Analysis of Mucopolysaccharidosis Type VI through Integrative Functional Metabolomics
Abstract
1. Introduction
2. Results
2.1. Untargeted Analysis
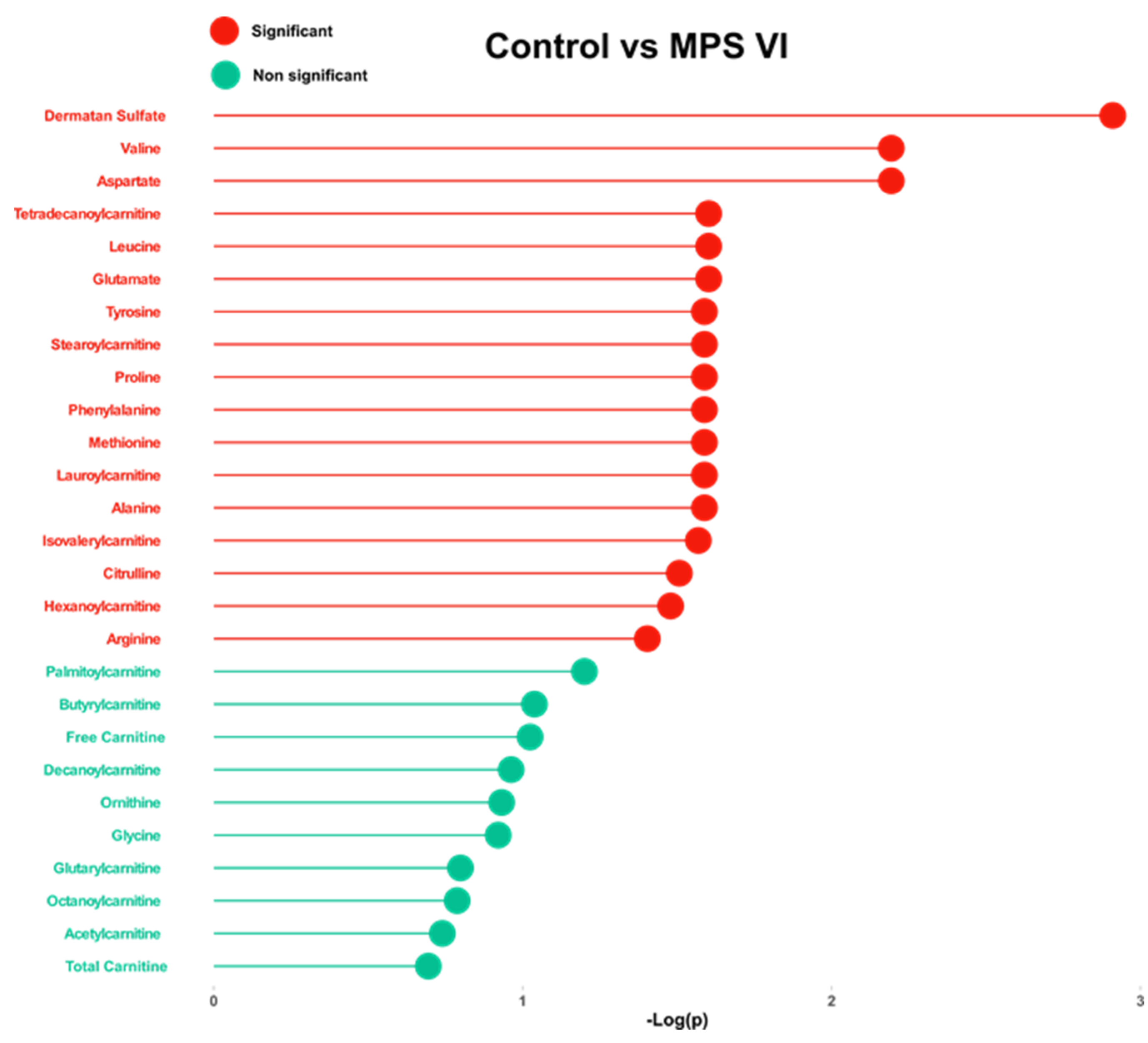
2.2. Targeted Analysis
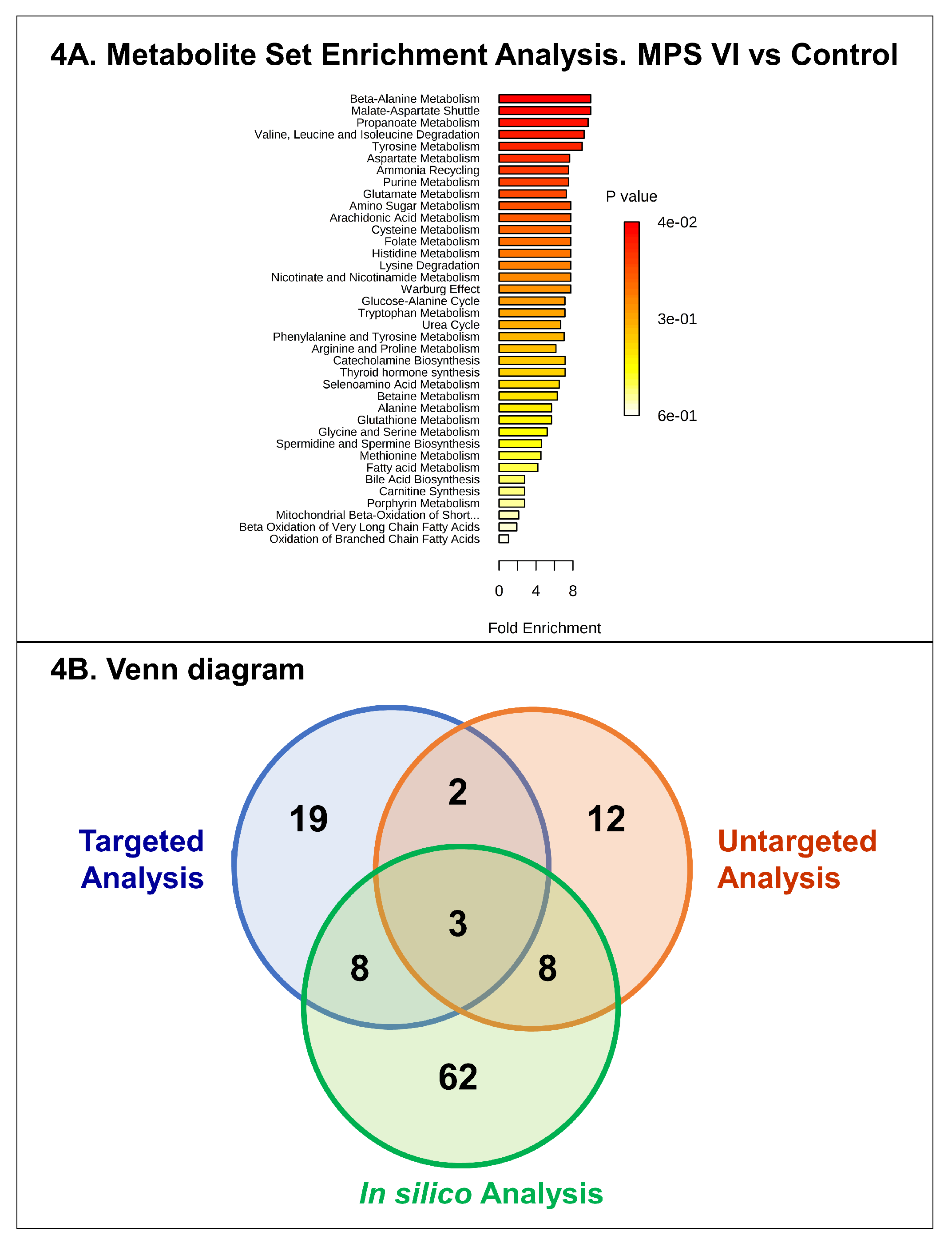
3. Discussion
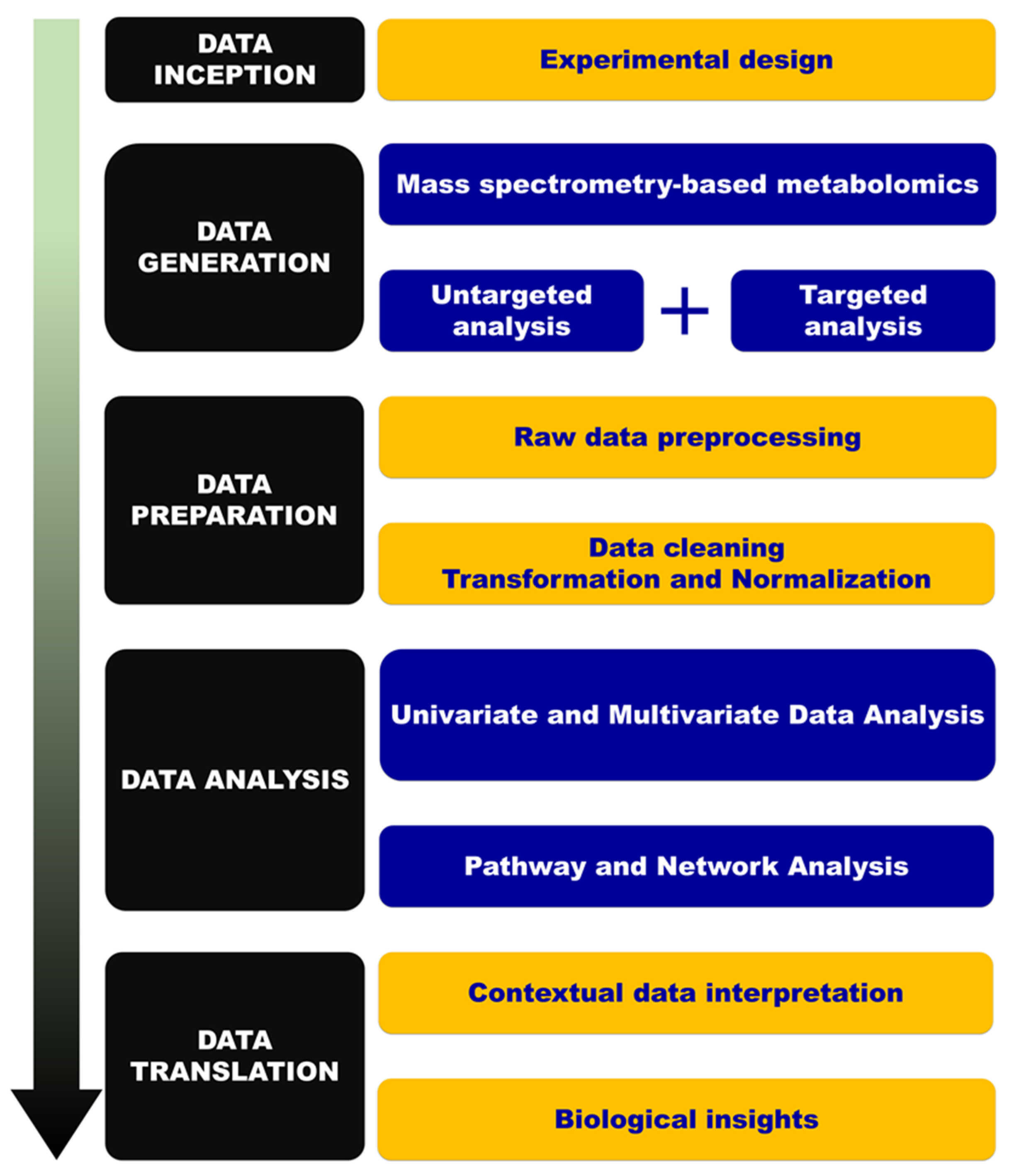
4. Material and Methods
4.1. Urine Samples
4.2. Reagents and Chemicals
4.3. Untargeted Metabolic Phenotyping
4.3.1. Sample Handling
4.3.2. Chromatographic Conditions
4.3.3. Ion Mobility and Mass Spectrometry
4.3.4. Raw Data Processing
4.3.5. Quality Control
4.4. Targeted Analysis
4.4.1. Amino Acids and Acylcarnitines Profiling
4.4.2. Dermatan Sulfate Assessment
4.5. Data Analysis and Modeling
4.6. Feature Selection, Annotation, and Network Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| IEM | Inborn errors of metabolism |
| LSD | Lysosomal storage diseases |
| MPS | Mucopolysaccharidoses |
| GAGs | Glycosaminoglycans |
| MPS VI | Mucopolysaccharidosis type VI |
| ERT | Enzyme replacement therapy |
| HS | Heparan sulfate |
| CCS | Collision cross section |
| DS | Dermatan sulfate |
| KS | Keratan sulfate |
| ROC | Receiver operating characteristic |
| FDR | False discovery rate |
| PCA | Principal component analysis |
| OPLS-DA | Orthogonal partial least-squares-discriminant analysis |
| VIP | Variable influence in projection |
| AUC | Area under curve |
References
- Bekri, S. The role of metabolomics in precision medicine. Expert Rev. Precis. Med. Drug Dev. 2016, 1, 517–532. [Google Scholar] [CrossRef]
- Tebani, A.; Afonso, C.; Marret, S.; Bekri, S. Omics-based strategies in precision medicine: Toward a paradigm shift in inborn errors of metabolism investigations. Int. J. Mol. Sci. 2016, 17, 1555. [Google Scholar] [CrossRef]
- Nicholson, J.K.; Lindon, J.C.; Holmes, E. ‘Metabonomics’: Understanding the metabolic responses of living systems to pathophysiological stimuli via multivariate statistical analysis of biological nmr spectroscopic data. Xenobiotica Fate Foreign Compd. Biol. Syst. 1999, 29, 1181–1189. [Google Scholar] [CrossRef] [PubMed]
- Fiehn, O. Metabolomics—The link between genotypes and phenotypes. Plant Mol. Biol. 2002, 48, 155–171. [Google Scholar] [CrossRef] [PubMed]
- Tebani, A.; Abily-Donval, L.; Afonso, C.; Marret, S.; Bekri, S. Clinical metabolomics: The new metabolic window for inborn errors of metabolism investigations in the post-genomic era. Int. J. Mol. Sci. 2016, 17, 1167. [Google Scholar] [CrossRef]
- Auray-Blais, C.; Maranda, B.; Lavoie, P. High-throughput tandem mass spectrometry multiplex analysis for newborn urinary screening of creatine synthesis and transport disorders, triple h syndrome and otc deficiency. Clin. Chim. Acta Int. J. Clin. Chem. 2014, 436, 249–255. [Google Scholar] [CrossRef] [PubMed]
- Pitt, J.J. Principles and applications of liquid chromatography-mass spectrometry in clinical biochemistry. Clin. Biochem. Rev. Aust. Assoc. Clin. Biochem. 2009, 30, 19–34. [Google Scholar]
- Pitt, J.J. Newborn screening. Clin. Biochem. Rev. Aust. Assoc. Clin. Biochem. 2010, 31, 57–68. [Google Scholar]
- Pitt, J.J.; Eggington, M.; Kahler, S.G. Comprehensive screening of urine samples for inborn errors of metabolism by electrospray tandem mass spectrometry. Clin. Chem. 2002, 48, 1970–1980. [Google Scholar]
- Spacil, Z.; Tatipaka, H.; Barcenas, M.; Scott, C.R.; Turecek, F.; Gelb, M.H. High-throughput assay of 9 lysosomal enzymes for newborn screening. Clin. Chem. 2013, 59, 502–511. [Google Scholar] [CrossRef]
- Therrell, B.L.; Padilla, C.D.; Loeber, J.G.; Kneisser, I.; Saadallah, A.; Borrajo, G.J.; Adams, J. Current status of newborn screening worldwide: 2015. Semin. Perinatol. 2015, 39, 171–187. [Google Scholar] [CrossRef] [PubMed]
- Tebani, A.; Afonso, C.; Bekri, S. Advances in metabolome information retrieval: Turning chemistry into biology. Part ii: Biological information recovery. J. Inherit. Metab. Dis. 2018, 41, 393–406. [Google Scholar] [CrossRef] [PubMed]
- Tebani, A.; Afonso, C.; Bekri, S. Advances in metabolome information retrieval: Turning chemistry into biology. Part i: Analytical chemistry of the metabolome. J. Inherit. Metab. Dis. 2018, 41, 379–391. [Google Scholar] [CrossRef] [PubMed]
- Ballabio, A.; Gieselmann, V. Lysosomal disorders: From storage to cellular damage. Biochim. Biophys. Acta 2009, 1793, 684–696. [Google Scholar] [CrossRef] [PubMed]
- Wraith, J.E. The mucopolysaccharidoses: A clinical review and guide to management. Arch. Dis. Child. 1995, 72, 263–267. [Google Scholar] [CrossRef]
- Neufeld, E.F.; Muenzer, J. The mucopolysaccharidoses. In The Metabolic and Molecular Basis of Inherited Disease; Scrive, C., Ballabio, A., Sly, W., Vaele, D., Eds.; Mc Graw-Hill: New York, NY, USA, 2001; pp. 3421–3452. [Google Scholar]
- Stewart, F.J.; Bentley, A.; Burton, B.K.; Guffon, N.; Hale, S.L.; Harmatz, P.R.; Kircher, S.G.; Kochhar, P.K.; Mitchell, J.J.; Plöckinger, U.; et al. Pregnancy in patients with mucopolysaccharidosis: A case series. Mol. Genet. Metab. Rep. 2016, 8, 111–115. [Google Scholar] [CrossRef]
- Montano, A.M.; Lock-Hock, N.; Steiner, R.D.; Graham, B.H.; Szlago, M.; Greenstein, R.; Pineda, M.; Gonzalez-Meneses, A.; Coker, M.; Bartholomew, D.; et al. Clinical course of sly syndrome (mucopolysaccharidosis type vii). J. Med. Genet. 2016, 53, 403–418. [Google Scholar] [CrossRef]
- Parenti, G.; Andria, G.; Ballabio, A. Lysosomal storage diseases: From pathophysiology to therapy. Annu. Rev. Med. 2015, 66, 471–486. [Google Scholar] [CrossRef]
- Maroteaux, P.; Leveque, B.; Marie, J.; Lamy, M. A new dysostosis with urinary elimination of chondroitin sulfate b. La Presse Med. 1963, 71, 1849–1852. [Google Scholar]
- Muenzer, J. Overview of the mucopolysaccharidoses. Rheumatology 2011, 50 (Suppl. 5), v4–v12. [Google Scholar] [CrossRef]
- Valayannopoulos, V.; Nicely, H.; Harmatz, P.; Turbeville, S. Mucopolysaccharidosis vi. Orphanet J. Rare Dis. 2010, 5, 5. [Google Scholar] [CrossRef] [PubMed]
- Hendriksz, C.J.; Giugliani, R.; Harmatz, P.; Lampe, C.; Martins, A.M.; Pastores, G.M.; Steiner, R.D.; Leão Teles, E.; Valayannopoulos, V. Design, baseline characteristics, and early findings of the MPS VI (mucopolysaccharidosis vi) clinical surveillance program (csp). J. Inherit. Metab. Dis. 2013, 36, 373–384. [Google Scholar] [CrossRef]
- Quartel, A.; Hendriksz, C.J.; Parini, R.; Graham, S.; Lin, P.; Harmatz, P. Growth charts for individuals with mucopolysaccharidosis vi (maroteaux–lamy syndrome). In JIMD Reports; Zschocke, J., Baumgartner, M., Morava, E., Patterson, M., Rahman, S., Peters, V., Eds.; Springer: Berlin/Heidelberg, Germany, 2015; Volume 18, pp. 1–11. [Google Scholar]
- Scarpa, M.; Buffone, E.; Marca, P.L.; Campello, M.; Rampazzo, A. Difficulties in diagnosing slowly progressive mucopolysaccharidosis vi: A case series. J. Pediatr. Rehabil. Med. 2010, 3, 71–75. [Google Scholar] [PubMed]
- Harmatz, P.; Hendriksz, C.J.; Lampe, C.; McGill, J.J.; Parini, R.; Leao-Teles, E.; Valayannopoulos, V.; Cole, T.J.; Matousek, R.; Graham, S.; et al. The effect of galsulfase enzyme replacement therapy on the growth of patients with mucopolysaccharidosis vi (maroteaux-lamy syndrome). Mol. Genet. Metab. 2017, 122, 107–112. [Google Scholar] [CrossRef]
- Furujo, M.; Kosuga, M.; Okuyama, T. Enzyme replacement therapy attenuates disease progression in two japanese siblings with mucopolysaccharidosis type vi: 10-year follow up. Mol. Genet. Metab Rep. 2017, 13, 69–75. [Google Scholar] [CrossRef] [PubMed]
- Jester, S.; Larsson, J.; Eklund, E.A.; Papadopoulou, D.; Månsson, J.-E.; Békássy, A.N.; Turkiewicz, D.; Toporski, J.; Øra, I. Haploidentical stem cell transplantation in two children with mucopolysaccharidosis vi: Clinical and biochemical outcome. Orphanet J. Rare Dis. 2013, 8, 134. [Google Scholar] [CrossRef] [PubMed]
- Salazar, D.A.; Rodriguez-Lopez, A.; Herreno, A.; Barbosa, H.; Herrera, J.; Ardila, A.; Barreto, G.E.; Gonzalez, J.; Almeciga-Diaz, C.J. Systems biology study of mucopolysaccharidosis using a human metabolic reconstruction network. Mol. Genet. Metab. 2016, 117, 129–139. [Google Scholar] [CrossRef]
- Fiorenza, M.T.; Moro, E.; Erickson, R.P. The pathogenesis of lysosomal storage disorders: Beyond the engorgement of lysosomes to abnormal development and neuroinflammation. Hum. Mol. Genet. 2018, 27, R119–R129. [Google Scholar] [CrossRef]
- Petrova, R.; Joyner, A.L. Roles for hedgehog signaling in adult organ homeostasis and repair. Development 2014, 141, 3445–3457. [Google Scholar] [CrossRef]
- Whalen, D.M.; Malinauskas, T.; Gilbert, R.J.; Siebold, C. Structural insights into proteoglycan-shaped hedgehog signaling. Proc. Natl. Acad. Sci. USA 2013, 110, 16420–16425. [Google Scholar] [CrossRef]
- Chang, S.C.; Mulloy, B.; Magee, A.I.; Couchman, J.R. Two distinct sites in sonic hedgehog combine for heparan sulfate interactions and cell signaling functions. J. Biol. Chem. 2011, 286, 44391–44402. [Google Scholar] [CrossRef] [PubMed]
- Donida, B.; Jacques, C.E.D.; Mescka, C.P.; Rodrigues, D.G.B.; Marchetti, D.P.; Ribas, G.; Giugliani, R.; Vargas, C.R. Oxidative damage and redox in lysosomal storage disorders: Biochemical markers. Clin. Chim. Acta Int. J. Clin. Chem. 2017, 466, 46–53. [Google Scholar] [CrossRef] [PubMed]
- Tebani, A.; Schmitz-Afonso, I.; Abily-Donval, L.; Heron, B.; Piraud, M.; Ausseil, J.; Brassier, A.; De Lonlay, P.; Zerimech, F.; Vaz, F.M.; et al. Urinary metabolic phenotyping of mucopolysaccharidosis type i combining untargeted and targeted strategies with data modeling. Clin. Chim. Acta Int. J. Clin. Chem. 2017, 475, 7–14. [Google Scholar] [CrossRef] [PubMed]
- Tebani, A.; Abily-Donval, L.; Schmitz-Afonso, I.; Héron, B.; Piraud, M.; Ausseil, J.; Zerimech, F.; Gonzalez, B.; Marret, S.; Afonso, C.; et al. Unveiling metabolic remodeling in mucopolysaccharidosis type iii through integrative metabolomics and pathway analysis. J. Transl. Med. 2018, 16, 248. [Google Scholar] [CrossRef] [PubMed]
- Nagesh Babu, G.; Gupta, M.; Paliwal, V.K.; Singh, S.; Chatterji, T.; Roy, R. Serum metabolomics study in a group of parkinson’s disease patients from northern india. Clin. Chim. Acta Int. J. Clin. Chem. 2018, 480, 214–219. [Google Scholar] [CrossRef] [PubMed]
- Bellia, F.; Vecchio, G.; Cuzzocrea, S.; Calabrese, V.; Rizzarelli, E. Neuroprotective features of carnosine in oxidative driven diseases. Mol. Aspects Med. 2011, 32, 258–266. [Google Scholar] [CrossRef] [PubMed]
- Cooper, K.F. Till death do us part: The marriage of autophagy and apoptosis. Oxid. Med. Cell. Longev. 2018, 2018, 4701275. [Google Scholar] [CrossRef]
- Donida, B.; Marchetti, D.P.; Jacques, C.E.D.; Ribas, G.; Deon, M.; Manini, P.; da Rosa, H.T.; Moura, D.J.; Saffi, J.; Giugliani, R.; et al. Oxidative profile exhibited by mucopolysaccharidosis type iva patients at diagnosis: Increased keratan urinary levels. Mol. Genet. Metab. Rep. 2017, 11, 46–53. [Google Scholar] [CrossRef]
- Paglia, G.; Angel, P.; Williams, J.P.; Richardson, K.; Olivos, H.J.; Thompson, J.W.; Menikarachchi, L.; Lai, S.; Walsh, C.; Moseley, A.; et al. Ion mobility-derived collision cross section as an additional measure for lipid fingerprinting and identification. Anal. Chem. 2015, 87, 1137–1144. [Google Scholar] [CrossRef]
- Auray-Blais, C.; Lavoie, P.; Tomatsu, S.; Valayannopoulos, V.; Mitchell, J.J.; Raiman, J.; Beaudoin, M.; Maranda, B.; Clarke, J.T.R. Uplc-ms/ms detection of disaccharides derived from glycosaminoglycans as biomarkers of mucopolysaccharidoses. Anal. Chim. Acta 2016, 936, 139–148. [Google Scholar] [CrossRef]
- Shen, X.; Gong, X.; Cai, Y.; Guo, Y.; Tu, J.; Li, H.; Zhang, T.; Wang, J.; Xue, F.; Zhu, Z.-J. Normalization and integration of large-scale metabolomics data using support vector regression. Metabolomics 2016, 12, 89. [Google Scholar] [CrossRef]
- Eriksson, L.; Trygg, J.; Wold, S. A chemometrics toolbox based on projections and latent variables. J. Chemom. 2014, 28, 332–346. [Google Scholar] [CrossRef]
- Eriksson, L.; Trygg, J.; Wold, S. Cv-anova for significance testing of pls and opls® models. J. Chemom. 2008, 22, 594–600. [Google Scholar] [CrossRef]
- Ritchie, M.E.; Phipson, B.; Wu, D.; Hu, Y.; Law, C.W.; Shi, W.; Smyth, G.K. Limma powers differential expression analyses for rna-sequencing and microarray studies. Nucleic Acids Res. 2015, 43, e47. [Google Scholar] [CrossRef] [PubMed]
- Hanley, J.A.; McNeil, B.J. The meaning and use of the area under a receiver operating characteristic (roc) curve. Radiology 1982, 143, 29–36. [Google Scholar] [CrossRef] [PubMed]
- Galindo-Prieto, B.; Eriksson, L.; Trygg, J. Variable influence on projection (vip) for orthogonal projections to latent structures (opls). J. Chemom. 2014, 28, 623–632. [Google Scholar] [CrossRef]
- Zhou, Z.; Xiong, X.; Zhu, Z.J. Metccs predictor: A web server for predicting collision cross-section values of metabolites in ion mobility-mass spectrometry based metabolomics. Bioinformatics 2017, 33, 2235–2237. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Park, Y.; Duraisingham, S.; Strobel, F.H.; Khan, N.; Soltow, Q.A.; Jones, D.P.; Pulendran, B. Predicting network activity from high throughput metabolomics. PLoS Comput. Biol. 2013, 9, e1003123. [Google Scholar] [CrossRef]
- Xia, J.; Sinelnikov, I.V.; Han, B.; Wishart, D.S. Metaboanalyst 3.0-making metabolomics more meaningful. Nucleic Acids Res. 2015, 43, W251–W257. [Google Scholar] [CrossRef]
- Wight, T.N. A role for proteoglycans in vascular disease. Matrix Biol. 2018, 71–72, 396–420. [Google Scholar] [CrossRef]
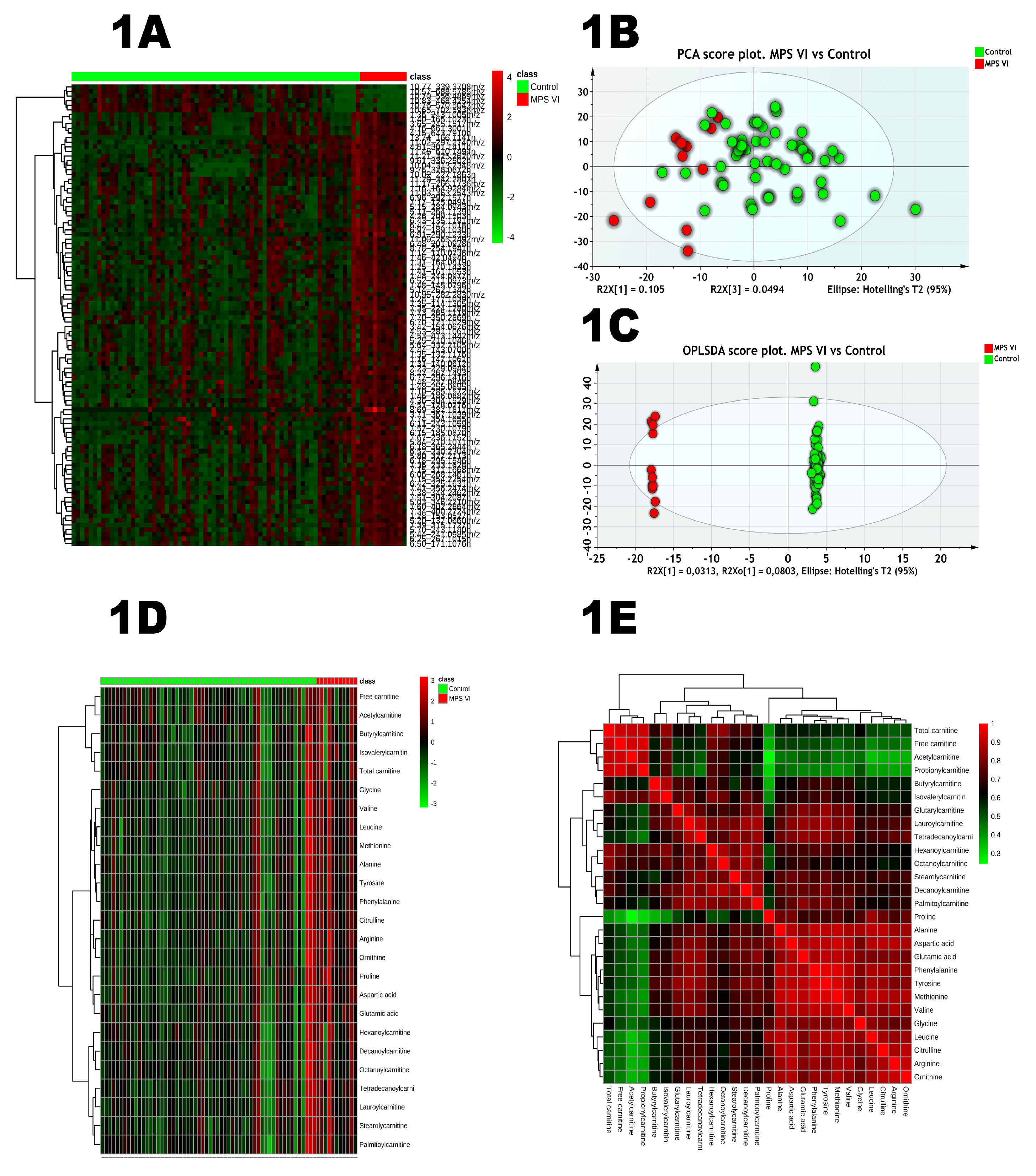
| HMDB | Putative Annotation | Formula | M | m/z | Adduct | Δm/z (ppm) | tR (min) | tD (ms) | CCS (A2) | %RSD | VIP | FDR | AUC |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| HMDB0003464 | 4-guanidinobutanoic acid | C5H11N3O2 | 145.0851 | 146.0932 | M + H | 5 | 1.48 | 1.89 | 124.4 | 4.60 | 0.97 | 5.18 × 10−4 | 0.88 |
| HMDB0001276 | N-acetylspermidine | C9H21N3O | 187.1685 | 188.1774 | M + H | 9 | 1.25 | 2.38 | 139.3 | 8.58 | 0.53 | 1.74 × 10−2 | 0.83 |
| HMDB00062 | Carnitine | C8H18N4O2 | 202.1430 | 203.1518 | M + can + H | 0.48 | 1.41 | 2.43 | 140.4 | 9.97 | 0.70 | 3.52 × 10−3 | 0.85 |
| HMDB0015444 | Phenylalaninylalanine | C12H16N2O3 | 236.1161 | 237.1225 | M + H | 4 | 7.67 | 2.7 | 147.8 | 10.51 | 1.55 | 1.95 × 10−4 | 0.94 |
| HMDB0002012 | Ubiquinone-1 | C14H18O4 | 250.1205 | 251.1291 | M + H | 5 | 7.17 | 2.86 | 152.4 | 5.69 | 0.16 | 2.74 × 10−2 | 0.80 |
| HMDB0000145 | Estrone | C18H22O2 | 270.1620 | 271.1675 | M + H | 6 | 6.50 | 3.19 | 161.5 | 4.26 | 0.27 | 2.83 × 10−2 | 0.79 |
| Pathway | Overlap Size | p-Value (FDR = 5%) |
|---|---|---|
| Vitamin B9 (folate) metabolism | 5 | 2.87 × 10−4 |
| Glycine, serine, alanine and threonine metabolism | 7 | 3.36 × 10−4 |
| Alanine and Aspartate metabolism | 4 | 4.68 × 10−4 |
| Histidine metabolism | 4 | 1.29 × 10−3 |
| Vitamin E metabolism | 5 | 2.21 × 10−3 |
| Carnitine shuttle | 5 | 2.21 × 10−3 |
| Glycosphingolipid metabolism | 3 | 3.61 × 10−3 |
| Vitamin B3 (nicotinate and nicotinamide) metabolism | 3 | 3.61 × 10−3 |
| Selenoamino acid metabolism | 2 | 4.15 × 10−03 |
| Glutathione Metabolism | 2 | 4.15 × 10−3 |
| CoA Catabolism | 2 | 4.15 × 10−3 |
| Electron transport chain | 2 | 4.15 × 10−3 |
| Vitamin B5–CoA biosynthesis from pantothenate | 2 | 4.15 × 10−3 |
| Methionine and cysteine metabolism | 6 | 4.66 × 10−3 |
| Aspartate and asparagine metabolism | 7 | 8.25 × 10−3 |
| Purine metabolism | 5 | 1.01 × 10−2 |
| Arginine and proline metabolism | 4 | 1.20 × 10−2 |
| Lysine metabolism | 4 | 1.70 × 10−2 |
| Linoleate metabolism | 4 | 1.70 × 10−2 |
| Aminosugar metabolism | 3 | 2.29 × 10−2 |
| Porphyrin metabolism | 3 | 2.29 × 10×2 |
| Pyruvate metabolism | 2 | 2.63 × 10−2 |
| Control vs. MPS VI | ||||
|---|---|---|---|---|
| AUC | p-Value (FDR) | Fold Change | Effect in MPS VI | |
| Dermatan sulfate | 0.90 | 1.23 × 10−3 | 10.0 | Increased |
| Aspartic acid | 0.85 | 6.41 × 10−3 | 1.61 | Increased |
| Valine | 0.83 | 6.41 × 10−3 | 1.74 | Increased |
| Glutamic acid | 0.79 | 2.50 × 10−2 | 1.42 | Increased |
| Leucine | 0.79 | 2.50 × 10−2 | 1.44 | Increased |
| Tetradecanoylcarnitine | 0.78 | 2.50 × 10−2 | 1.36 | Increased |
| Alanine | 0.75 | 2.58 × 10−2 | 1.32 | Increased |
| Lauroylcarnitine | 0.75 | 2.58 × 10−2 | 1.18 | Increased |
| Methionine | 0.77 | 2.58 × 10−2 | 1.28 | Increased |
| Phenylalanine | 0.77 | 2.58 × 10−2 | 1.28 | Increased |
| Proline | 0.79 | 2.58 × 10−2 | 1.48 | Increased |
| Stearoylcarnitine | 0.76 | 2.58 × 10−2 | 1.07 | Increased |
| Tyrosine | 0.76 | 2.58 × 10−2 | 1.30 | Increased |
| Isovalerylcarnitine | 0.71 | 2.70 × 10−2 | 1.42 | Increased |
| Citrulline | 0.79 | 3.11 × 10−2 | 1.26 | Increased |
| Hexanoylcarnitine | 0.71 | 3.32 × 10−2 | 1.20 | Increased |
| Arginine | 0.80 | 3.95 × 10−2 | 1.52 | Increased |
| Palmitoylcarnitine | 0.68 | 6.31 × 10−2 | 0.00 | / |
| Butyrylcarnitine | 0.68 | 9.16 × 10−2 | 1.15 | Increased |
| Free carnitine | 0.67 | 9.46 × 10−2 | 1.27 | Increased |
| Decanoylcarnitine | 0.67 | 1.09 × 10−1 | 0.90 | Decreased |
| Ornithine | 0.75 | 1.17 × 10−1 | 1.05 | Increased |
| Glycine | 0.70 | 1.20 × 10−1 | 0.95 | Decreased |
| Glutarylcarnitine | 0.66 | 1.59 × 10−1 | 0.95 | Decreased |
| Octanoylcarnitine | 0.65 | 1.63 × 10−1 | 0.79 | Decreased |
| Acetylcarnitine | 0.62 | 1.82 × 10−1 | 1.04 | Increased |
| Total carnitine | 0.62 | 2.02 × 10−1 | 0.89 | Decreased |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tebani, A.; Abily-Donval, L.; Schmitz-Afonso, I.; Piraud, M.; Ausseil, J.; Zerimech, F.; Pilon, C.; Pereira, T.; Marret, S.; Afonso, C.; et al. Analysis of Mucopolysaccharidosis Type VI through Integrative Functional Metabolomics. Int. J. Mol. Sci. 2019, 20, 446. https://doi.org/10.3390/ijms20020446
Tebani A, Abily-Donval L, Schmitz-Afonso I, Piraud M, Ausseil J, Zerimech F, Pilon C, Pereira T, Marret S, Afonso C, et al. Analysis of Mucopolysaccharidosis Type VI through Integrative Functional Metabolomics. International Journal of Molecular Sciences. 2019; 20(2):446. https://doi.org/10.3390/ijms20020446
Chicago/Turabian StyleTebani, Abdellah, Lenaig Abily-Donval, Isabelle Schmitz-Afonso, Monique Piraud, Jérôme Ausseil, Farid Zerimech, Carine Pilon, Tony Pereira, Stéphane Marret, Carlos Afonso, and et al. 2019. "Analysis of Mucopolysaccharidosis Type VI through Integrative Functional Metabolomics" International Journal of Molecular Sciences 20, no. 2: 446. https://doi.org/10.3390/ijms20020446
APA StyleTebani, A., Abily-Donval, L., Schmitz-Afonso, I., Piraud, M., Ausseil, J., Zerimech, F., Pilon, C., Pereira, T., Marret, S., Afonso, C., & Bekri, S. (2019). Analysis of Mucopolysaccharidosis Type VI through Integrative Functional Metabolomics. International Journal of Molecular Sciences, 20(2), 446. https://doi.org/10.3390/ijms20020446






