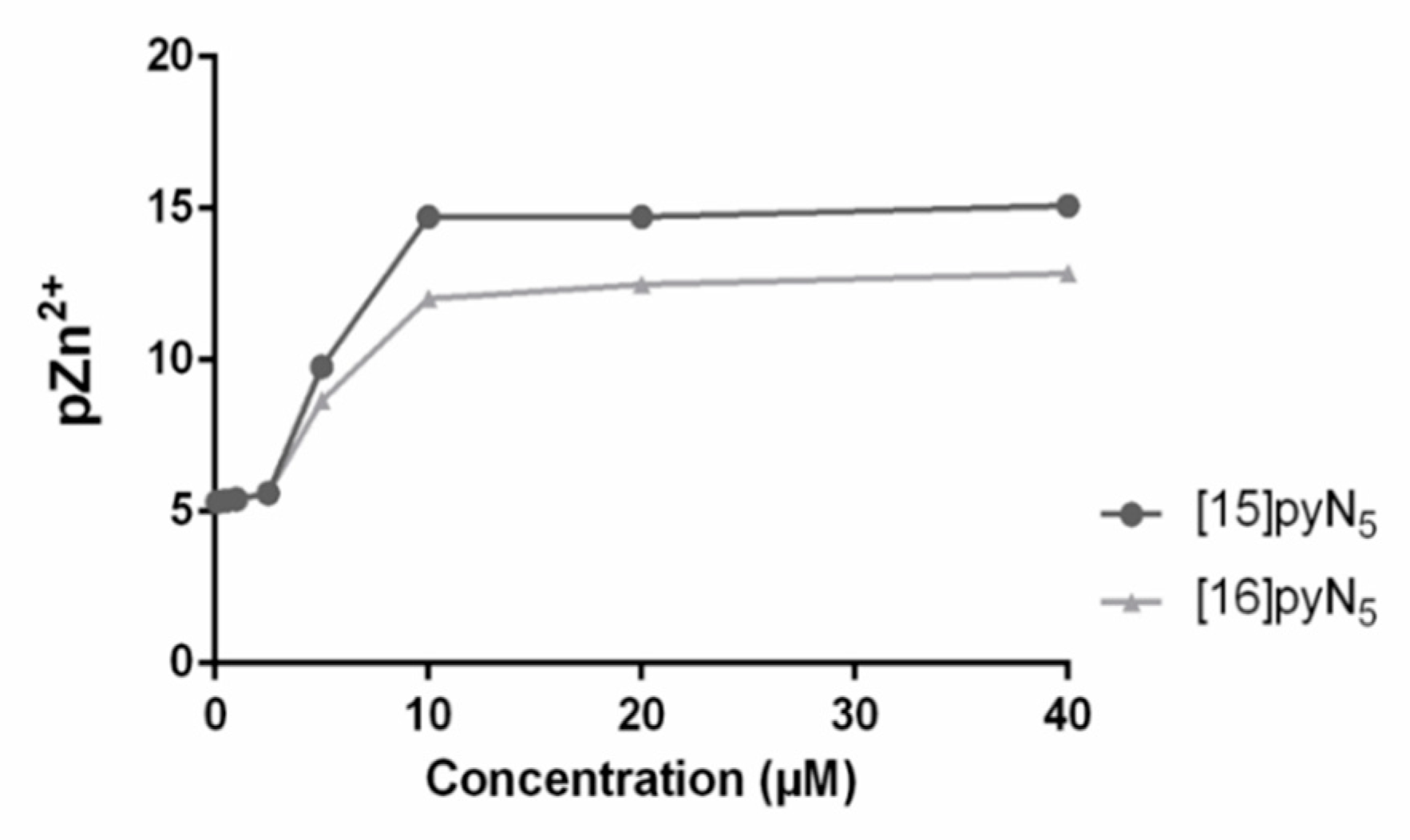
2.4. MMP Gelatinase Activity is Inhibited by the Macrocycles [15]pyN5 and [16]pyN5
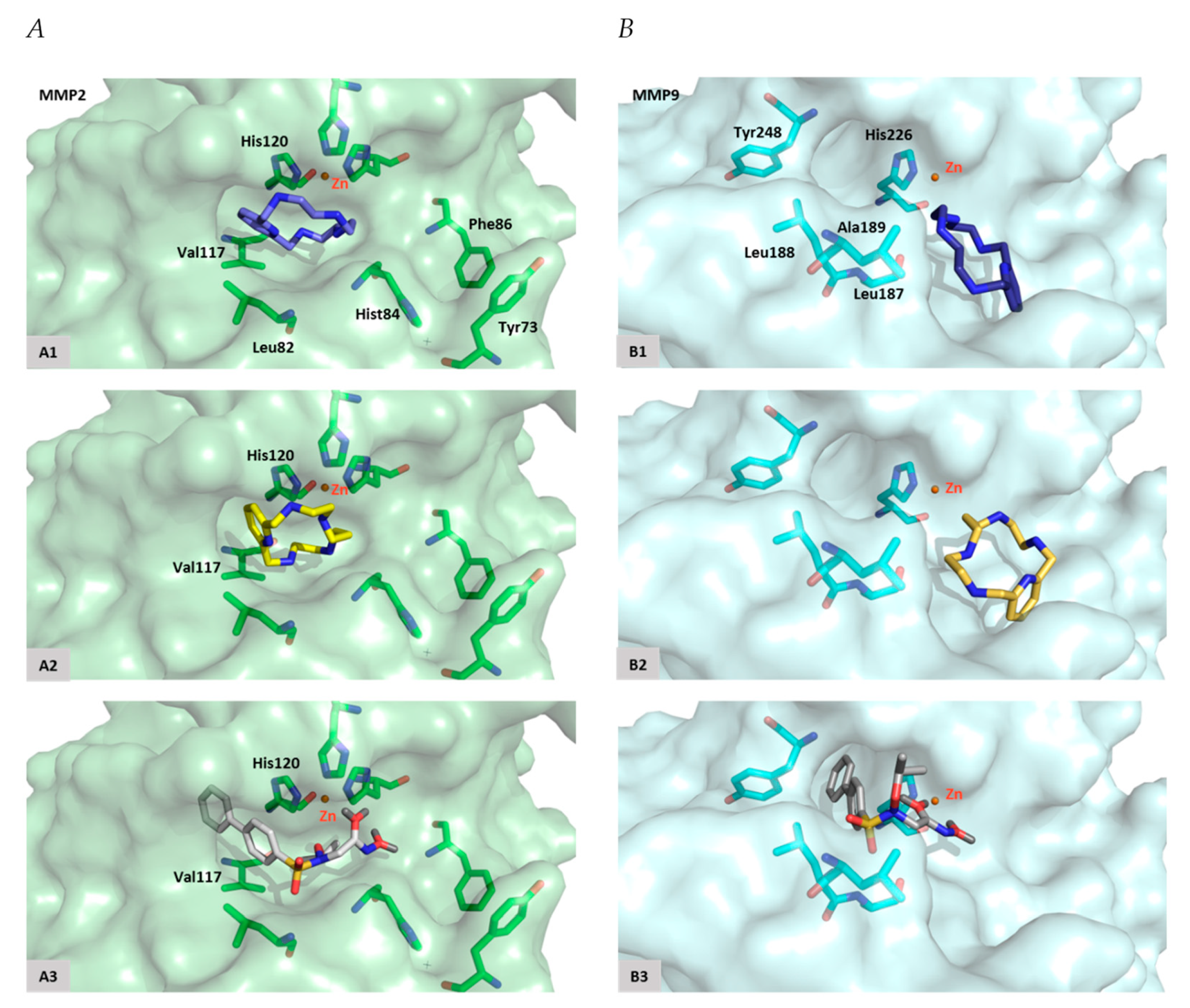
2.5. Macrocycles [15]pyN5 and [16]pyN5 Interact with the Catalytic Zinc and its Coordinating Amino Acids
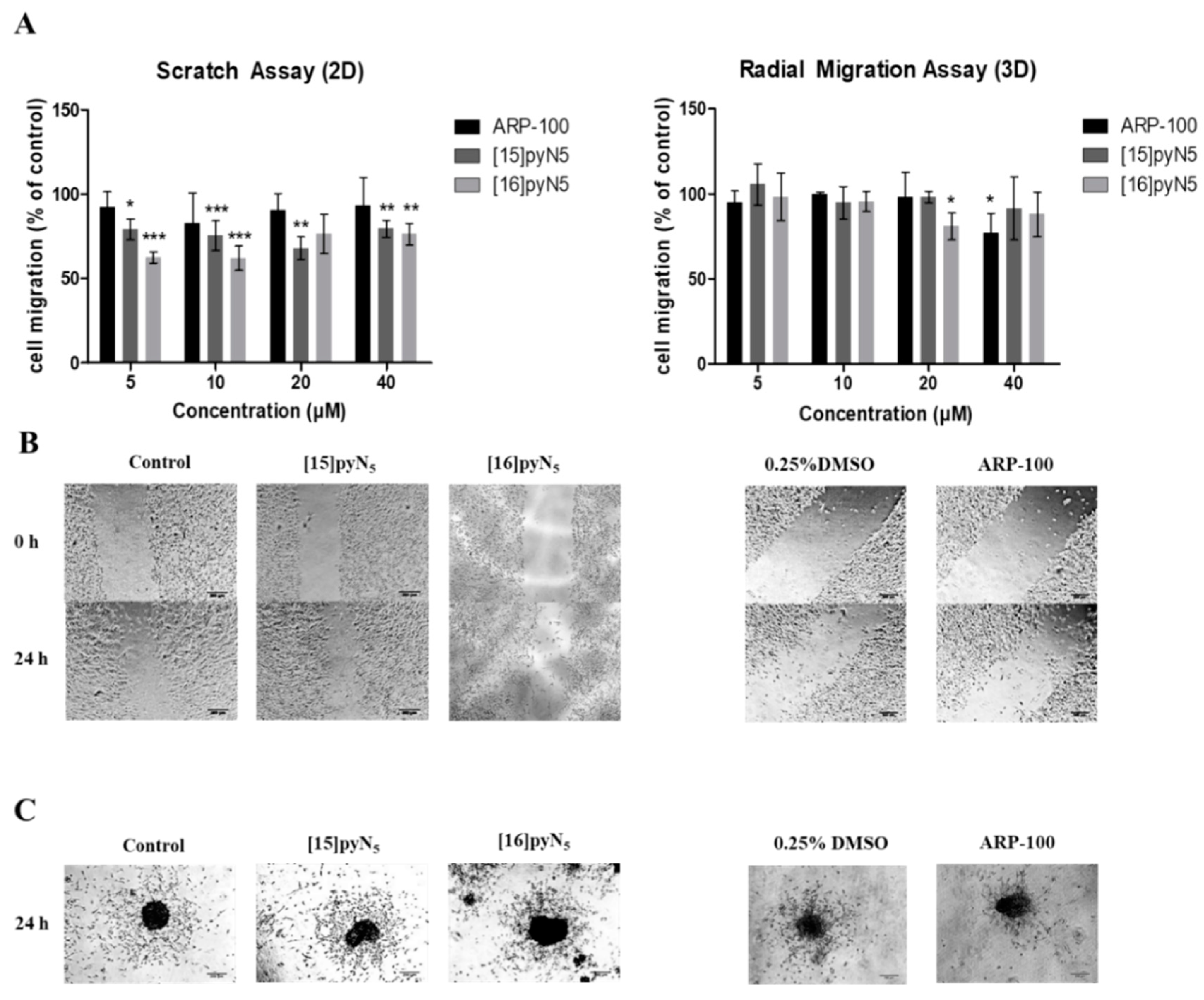
2.6. Cell migration is Significantly Impaired by [15]pyN5 and [16]pyN5
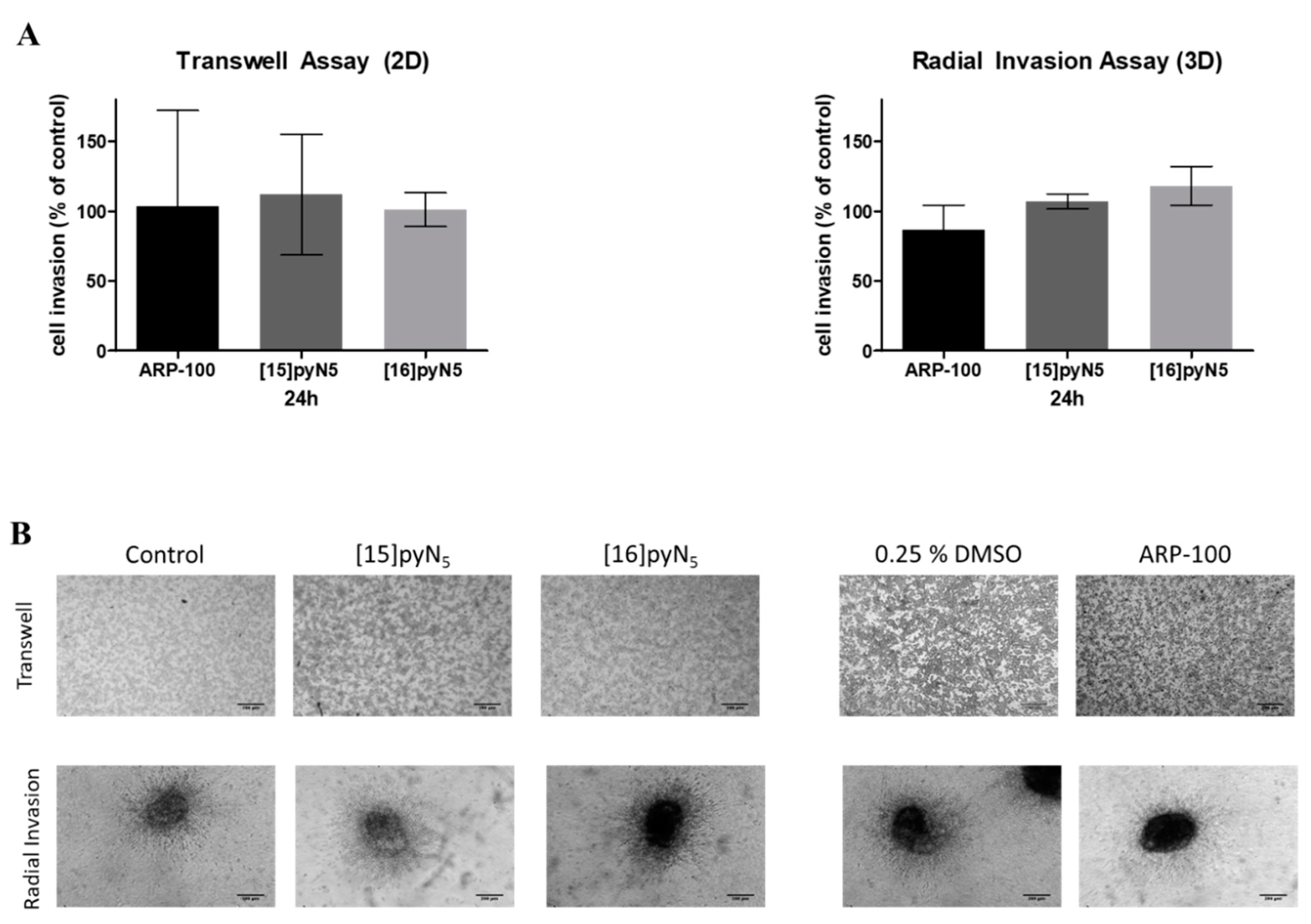
2.7. Breast Cancer Cell Invasion Ability is Maintained upon Exposure to [15]pyN5 and [16]pyN5
3. Discussion and Conclusions
4. Methods
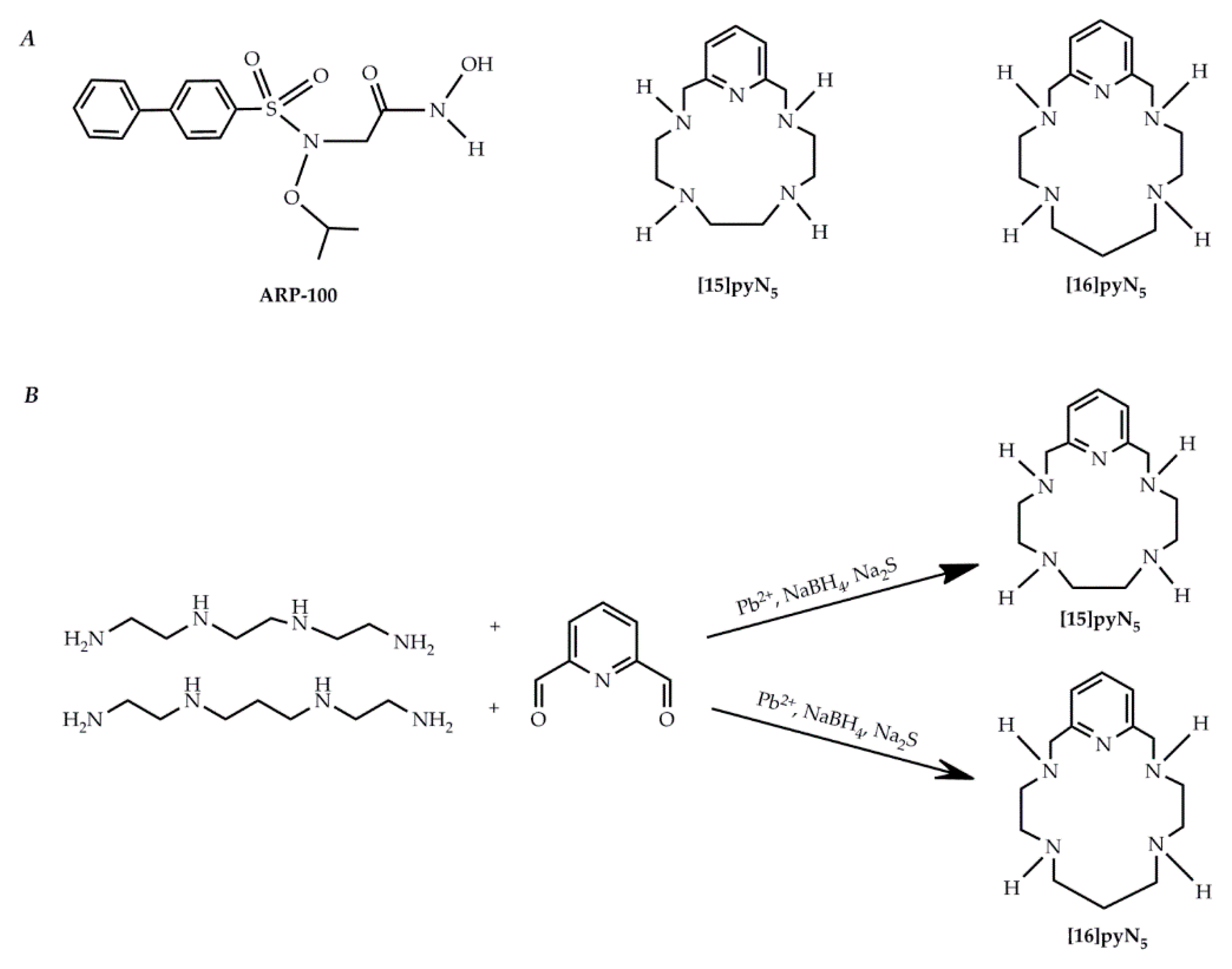
4.1. Chemicals
4.2. Cell Culture
4.3. Histology
4.3.1. Haematoxylin and Eosin (H&E) Staining
4.3.2. Ki-67 Staining
4.4. Cell Viability Assays
4.5. Production of Conditioned Media
4.6. Docking Studies
4.6.1. Protein and Ligand Structures
4.6.2. MMP-2 and 9 Binding Sites Definition
4.7. Modified Zymography Assay
4.8. Migration Assays
4.8.1. Scratch Assay
4.8.2. Radial Migration
4.9. Invasion Assays
4.9.1. Chemotaxis/Chemoinvasion Assay
4.9.2. Radial Invasion
4.10. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| 3D | three-dimensional |
| [15]pyN5 | 3,6,9,12,18-pentaazabicyclo[1 2.3.1]octadeca-1(18),14,16-triene |
| [16]pyN5 | 3,6,10,13,19-pentaazabicyclo[13.3.1]nonadeca-1(19),15,17-triene |
| AHA | acetohydroxamic acid |
| ARP-100 | 2-[([1,1′-biphenyl]-4-ylsulfonyl)(1-methylethoxy)amino]-N-hydroxyacetamide |
| CM2D | Conditioned medium from 2D models |
| CM3D | Conditioned medium from 3D models |
| DMEM | Dulbecco modified eagle modified medium |
| DMSO | dimethylsulfoxide |
| EDTA | ethylenediaminetetraacetic acid |
| ECM | extracellular matrix |
| EMT | epithelial to mesenchymal transition |
| FBS | fetal bovine serum |
| H&E | hematoxylin and Eosin |
| MMPi | MMP inhibitor(s) |
| MMPs | matrix metalloproteinases |
| MSS | musculoskeletal syndrome |
| MTS | tetrazolium salt 3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4- sulfophenyl)-2H-tetrazolium |
| PAGE | polyacrylamide gel electrophoresis |
| Py-macrocyles | pyridine containing macrocycles |
| SDS | sodium dodecyl sulfate |
| TIMPs | tissue inhibitors of matrix metalloproteinase |
| ULA | ultra low attachment |
References
- Egeblad, M.; Werb, Z. New functions for the matrix metalloproteinases in cancer progression. Nat. Rev. Cancer 2002, 2, 161–174. [Google Scholar] [CrossRef] [PubMed]
- Roy, R.; Yang, J.; Moses, M.A. Matrix Metalloproteinases As Novel Biomarker s and Potential Therapeutic Targets in Human Cancer. J. Clin. Oncol. 2009, 27, 5287–5297. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, H.; Cao, D.-C.; Liu, Y.; Hou, Y.-F.; Wu, J.; Lu, J.-S.; Di, G.-H.; Liu, G.; Li, F.-M.; Ou, Z.-L.; et al. Prognostic value of matrix metalloproteinases (MMP-2 and MMP-9) in patients with lymph node-negative breast carcinoma. Breast Cancer Res. Treat. 2004, 88, 75–85. [Google Scholar] [CrossRef] [PubMed]
- Leppä, S.; Saarto, T.; Vehmanen, L.; Blomqvist, C.; Elomaa, I. A high serum matrix metalloproteinase-2 level is associated with an adverse prognosis in node-positive breast carcinoma. Clin. Cancer Res. 2004, 10, 1057–1063. [Google Scholar] [CrossRef]
- Tauro, M.; Lynch, C. Cutting to the Chase: How Matrix Metalloproteinase-2 Activity Controls Breast-Cancer-to-Bone Metastasis. Cancers 2018, 10, 185. [Google Scholar] [CrossRef]
- Bode, W.; Fernandez-Catalan, C.; Grams, H.; Tschesche, F.; Maskos, H.; Nagase, K. Structural properties of matrix metalloproteinases. Cell. Mol. Life Sci. 1999, 55, 639–652. [Google Scholar] [CrossRef]
- Köhrmann, A.; Kammerer, U.; Kapp, M.; Dietl, J.; Anacker, J. Expression of matrix metalloproteinases (MMPs) in primary human breast cancer and breast cancer cell lines: New findings and review of the literature. BMC Cancer 2009, 9, 188. [Google Scholar] [CrossRef]
- Davies, K.J. The Complex Interaction of Matrix Metalloproteinases in the Migration of Cancer Cells through Breast Tissue Stroma. Int. J. Breast Cancer 2014, 2014, 839094. [Google Scholar] [CrossRef]
- Bauvois, B. New facets of matrix metalloproteinases MMP-2 and MMP-9 as cell surface transducers: Outside-in signaling and relationship to tumor progression. Biochim. Biophys. Acta Rev. Cancer 2012, 1825, 29–36. [Google Scholar] [CrossRef]
- Bai, X.-Y.; Li, S.; Wang, M.; Li, X.; Yang, Y.; Xu, Z.; Li, B.; Li, Y.; Xia, K.; Chen, H.; et al. Krüppel-like factor 9 down-regulates matrix metalloproteinase 9 transcription and suppresses human breast cancer invasion. Cancer Lett. 2018, 412, 224–235. [Google Scholar] [CrossRef]
- Radisky, E.S.; Radisky, D.C. Matrix metalloproteinases as breast cancer drivers and therapeutic targets. Front. Biosci. (Landmark Ed.) 2015, 20, 1144–1163. [Google Scholar] [CrossRef] [PubMed]
- Macaulay, V.M.; O’Byrne, K.J.; Saunders, M.P.; Braybrooke, J.P.; Long, L.; Gleeson, F.; Mason, C.S.; Harris, A.L.; Brown, P.; Talbot, D.C. Phase I study of intrapleural batimastat (BB-94), a matrix metalloproteinase inhibitor, in the treatment of malignant pleural effusions. Clin. Cancer Res. 1999, 5, 513–520. [Google Scholar] [PubMed]
- Renkiewicz, R.; Qiu, L.; Lesch, C.; Sun, X.; Devalaraja, R.; Cody, T.; Kaldjian, E.; Welgus, H.; Baragi, V. Broad-spectrum matrix metalloproteinase inhibitor marimastat-induced musculoskeletal side effects in rats. Arthritis Rheum. 2003, 48, 1742–1749. [Google Scholar] [CrossRef] [PubMed]
- Fingleton, B. MMPs as therapeutic targets—Still a viable option? Semin. Cell Dev. Biol. 2008, 19, 61–68. [Google Scholar] [CrossRef] [PubMed]
- Jacobsen, J.A.; Major Jourden, J.L.; Miller, M.T.; Cohen, S.M. To bind zinc or not to bind zinc: An examination of innovative approaches to improved metalloproteinase inhibition. Biochim. Biophys. Acta - Mol. Cell Res. 2010, 1803, 72–94. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ndinguri, M.; Bhowmick, M.; Tokmina-Roszyk, D.; Robichaud, T.; Fields, G. Peptide-Based Selective Inhibitors of Matrix Metalloproteinase-Mediated Activities. Molecules 2012, 17, 14230–14248. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tuccinardi, T.; Martinelli, A.; Nuti, E.; Carelli, P.; Balzano, F.; Uccello-Barretta, G.; Murphy, G.; Rossello, A. Amber force field implementation, molecular modelling study, synthesis and MMP-1/MMP-2 inhibition profile of (R)- and (S)-N-hydroxy-2-(N-isopropoxybiphenyl-4-ylsulfonamido)-3-methylbutanamides. Bioorg. Med. Chem. 2006, 14, 4260–4276. [Google Scholar] [CrossRef]
- Jacobsen, F.E.; Lewis, J.A.; Cohen, S.M. A New Role for Old Ligands: Discerning Chelators for Zinc Metalloproteinases. J. Am. Chem. Soc. 2006, 128, 3156–3157. [Google Scholar] [CrossRef]
- Hancock, R.D. Chelate ring size and metal ion selection. The basis of selectivity for metal ions in open-chain ligands and macrocycles. J. Chem. Educ. 1992, 69, 615. [Google Scholar] [CrossRef]
- Fernandes, A.S.; Cabral, M.F.; Costa, J.; Castro, M.; Delgado, R.; Drew, M.G.B.; Félix, V. Two macrocyclic pentaaza compounds containing pyridine evaluated as novel chelating agents in copper(II) and nickel(II) overload. J. Inorg. Biochem. 2011, 105, 410–419. [Google Scholar] [CrossRef]
- Gonçalves, S.; Fernandes, A.S.; Oliveira, N.G.; Marques, J.; Costa, J.; Fátima Cabral, M.; Miranda, J.; Cipriano, M.; Guerreiro, P.S.; Castro, M. Cytotoxic effects of cadmium in mammary epithelial cells: Protective role of the macrocycle [15]pyN5. Food Chem. Toxicol. 2012, 50, 2180–2187. [Google Scholar] [CrossRef] [PubMed]
- Fernandes, A.S.; Flórido, A.; Saraiva, N.; Cerqueira, S.; Ramalhete, S.; Cipriano, M.; Cabral, M.F.; Miranda, J.P.; Castro, M.; Costa, J.; et al. Role of the Copper(II) Complex Cu[15]pyN 5 in Intracellular ROS and Breast Cancer Cell Motility and Invasion. Chem. Biol. Drug Des. 2015, 86, 578–588. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.B.; Stein, R.; O’Hare, M.J. Three-dimensional in vitro tissue culture models of breast cancer—A review. Breast Cancer Res. Treat. 2004, 85, 281–291. [Google Scholar] [CrossRef]
- dit Faute, M.A.; Laurent, L.; Ploton, D.; Poupon, M.; Jardillier, J.-C.; Bobichon, H. Distinctive alterations of invasiveness, drug resistance and cell-cell organization in 3D-cultures of MCF-7, a human breast cancer cell line, and its multidrug resistant variant. Clin. Exp. Metastasis 2002, 19, 161–167. [Google Scholar] [CrossRef]
- Mellor, H.R.; Ferguson, D.J.P.; Callaghan, R. A model of quiescent tumor microregions for evaluating multicellular resistance to chemotherapeutic drugs. Br. J. Cancer 2005, 93, 302–309. [Google Scholar] [CrossRef]
- Vinci, M.; Gowan, S.; Boxall, F.; Patterson, L.; Zimmermann, M.; Court, W.; Lomas, C.; Mendiola, M.; Hardisson, D.; Eccles, S. a Advances in establishment and analysis of three-dimensional tumor spheroid-based functional assays for target validation and drug evaluation. BMC Biol. 2012, 10, 29. [Google Scholar] [CrossRef]
- Francia, G.; Man, S.; Teicher, B.; Grasso, L.; Kerbel, R.S. Gene Expression Analysis of Tumor Spheroids Reveals a Role for Suppressed DNA Mismatch Repair in Multicellular Resistance to Alkylating Agents. Mol. Cell. Biol. 2004, 24, 6837–6849. [Google Scholar] [CrossRef] [Green Version]
- Cipriano, M.; Freyer, N.; Knöspel, F.; Oliveira, N.G.; Barcia, R.; Cruz, P.E.; Cruz, H.; Castro, M.; Santos, J.M.; Zeilinger, K.; et al. Self-assembled 3D spheroids and hollow-fibre bioreactors improve MSC-derived hepatocyte-like cell maturation in vitro. Arch. Toxicol. 2017, 91, 1815–1832. [Google Scholar] [CrossRef]
- Pinheiro, P.F.; Pereira, S.A.; Harjivan, S.G.; Martins, I.L.; Marinho, A.T.; Cipriano, M.; Jacob, C.C.; Oliveira, N.G.; Castro, M.F.; Marques, M.M.; et al. Hepatocyte spheroids as a competent in vitro system for drug biotransformation studies: Nevirapine as a bioactivation case study. Arch. Toxicol. 2017, 91, 1199–1211. [Google Scholar] [CrossRef]
- Li, Q.; Chen, C.; Kapadia, A.; Zhou, Q.; Harper, M.K.; Schaack, J.; Labarbera, D. V 3D Models of Epithelial-Mesenchymal Transition in Breast Cancer Metastasis. J. Biomol. Screen. 2011, 16, 141–154. [Google Scholar] [CrossRef]
- Vidi, P.-A.; Bissell, M.J.; Lelièvre, S.A. Three-Dimensional Culture of Human Breast Epithelial Cells: The How and the Why. In Epithelial Cell Culture Protocols; Randell, S., Fulcher, M., Eds.; Humana Press: New Jersey, NY, USA, 2012; Volume 945, pp. 193–219. [Google Scholar] [Green Version]
- Imamura, Y.; Mukohara, T.; Shimono, Y.; Funakoshi, Y.; Chayahara, N.; Toyoda, M.; Kiyota, N.; Takao, S.; Kono, S.; Nakatsura, T.; et al. Comparison of 2D- and 3D-culture models as drug-testing platforms in breast cancer. Oncol. Rep. 2015, 33, 1837–1843. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, B.J.; Zhao, S.; Bunaciu, R.P.; Yen, A.; Wu, M. A 3D in situ cell counter reveals that breast tumor cell (MDA-MB-231) proliferation rate is reduced by the collagen matrix density. Biotechnol. Prog. 2015, 31, 990–996. [Google Scholar] [CrossRef] [PubMed]
- Chetty, C.; Lakka, S.S.; Bhoopathi, P.; Rao, J.S. MMP-2 alters VEGF expression via αVβ3 integrin-mediated PI3K/AKT signaling in A549 lung cancer cells. Int. J. Cancer 2009, 127, 1081–1095. [Google Scholar] [CrossRef] [PubMed]
- Cai, J.; Tang, H.; Xu, L.; Wang, X.; Yang, C.; Ruan, S.; Guo, J.; Hu, S.; Wang, Z. Fibroblasts in omentum activated by tumor cells promote ovarian cancer growth, adhesion and invasiveness. Carcinogenesis 2012, 33, 20–29. [Google Scholar] [CrossRef]
- Toth, M.; Sohail, A.; Fridman, R. Assessment of Gelatinases (MMP-2 and MMP-9) by Gelatin Zymography. In Metastasis Research Protocols; Dwek, M., Brooks, S., Schumacher, U., Eds.; Humana Press: New Jersey, NY, USA, 2012; Volume 878, pp. 121–135. [Google Scholar] [Green Version]
- Gu, Y.; Lee, H.; Golub, L.M.; Sorsa, T.; Konttinen, Y.T.; Simon, S.R. Inhibition of breast cancer cell extracellular matrix degradative activity by chemically modified tetracyclines. Ann. Med. 2005, 37, 450–460. [Google Scholar] [CrossRef]
- Alderighi, L.; Gans, P.; Ienco, A.; Peters, D.; Sabatini, A.; Vacca, A. Hyperquad simulation and speciation (HySS): A utility program for the investigation of equilibria involving soluble and partially soluble species. Coord. Chem. Rev. 1999, 184, 311–318. [Google Scholar] [CrossRef]
- Subik, K.; Lee, J.-F.; Baxter, L.; Strzepek, T.; Costello, D.; Crowley, P.; Xing, L.; Hung, M.; Bonfiglio, T.; Hicks, D.G.; et al. The Expression Patterns of ER, PR, HER2, CK5/6, EGFR, Ki-67 and AR by Immunohistochemical Analysis in Breast Cancer Cell Lines. Breast Cancer (Auckl). 2010, 4, 35–41. [Google Scholar] [CrossRef]
- Prat, A.; Parker, J.S.; Karginova, O.; Fan, C.; Livasy, C.; Herschkowitz, J.I.; He, X.; Perou, C.M. Phenotypic and molecular characterization of the claudin-low intrinsic subtype of breast cancer. Breast Cancer Res. 2010, 12, R68. [Google Scholar] [CrossRef]
- Ivascu, A.; Kubbies, M. Diversity of cell-mediated adhesions in breast cancer spheroids. Int. J. Oncol. 2007, 31, 1403–1413. [Google Scholar] [CrossRef] [Green Version]
- Sethi, T.; Rintoul, R.C.; Moore, S.M.; MacKinnon, A.C.; Salter, D.; Choo, C.; Chilvers, E.R.; Dransfield, I.; Donnelly, S.C.; Strieter, R.; et al. Extracellular matrix proteins protect small cell lung cancer cells against apoptosis: A mechanism for small cell lung cancer growth and drug resistance in vivo. Nat. Med. 1999, 5, 662–668. [Google Scholar] [CrossRef]
- Weigelt, B.; Lo, A.T.; Park, C.C.; Gray, J.W.; Bissell, M.J. HER2 signaling pathway activation and response of breast cancer cells to HER2-targeting agents is dependent strongly on the 3D microenvironment. Breast Cancer Res. Treat. 2010, 122, 35–43. [Google Scholar] [CrossRef] [PubMed]
- Doublier, S.; Belisario, D.C.; Polimeni, M.; Annaratone, L.; Riganti, C.; Allia, E.; Ghigo, D.; Bosia, A.; Sapino, A. HIF-1 activation induces doxorubicin resistance in MCF7 3-D spheroids via P-glycoprotein expression: A potential model of the chemo-resistance of invasive micropapillary carcinoma of the breast. BMC Cancer 2012, 12, 4. [Google Scholar] [CrossRef] [PubMed]
- Patel, N.R.; Aryasomayajula, B.; Abouzeid, A.H.; Torchilin, V.P. Cancer cell spheroids for screening of chemotherapeutics and drug-delivery systems. Ther. Deliv. 2015, 6, 509–520. [Google Scholar] [CrossRef] [PubMed]
- Minchinton, A.I.; Tannock, I.F. Drug penetration in solid tumors. Nat. Rev. Cancer 2006, 6, 583–592. [Google Scholar] [CrossRef]
- Maity, G.; Choudhury, P.R.; Sen, T.; Ganguly, K.K.; Sil, H.; Chatterjee, A. Culture of human breast cancer cell line (MDA-MB-231) on fibronectin-coated surface induces pro-matrix metalloproteinase-9 expression and activity. Tumor Biol. 2011, 32, 129–138. [Google Scholar] [CrossRef]
- McCawley, L.J.; Matrisian, L.M. Matrix metalloproteinases: they’re not just for matrix anymore! Curr. Opin. Cell Biol. 2001, 13, 534–540. [Google Scholar] [CrossRef]
- Ardi, V.C.; Van den Steen, P.E.; Opdenakker, G.; Schweighofer, B.; Deryugina, E.I.; Quigley, J.P. Neutrophil MMP-9 Proenzyme, Unencumbered by TIMP-1, Undergoes Efficient Activation in Vivo and Catalytically Induces Angiogenesis via a Basic Fibroblast Growth Factor (FGF-2)/FGFR-2 Pathway. J. Biol. Chem. 2009, 284, 25854–25866. [Google Scholar] [CrossRef] [Green Version]
- Kajita, M.; Itoh, Y.; Chiba, T.; Mori, H.; Okada, A.; Kinoh, H.; Seiki, M. Membrane-Type 1 Matrix Metalloproteinase Cleaves Cd44 and Promotes Cell Migration. J. Cell Biol. 2001, 153, 893–904. [Google Scholar] [CrossRef]
- Correia, A.L.; Mori, H.; Chen, E.I.; Schmitt, F.C.; Bissell, M.J. The hemopexin domain of MMP3 is responsible for mammary epithelial invasion and morphogenesis through extracellular interaction with HSP90. Genes Dev. 2013, 27, 805–817. [Google Scholar] [CrossRef]
- Basu, B.; Correa de Sampaio, P.; Mohammed, H.; Fogarasi, M.; Corrie, P.; Watkins, N.A.; Smethurst, P.A.; English, W.R.; Ouwehand, W.H.; Murphy, G. Inhibition of MT1-MMP activity using functional antibody fragments selected against its hemopexin domain. Int. J. Biochem. Cell Biol. 2012, 44, 393–403. [Google Scholar] [CrossRef]
- Zaremba-Czogalla, M.; Hryniewicz-Jankowska, A.; Tabola, R.; Nienartowicz, M.; Stach, K.; Wierzbicki, J.; Cirocchi, R.; Ziolkowski, P.; Tabaczar, S.; Augoff, K. A novel regulatory function of CDKN1A/p21 in TNFα-induced matrix metalloproteinase 9-dependent migration and invasion of triple-negative breast cancer cells. Cell Signal. 2018, 47, 27–36. [Google Scholar] [CrossRef] [PubMed]
- Dufour, A.; Sampson, N.S.; Zucker, S.; Cao, J. Role of the hemopexin domain of matrix metalloproteinases in cell migration. J. Cell. Physiol. 2008, 217, 643–651. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Millerot-Serrurot, E.; Guilbert, M.; Fourré, N.; Witkowski, W.; Said, G.; Van Gulick, L.; Terryn, C.; Zahm, J.-M.; Garnotel, R.; Jeannesson, P. 3D collagen type I matrix inhibits the antimigratory effect of doxorubicin. Cancer Cell Int. 2010, 10, 26. [Google Scholar] [CrossRef] [PubMed]
- Sodek, K.L.; Brown, T.J.; Ringuette, M.J. Collagen I but not Matrigel matrices provide an MMP-dependent barrier to ovarian cancer cell penetration. BMC Cancer 2008, 8, 223. [Google Scholar] [CrossRef] [PubMed]
- Menyhárt, O.; Harami-Papp, H.; Sukumar, S.; Schäfer, R.; Magnani, L.; de Barrios, O.; Győrffy, B. Guidelines for the selection of functional assays to evaluate the hallmarks of cancer. Biochim. Biophys. Acta Rev. Cancer 2016, 1866, 300–319. [Google Scholar] [CrossRef] [Green Version]
- Guerreiro, P.S.; Corvacho, E.; Costa, J.G.; Saraiva, N.; Fernandes, A.S.; Castro, M.; Miranda, J.P.; Oliveira, N.G. The APE1 redox inhibitor E3330 reduces collective cell migration of human breast cancer cells and decreases chemoinvasion and colony formation when combined with docetaxel. Chem. Biol. Drug Des. 2017, 90, 561–571. [Google Scholar] [CrossRef]
- Fisher, J.F.; Mobashery, S. Recent advances in MMP inhibitor design. Cancer Metastasis Rev. 2006, 25, 115–136. [Google Scholar] [CrossRef]
- Scatena, R. Prinomastat, a hydroxamate-based matrix metalloproteinase inhibitor. A novel pharmacological approach for tissue remodelling-related diseases. Expert Opin. Investig. Drugs 2000, 9, 2159–2165. [Google Scholar] [CrossRef]
- Levin, J.I.; Chen, J.; Du, M.; Hogan, M.; Kincaid, S.; Nelson, F.C.; Venkatesan, A.M.; Wehr, T.; Zask, A.; DiJoseph, J.; et al. The discovery of anthranilic acid-Based MMP inhibitors. Part 2: SAR of the 5-position and P11 groups. Bioorg. Med. Chem. Lett. 2001, 11, 2189–2192. [Google Scholar] [CrossRef]
- Santos, J.M.; Camões, S.P.; Filipe, E.; Cipriano, M.; Barcia, R.N.; Filipe, M.; Teixeira, M.; Simões, S.; Gaspar, M.; Mosqueira, D.; et al. Three-dimensional spheroid cell culture of umbilical cord tissue-derived mesenchymal stromal cells leads to enhanced paracrine induction of wound healing. Stem Cell Res. Ther. 2015, 6, 90. [Google Scholar] [CrossRef]
- Liang, C.-C.; Park, A.Y.; Guan, J.-L. In vitro scratch assay: A convenient and inexpensive method for analysis of cell migration in vitro. Nat. Protoc. 2007, 2, 329–333. [Google Scholar] [CrossRef] [PubMed]
- Flórido, A.; Saraiva, N.; Cerqueira, S.; Almeida, N.; Parsons, M.; Batinic-Haberle, I.; Miranda, J.P.; Costa, J.G.; Carrara, G.; Castro, M.; et al. The manganese(III) porphyrin MnTnHex-2-PyP5+ modulates intracellular ROS and breast cancer cell migration: Impact on doxorubicin-treated cells. Redox Biol. 2019, 20, 367–378. [Google Scholar] [CrossRef] [PubMed]
- Rajan, N.; Habermehl, J.; Coté, M.-F.; Doillon, C.J.; Mantovani, D. Preparation of ready-to-use, storable and reconstituted type I collagen from rat tail tendon for tissue engineering applications. Nat. Protoc. 2007, 1, 2753–2758. [Google Scholar] [CrossRef] [PubMed]

| [15]pyN5 | [16]pyN5 | ARP-100 | ||
|---|---|---|---|---|
| MMP-2 | Score | 53.1 | 53.3 | 93.2 |
| Zn2+ | 2.1 | 2.6 | 2.4 | |
| His120 | 3.6 | 3.8 | 3.5 | |
| Leu82 | 4.7 | 4.4 | 2.9 | |
| Val117 | 3.8 | 3.0 | 3.7 | |
| His84 | 3.7 | 3.3 | 3.4 | |
| Tyr73 | 9.0 | 10.0 | 8.9 | |
| Phe86 | 8.3 | 8.8 | 6.3 | |
| MMP-9 | Score | 34.8 | 42.0 | 90.0 |
| Zn2+ | 4.0 | 4.4 | 2.6 | |
| Leu187 | 3.1 | 4.0 | 3.9 | |
| Leu188 | 6.1 | 6.4 | 3.2 | |
| Ala189 | 3.0 | 4.0 | 3.1 | |
| His226 | 5.2 | 6.5 | 3.4 | |
| Try248 | 9.8 | 10.8 | 3.6 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Proença, S.; Antunes, B.; Guedes, R.C.; Ramilo-Gomes, F.; Cabral, M.F.; Costa, J.; Fernandes, A.S.; Castro, M.; Oliveira, N.G.; Miranda, J.P. Pyridine-Containing Macrocycles Display MMP-2/9 Inhibitory Activity and Distinct Effects on Migration and Invasion of 2D and 3D Breast Cancer Models. Int. J. Mol. Sci. 2019, 20, 5109. https://doi.org/10.3390/ijms20205109
Proença S, Antunes B, Guedes RC, Ramilo-Gomes F, Cabral MF, Costa J, Fernandes AS, Castro M, Oliveira NG, Miranda JP. Pyridine-Containing Macrocycles Display MMP-2/9 Inhibitory Activity and Distinct Effects on Migration and Invasion of 2D and 3D Breast Cancer Models. International Journal of Molecular Sciences. 2019; 20(20):5109. https://doi.org/10.3390/ijms20205109
Chicago/Turabian StyleProença, Susana, Bernardo Antunes, Rita C. Guedes, Filipa Ramilo-Gomes, M. Fátima Cabral, Judite Costa, Ana S. Fernandes, Matilde Castro, Nuno G. Oliveira, and Joana P. Miranda. 2019. "Pyridine-Containing Macrocycles Display MMP-2/9 Inhibitory Activity and Distinct Effects on Migration and Invasion of 2D and 3D Breast Cancer Models" International Journal of Molecular Sciences 20, no. 20: 5109. https://doi.org/10.3390/ijms20205109







