Identification of miRNAs Involved in Bacillus velezensis FZB42-Activated Induced Systemic Resistance in Maize
Abstract
:1. Introduction
2. Results
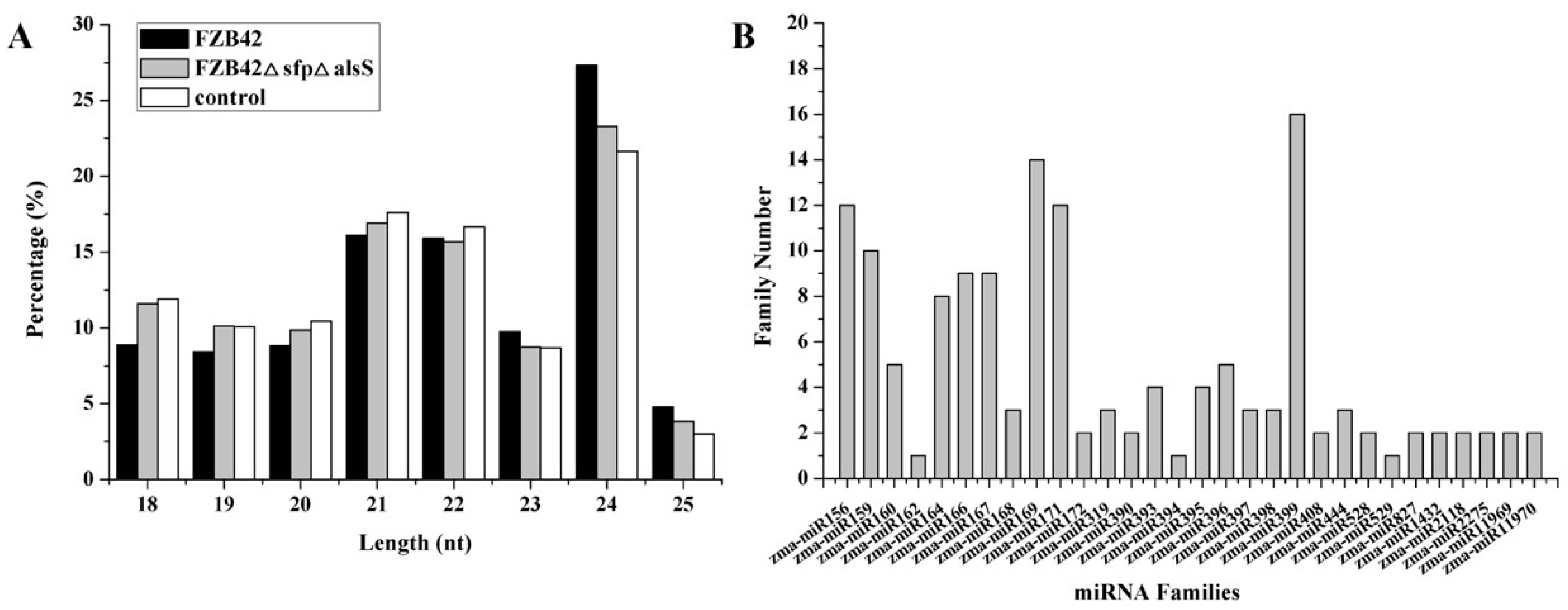
2.1. Global miRNA Profile Analysis
2.2. Identification of miRNAs in Maize
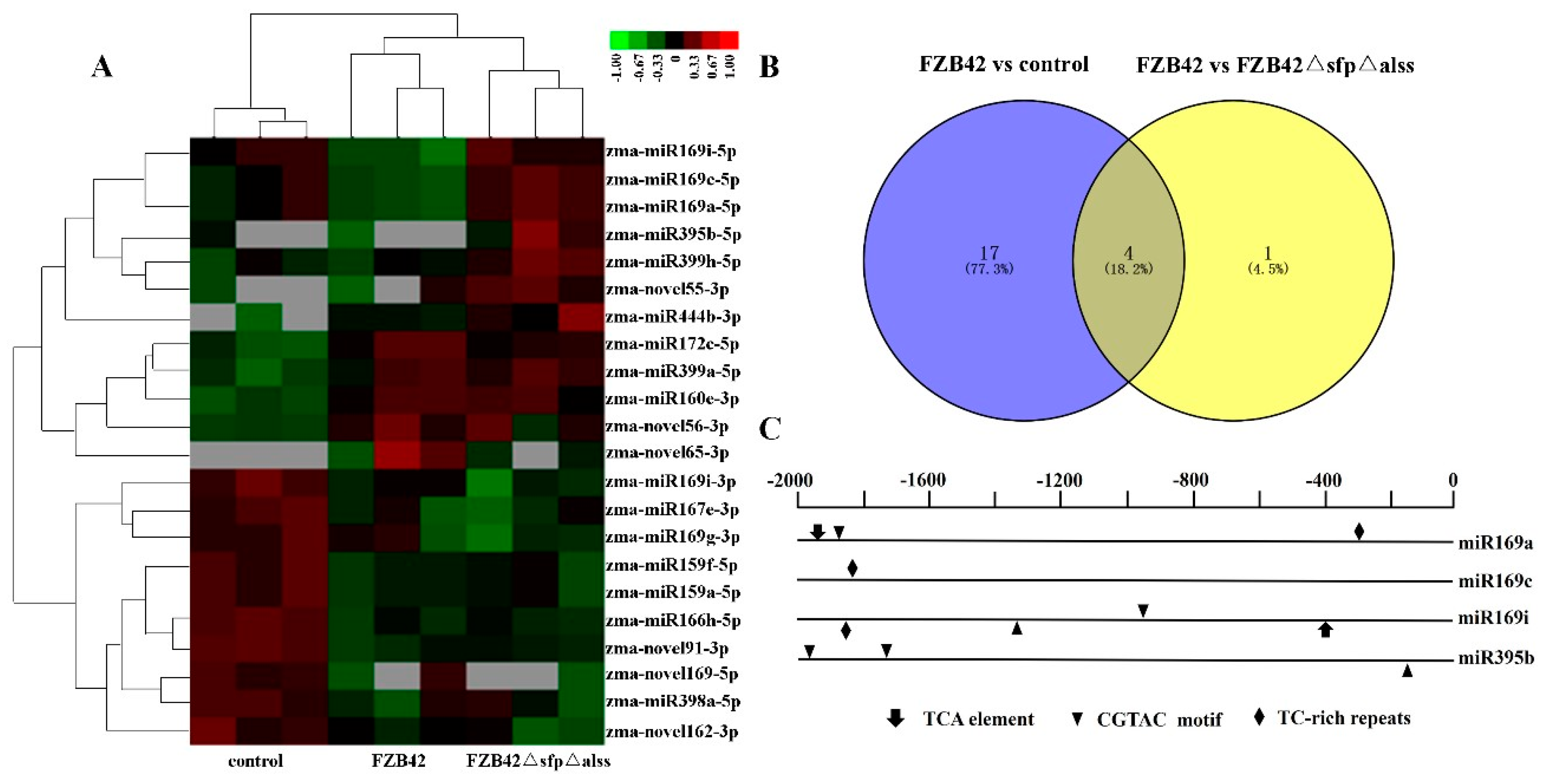
2.3. Differentially Expressed miRNAs Responsive to Bacillus
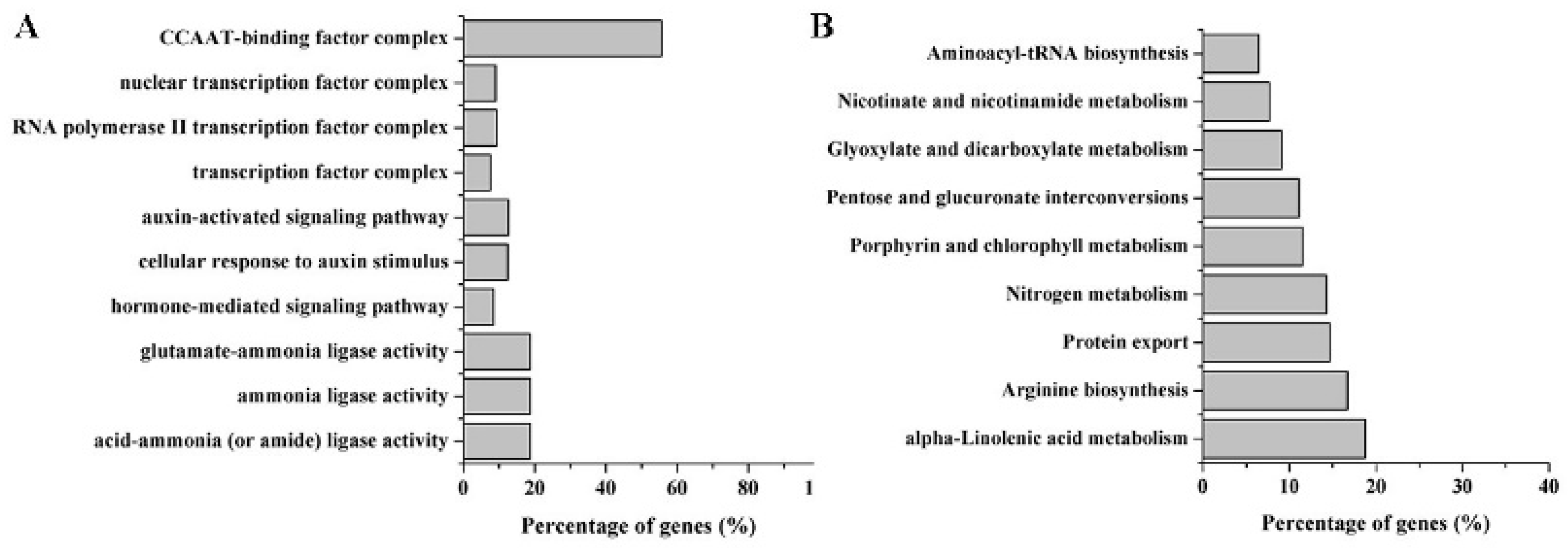
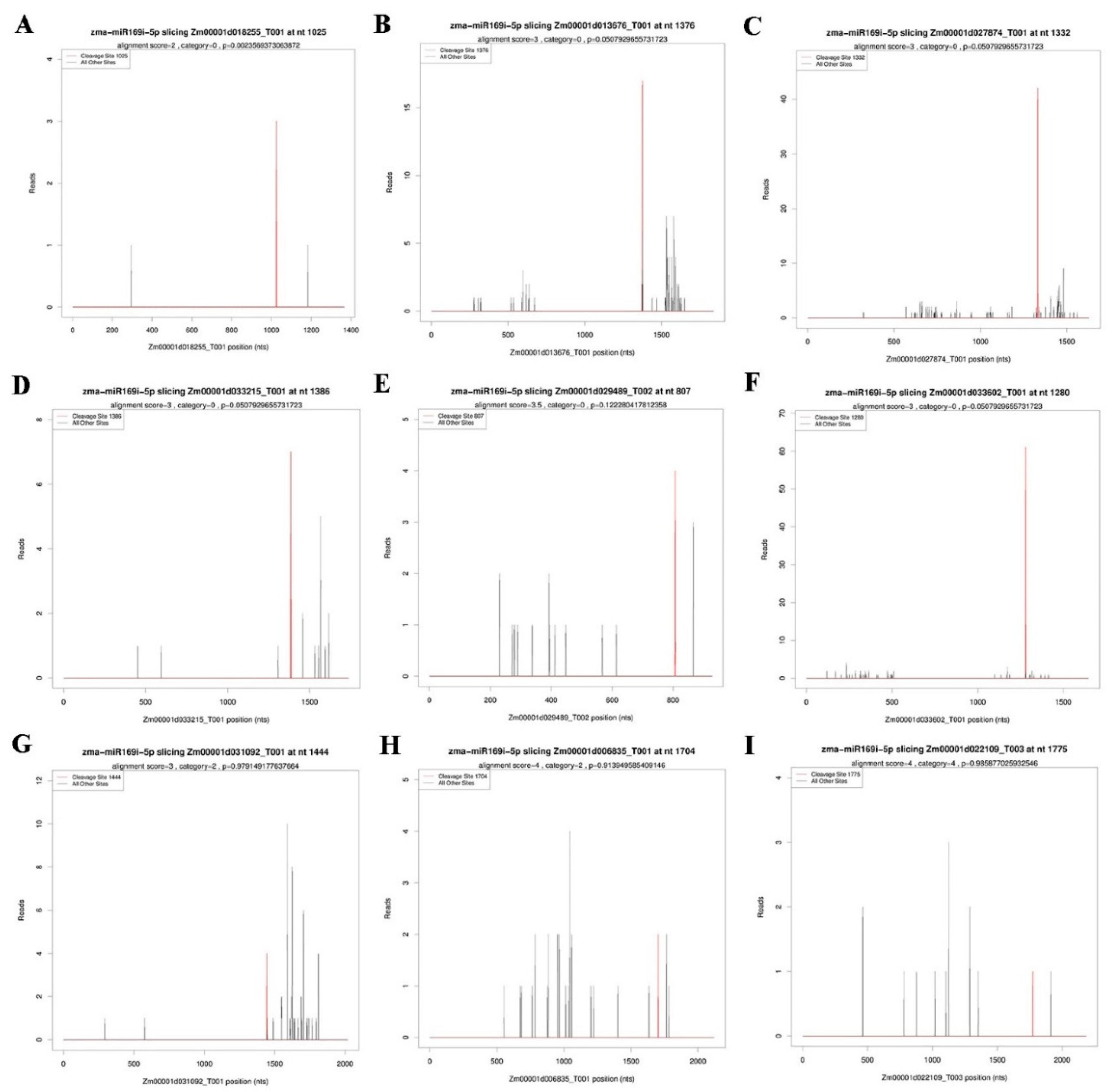
2.4. Target Prediction of Differentially Expressed miRNAs
3. Discussion
4. Methods
4.1. Microbial Strains, Plant and Growth Conditions
4.2. Construction and Analysis of Small RNA Libraries
4.3. Identification of Induced Systemic Resistance-Associated miRNAs
4.4. Analysis of miRNA Promoter
4.5. cDNA Library Construction for Degradome Sequencing
4.6. Prediction of miRNA Targets
4.7. qRT-PCR Analysis
4.8. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Jones-Rhoades, M.W.; Bartel, D.P.; Bartel, B. MicroRNAs and their regulatory roles in plants. Annu. Rev. Plant Biol. 2006, 57, 19–53. [Google Scholar] [CrossRef] [PubMed]
- Rogers, K.; Chen, X. Biogenesis, turnover, and mode of action of plant microRNAs. Plant Cell 2013, 25, 2383–2399. [Google Scholar] [CrossRef] [PubMed]
- Chen, X. A microRNA as a translational repressor of APETALA2 in Arabidopsis flower development. Science 2004, 303, 2022–2025. [Google Scholar] [CrossRef] [PubMed]
- Chen, X. Small RNAs and their roles in plant development. Annu. Rev. Cell Dev. Biol. 2009, 25, 21–44. [Google Scholar] [CrossRef] [PubMed]
- Navarro, L.; Dunoyer, P.; Jay, F.; Arnold, B.; Dharmasiri, N.; Estelle, M.; Voinnet, O.; Jones, J.D.G. A plant miRNA contributes to antibacterial resistance by repressing auxin signaling. Science 2006, 312, 436–439. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Zhu, S.; Liu, F.; Wang, W.; Wang, X.; Han, G.; Cheng, B. Identification of arbuscular mycorrhiza fungi responsive microRNAs and their regulatory network in maize. Int. J. Mol. Sci. 2018, 19, 3201. [Google Scholar] [CrossRef] [PubMed]
- Bárbara, N.F.; Luis, P.Í.; Oswaldo, V.L.; Xochitl, A.A.; Alfonso, L.; Fuentes, S.I.; Mario, R.; Sujay, P.; Reyes, J.L.; Lourdes, G. The micro-RNA72c-APETALA2-1 node as a key regulator of the common bean-Rhizobium etli nitrogen fixation symbiosis. Plant Physiol. 2015, 168, 273–291. [Google Scholar]
- Daniela, T.; Zhe, Y.; Dennis, B.H.; Nikolaj, B.A.; Dugald, E.R.; Lene, H.M.; Hemal, B.; Moritz, S.; Jens, S.; Katharina, M. Systemic control of legime susceptibility to rhizobial infection by a mobile microRNA. Science 2018, 362, 233–236. [Google Scholar]
- Lauressergues, D.; Delaux, P.-M.; Formey, D.; Lelandais Brière, C.; Fort, S.; Cottaz, S.; Bécard, G.; Niebel, A.; Roux, C.; Combier, J.-P. The microRNA miR171h modulates arbuscular mycorrhizal colonization of Medicago truncatula by targeting NSP2. Plant J. 2012, 72, 512–522. [Google Scholar] [CrossRef]
- Bazin, J.; Khan, G.A.; Combier, J.-P.; Bustos Sanmamed, P.; Debernardi, J.M.; Rodriguez, R.; Sorin, C.; Palatnik, J.; Hartmann, C.; Crespi, M. miR396 affects mycorrhization and root meristem activity in the legume Medicago truncatula. Plant J. 2013, 74, 920–934. [Google Scholar] [CrossRef]
- Lugtenberg, B.; Kamilova, F. Plant-growth-promoting rhizobacteria. Annu. Rev. Microbiol. 2009, 63, 541–556. [Google Scholar] [CrossRef] [PubMed]
- Rudrappa, T.; Czymmek, K.J.; Paré, P.W.; Bais, H.P. Root-secreted malic acid recruits beneficial soil bacteria. Plant Physiol. 2008, 148, 1547–1556. [Google Scholar] [CrossRef] [PubMed]
- Idris, E.E.; Iglesias, D.J.; Talon, M.; Borriss, R. Tryptophan-dependent production of indole-3-acetic acid (IAA) affects level of plant growth promotion by Bacillus amyloliquefaciens FZB42. Mol. Plant Microbe Interact. 2007, 20, 619–626. [Google Scholar] [CrossRef] [PubMed]
- Ongena, M.; Jourdan, E.; Adam, A.; Paquot, M.; Brans, A.; Joris, B.; Arpigny, J.; Thonart, P. Surfactin and fengycin lipopeptides of Bacillus subtilis as elicitors of induced systemic resistance in plants. Environ. Microbiol. 2010, 9, 1084–1090. [Google Scholar] [CrossRef] [PubMed]
- Xie, S.; Jiang, H.; Ding, T.; Xu, Q.; Chai, W.; Cheng, B. Bacillus amyloliquefaciens FZB42 represses plant miR846 to induce systemic resistance via a jasmonic acid-dependent signalling pathway. Mol. Plant Pathol. 2018, 19, 1612–1623. [Google Scholar] [CrossRef] [PubMed]
- Niu, D.; Xia, J.; Jiang, C.; Qi, B.; Ling, X.; Lin, S.; Zhang, W.; Guo, J.; Jin, H.; Zhao, H. Bacillus cereus AR156 primes induced systemic resistance by suppressing miR825/825 and activating defense-related genes in Arabidopsis. J. Integr. Plant Biol. 2016, 58, 426–439. [Google Scholar] [CrossRef] [PubMed]
- Fan, B.; Borriss, R.; Bleiss, W.; Wu, X. Gram-positive rhizobacterium Bacillus amyloliquefaciens FZB42 colonizes three types of plants in different patterns. J. Microbiol. 2012, 50, 38–44. [Google Scholar] [CrossRef] [PubMed]
- Mauch-Mani, B.; Baccelli, I.; Luna, E.; Flors, V. Defense priming: An adaptive part of induced resistance. Annu. Rev. Plant Biol. 2017, 68, 485–512. [Google Scholar] [CrossRef]
- Fu, Z.; Dong, X. Systemic acquired resistance: Turning local infection into global defense. Annu. Rev. Plant Biol. 2013, 64, 839–863. [Google Scholar] [CrossRef]
- Shoresh, M.; Harman, G.E.; Mastouri, F. Induced systemic resistance and plant responses to fungal biocontrol agents. Annu. Rev. Phytopathol. 2010, 48, 21–43. [Google Scholar] [CrossRef]
- Sunkar, R.; Zhu, J.-K. Novel and stress-regulated microRNAs and other small RNAs from Arabidopsis. Plant Cell 2004, 16, 2001–2019. [Google Scholar] [CrossRef] [PubMed]
- Boccara, M.; Sarazin, A.; Thiébeauld, O.; Jay, F.; Voinnet, O.; Navarro, L.; Colot, V. The Arabidopsis miR472-RDR6 silencing pathway modulates PAMP- and effector-triggered immunity through the post-transcriptional control of disease resistance genes. PLoS Pathog. 2014, 10, e1003883. [Google Scholar] [CrossRef] [PubMed]
- Shivaprasad, P.V.; Ho-Ming, C.; Kanu, P.; Bond, D.M.; Santos, B.A.C.M.; Baulcombe, D.C. A microRNA superfamily regulates nucleotide binding site-leucine-rich repeats and other mRNAs. Plant Cell 2012, 24, 859–874. [Google Scholar] [CrossRef] [PubMed]
- Zhai, J.; Jeong, D.-H.; De Paoli, E.; Park, S.; Benjamin, D.R.; Li, Y.; Alvaro, J.G.; Yan, Z.; Sherry, L.K.; Michael, A.G. MicroRNAs as master regulators of the plant NB-LRR defense gene family via the production of phased, trans-acting siRNAs. Genes Dev. 2011, 25, 2540–2553. [Google Scholar] [CrossRef] [PubMed]
- Baldrich, P.; San Segundo, B. MicroRNAs in rice innate immunity. Rice 2016, 9, 6. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Zhang, Q.; Zhang, J.; Wu, L.; Qi, Y.; Zhou, J.-M. Identification of microRNAs involved in pathogen-associated molecular pattern-triggered plant innate immunity. Plant Physiol. 2010, 152, 2222–2231. [Google Scholar] [CrossRef] [PubMed]
- Park, Y.J.; Lee, H.J.; Kwak, K.J.; Lee, K.; Hong, S.W.; Kang, H. MicroRNA400-guided cleavage of pentatricopeptide repeat protein mRNAs renders Arabidopsis thaliana more susceptible to pathogenic bacteria and fungi. Plant Cell Physiol. 2014, 55, 1660–1668. [Google Scholar] [CrossRef]
- Chowdhury, S.P.; Uhl, J.; Grosch, R.; Alquéres, S.; Pittroff, S.; Dietel, K.; Schmitt-Kopplin, P.; Borriss, R.; Hartmann, A. Cyclic Lipopeptides of Bacillus amyloliquefaciens subsp. plantarum colonizing the lettuce rhizosphere enhance plant defense responses toward the bottom rot pathogen Rhizoctonia solani. Mol. Plant Microbe Interact. 2015, 28, 984–995. [Google Scholar]
- Ryu, C.-M.; Mohamed, A.F.; Hu, C.-H.; Munagala, S.R.; Joseph, W.K.; Paul, W.P. Bacterial volatiles induce systemic resistance in Arabidopsis. Plant Physiol. 2004, 134, 1017–1026. [Google Scholar] [CrossRef]
- Li, W.X.; Oono, Y.; Zhu, J.; He, X.J.; Wu, J.M.; Iida, K.; Lu, X.Y.; Cui, X.; Jin, H.; Zhu, J.K. The Arabidopsis NFYA5 transcription factor is regulated transcriptionally and posttranscriptionally to promote drought resistance. Plant Cell 2008, 20, 2238–2251. [Google Scholar] [CrossRef]
- Meng, Z.; Hong, D.; Jian-Kang, Z.; Fusuo, Z.; Wen-Xue, L. Involvement of miR169 in the nitrogen-starvation responses in Arabidopsis. New Phytol. 2011, 190, 906–915. [Google Scholar]
- Mingda, L.; Miaoyun, X.; Yunming, L.; Lan, Z.; Yunliu, F.; Lei, W. Expression of zma-miR169 miRNAs and their target ZmNF-YA genes in response to abiotic stress in maize leaves. Gene 2015, 555, 178–185. [Google Scholar]
- Hanemian, M.; Barlet, X.; Sorin, C.; Yadeta, K.A.; Keller, H.; Favery, B.; Simon, R.; Thomma, B.P.H.J.; Hartmann, C.; Crespi, M. Arabidopsis CLAVATA1 and CLAVATA2 receptors contribute to Ralstonia solanacearum pathogenicity through a miR169-dependent pathway. New Phytol. 2016, 211, 502–515. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Zhao, S.L.; Li, J.L.; Hu, X.H.; Wang, H.; Cao, X.L.; Xu, Y.J.; Zhao, Z.X.; Xiao, Z.Y.; Yang, N. Osa-miR169 negatively regulates rice immunity against the blast fungus Magnaporthe oryzae. Front. Plant Sci. 2017, 8, 2. [Google Scholar] [CrossRef] [PubMed]
- Song, S.; Xu, Y.; Huang, D.; Ashraf, M.A.; Li, J.; Hu, W.; Jin, Z.; Zeng, C.; Tang, F.; Xu, B.; et al. Identification and characterization of miRNA169 family members in banana (Musa acuminata L.) that respond to fusarium oxysporum f. sp. cubense infection in banana cultivars. PeerJ 2018, 6, e6209. [Google Scholar]
- Kazuhiko, Y.; Takanori, K.; Motoaki, S.; Kazuo, S.; Shigeyuki, Y. DNA-binding domains of plant-specific transcription factors: Structure, function, and evolution. Trends Plant Sci. 2013, 18, 267–276. [Google Scholar]
- Matuoka, K.; Chen, K.Y. Transcriptional regulation of cellular ageing by the CCAAT box-binding factor CBF/NF-Y. Ageing Res. Rev. 2002, 1, 639–650. [Google Scholar] [CrossRef]
- Katia, P.; Kumimoto, R.W.; Nerina, G.; Valentina, C.; Monica, F.; Chiara, T.; Holt, B.F.; Roberto, M. The promiscuous life of plant nuclear factor Y transcription factors. Plant Cell 2012, 24, 4777–4792. [Google Scholar]
- Laloum, T.; De Mita, S.; Gamas, P.; Baudin, M.; Niebel, A. CCAAT-box binding transcription factors in plants: Y so many? Trends Plant Sci. 2013, 18, 157–166. [Google Scholar] [CrossRef]
- Zanetti, M.E.; Blanco, F.A.; Beker, M.P.; Battaglia, M.; Aguilar, O.M. AC subunit of the plant nuclear factor NF-Y required for rhizobial infection and nodule development affects partner selection in the common bean-Rhizobium etli symbiosis. Plant Cell 2010, 22, 4142–4157. [Google Scholar] [CrossRef]
- Alam, M.M.; Nakamura, H.; Ichikawa, H.; Kobayashi, K.; Yaeno, T.; Yamaoka, N.; Nishiguchi, M. Overexpression of OsHAP2E for a CCAAT-binding factor confers resistance to Cucumber mosaic virus and Rice necrosis mosaic virus. J. Gen. Plant Pathol. 2015, 81, 32–41. [Google Scholar] [CrossRef]
- Rey, T.; Balzergue, S.; Laporte, P.; Bonhomme, M.; Jardinaud, M.F.; Huguet, S.; Dumas, B.; Niebel, A.; Jacquet, C. MtNF-YA1, A central transcriptional regulator of symbiotic nodule development, is also a determinant of Medicago truncatula susceptibility toward a root pathogen. Front. Plant Sci. 2016, 7, 1837. [Google Scholar] [CrossRef] [PubMed]
- Ding, Y.; Chen, Z.; Zhu, C. Microarray-based analysis of cadmium-responsive microRNAs in rice (Oryza sativa). J. Exp. Bot. 2011, 62, 3563–3573. [Google Scholar] [CrossRef] [PubMed]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef] [PubMed]


© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xie, S.; Yu, H.; Li, E.; Wang, Y.; Liu, J.; Jiang, H. Identification of miRNAs Involved in Bacillus velezensis FZB42-Activated Induced Systemic Resistance in Maize. Int. J. Mol. Sci. 2019, 20, 5057. https://doi.org/10.3390/ijms20205057
Xie S, Yu H, Li E, Wang Y, Liu J, Jiang H. Identification of miRNAs Involved in Bacillus velezensis FZB42-Activated Induced Systemic Resistance in Maize. International Journal of Molecular Sciences. 2019; 20(20):5057. https://doi.org/10.3390/ijms20205057
Chicago/Turabian StyleXie, Shanshan, Hengguo Yu, Enze Li, Yu Wang, Juan Liu, and Haiyang Jiang. 2019. "Identification of miRNAs Involved in Bacillus velezensis FZB42-Activated Induced Systemic Resistance in Maize" International Journal of Molecular Sciences 20, no. 20: 5057. https://doi.org/10.3390/ijms20205057
APA StyleXie, S., Yu, H., Li, E., Wang, Y., Liu, J., & Jiang, H. (2019). Identification of miRNAs Involved in Bacillus velezensis FZB42-Activated Induced Systemic Resistance in Maize. International Journal of Molecular Sciences, 20(20), 5057. https://doi.org/10.3390/ijms20205057




