Novel DnaJ Protein Facilitates Thermotolerance of Transgenic Tomatoes
Abstract
1. Introduction
2. Results
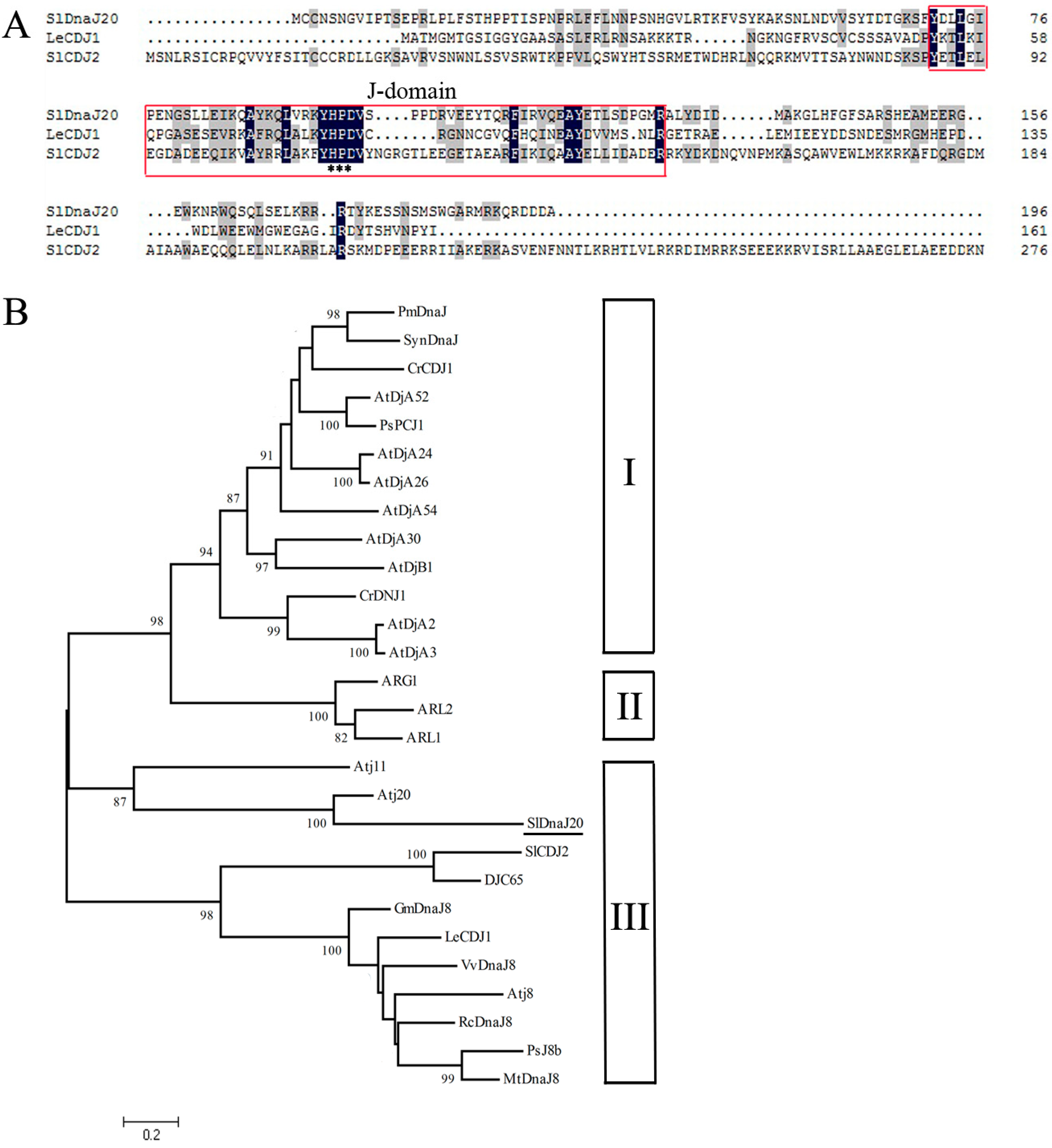
2.1. Identification and Bioinformatics Analysis of SlDnaJ20
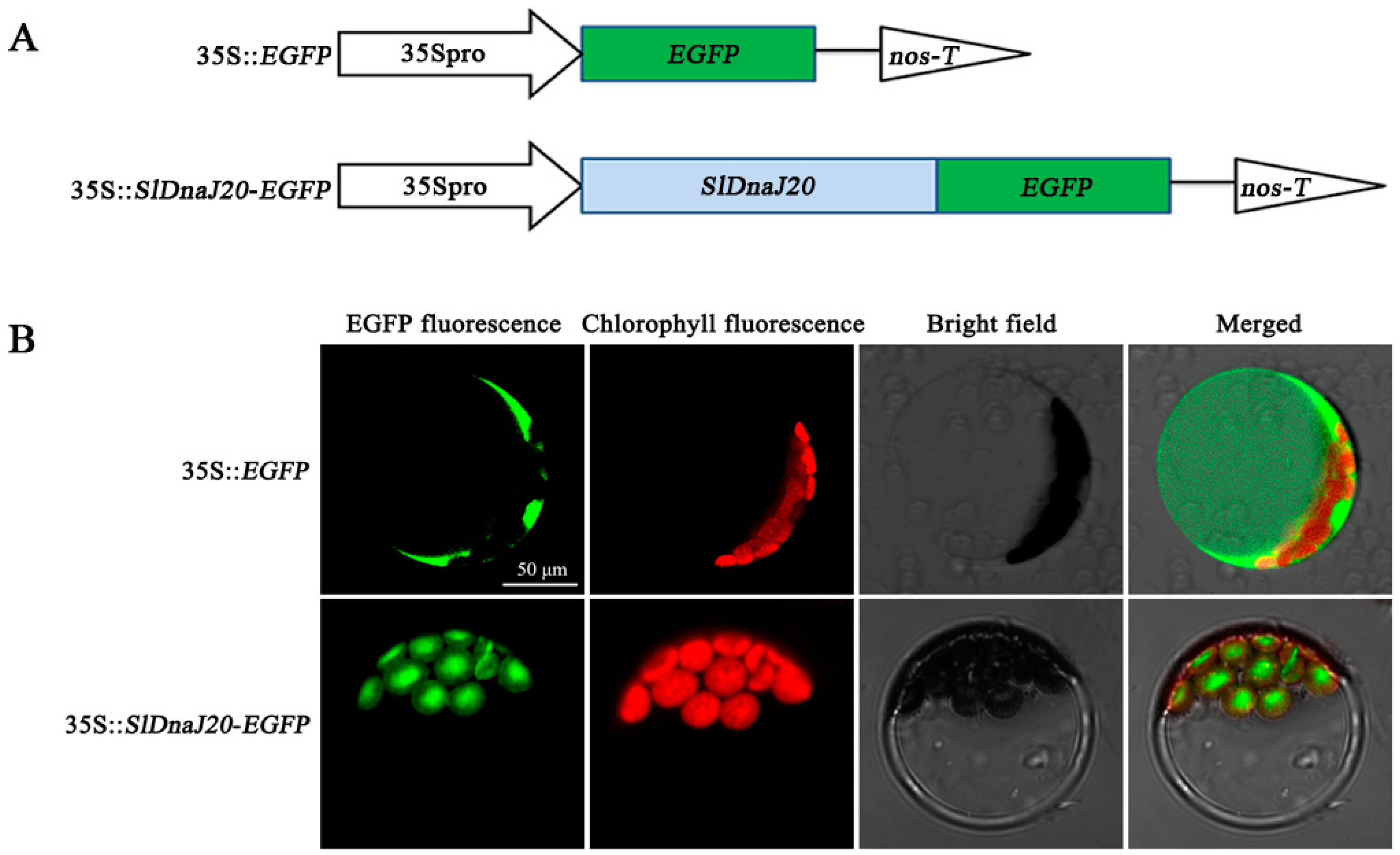
2.2. Subcellular Localization of SlDnaJ20
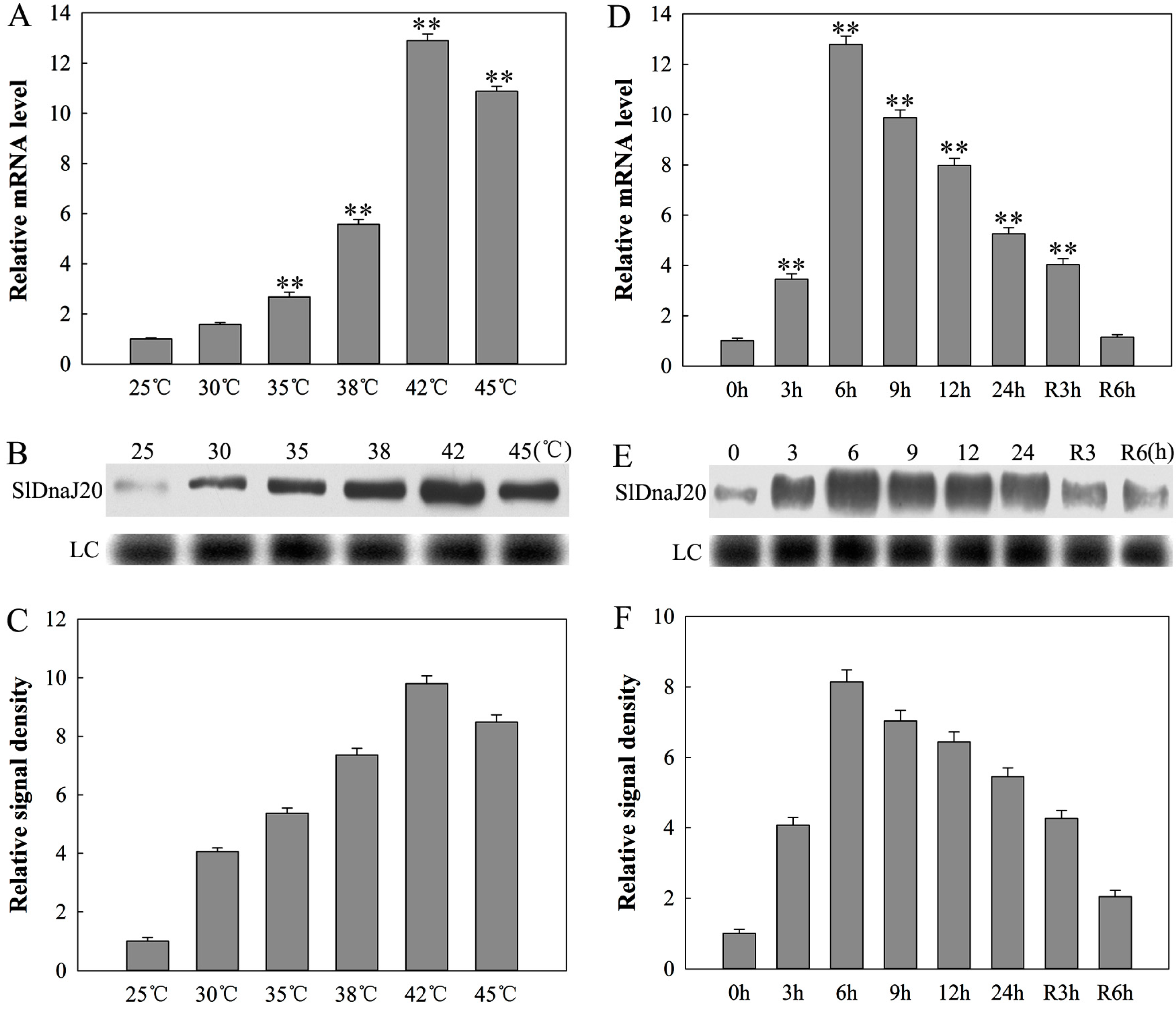
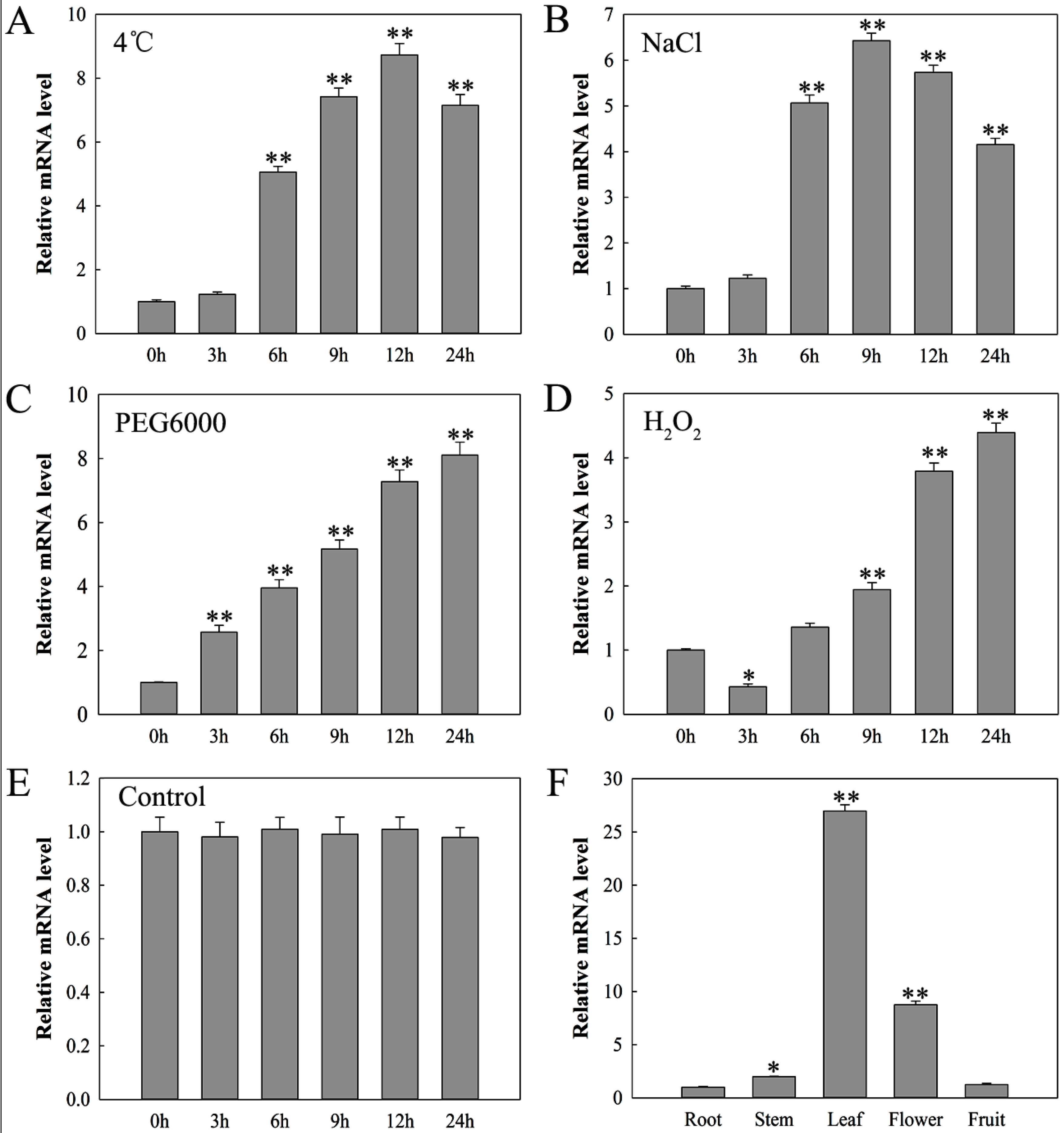
2.3. Expression Analysis of SlDnaJ20 in Tomato
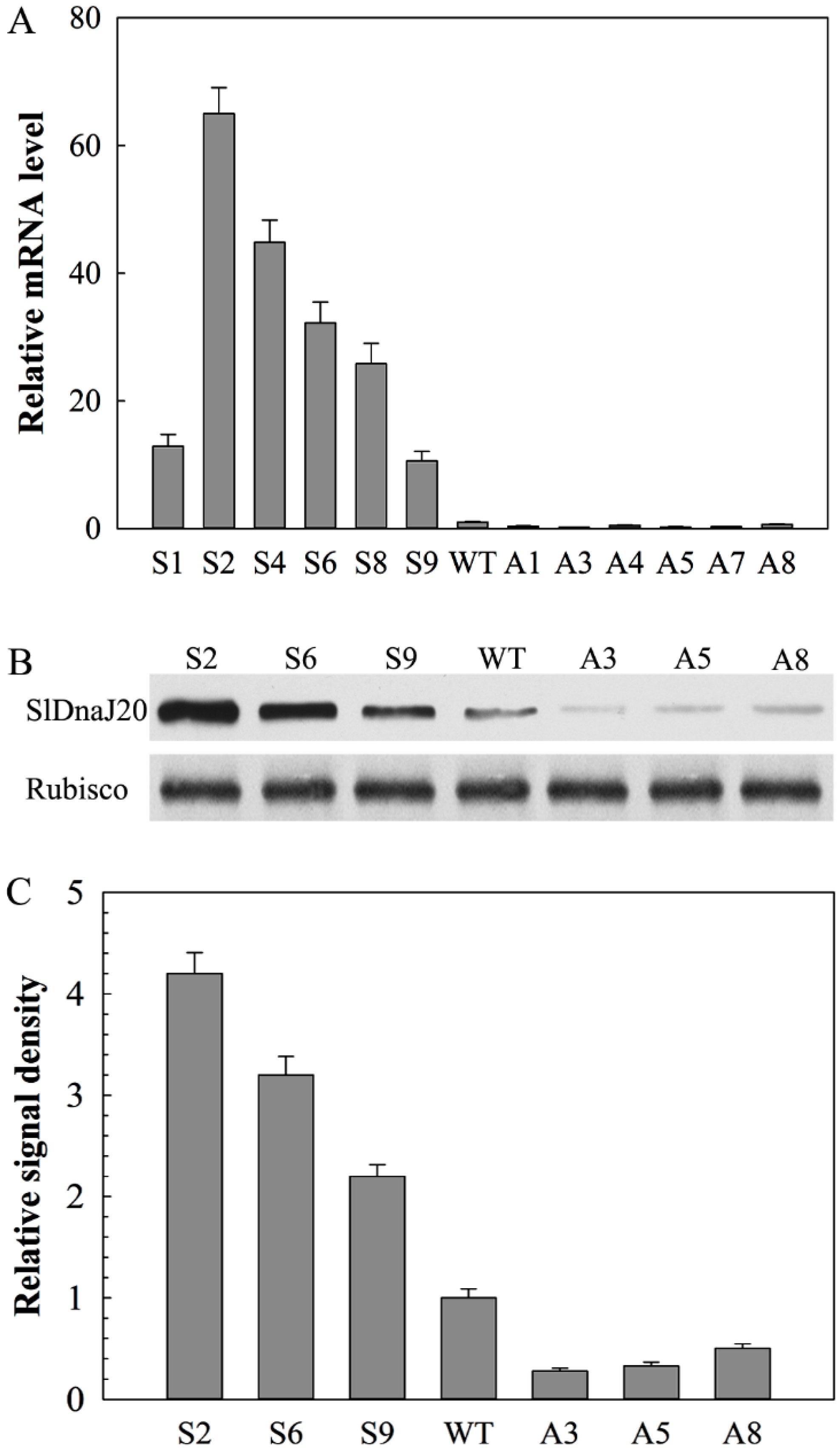
2.4. Identification of Transgenic Plants
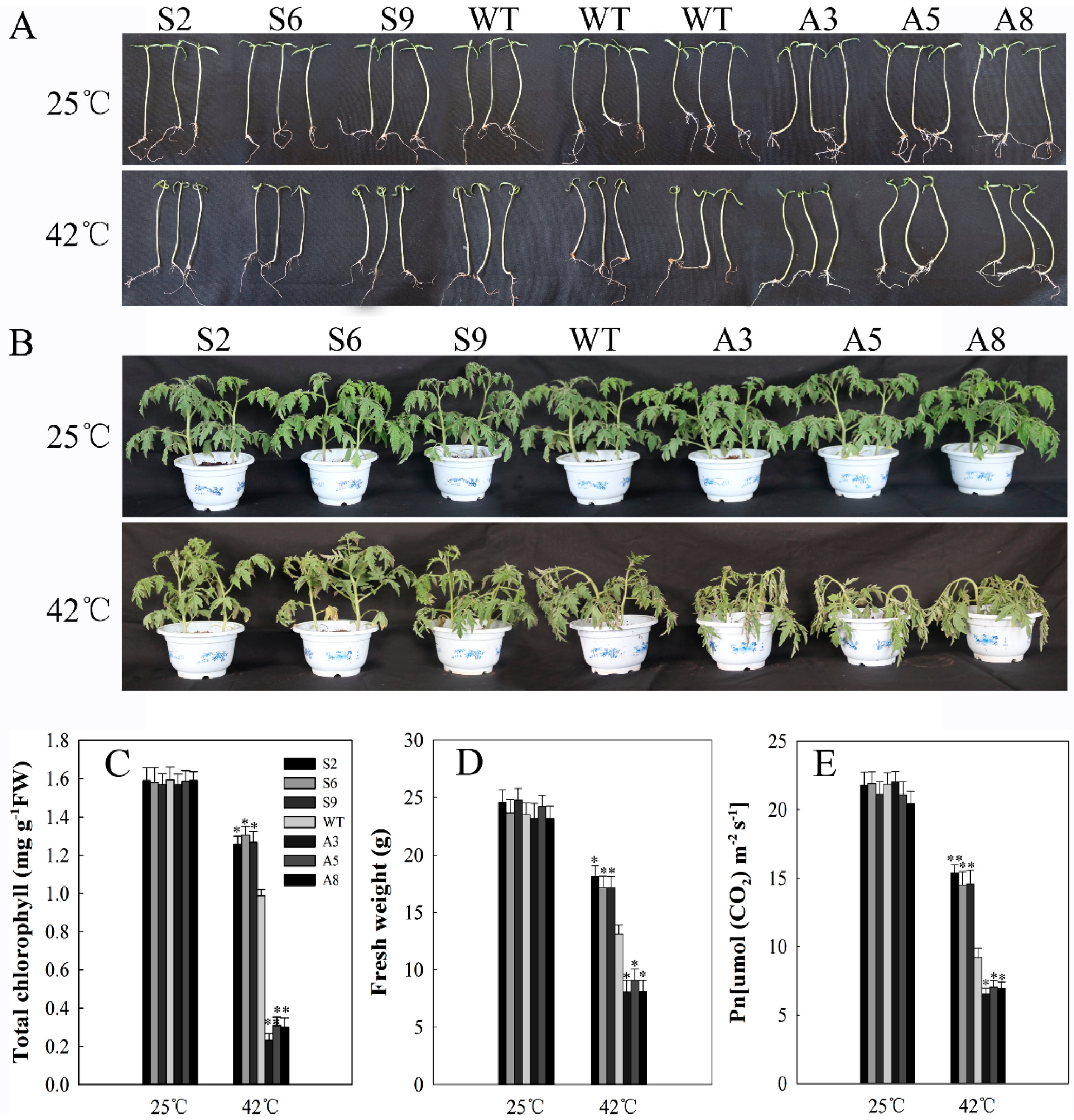
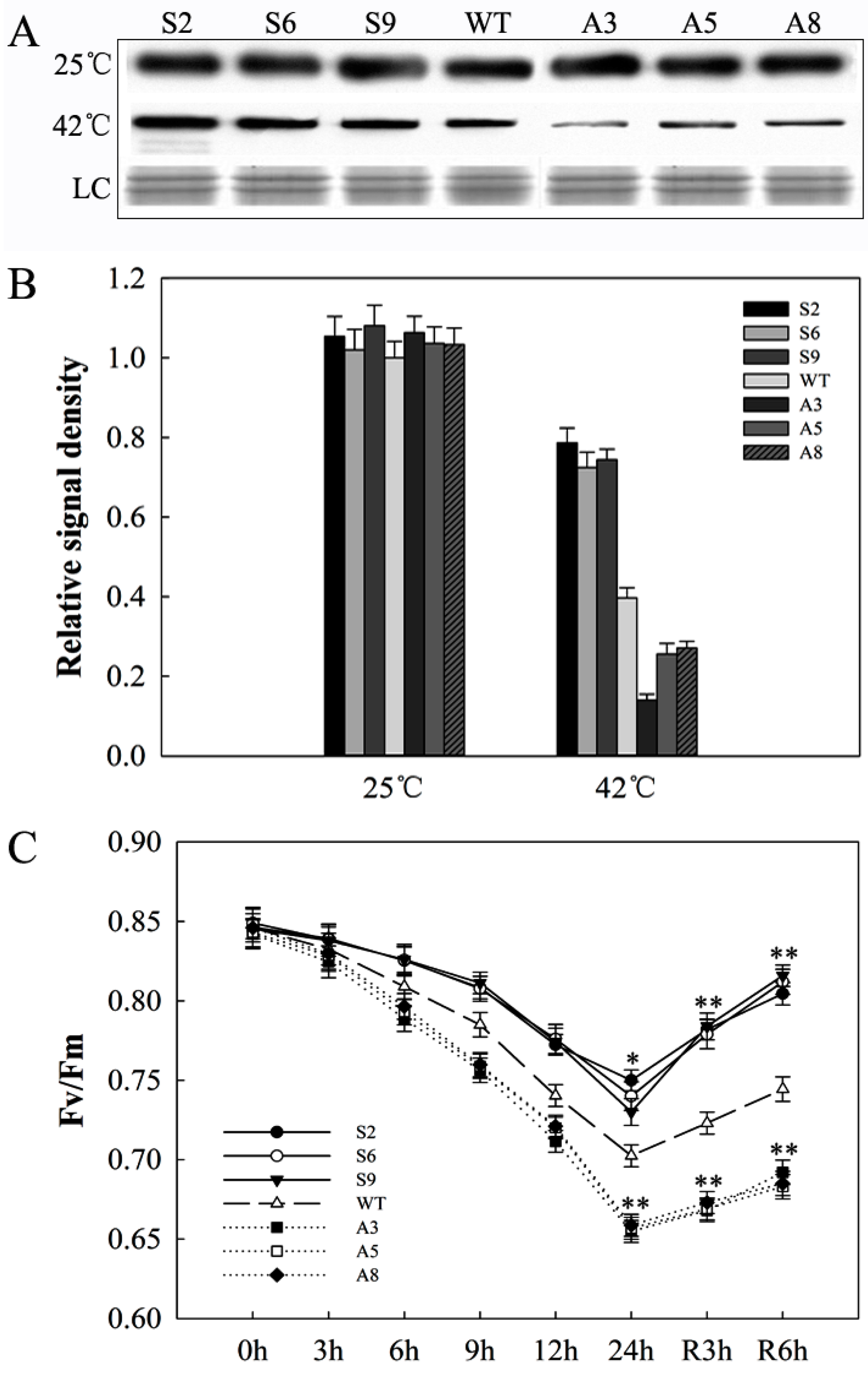
2.5. SlDnaJ20 Overexpression Enhanced Heat Stress Resistance
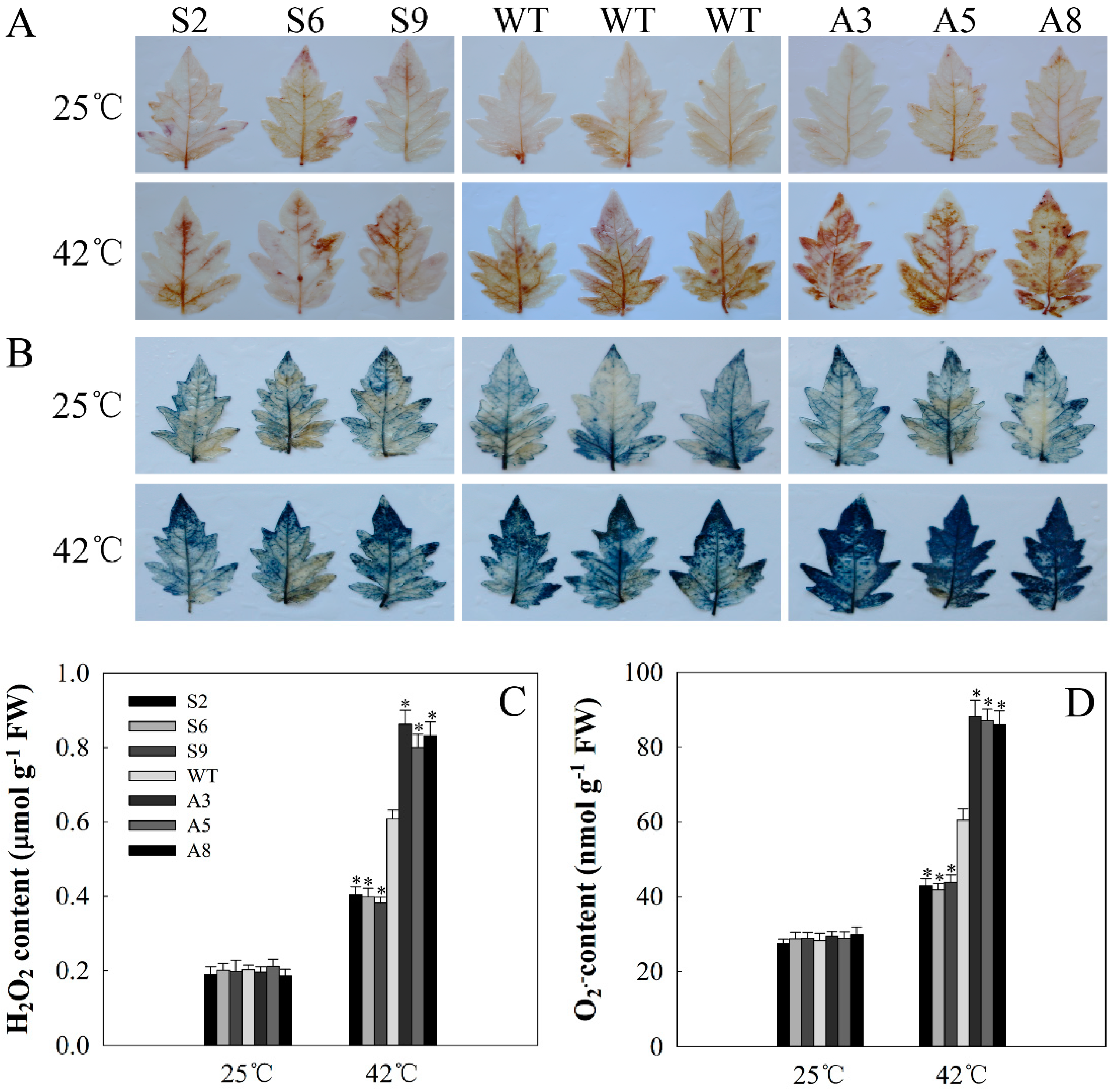
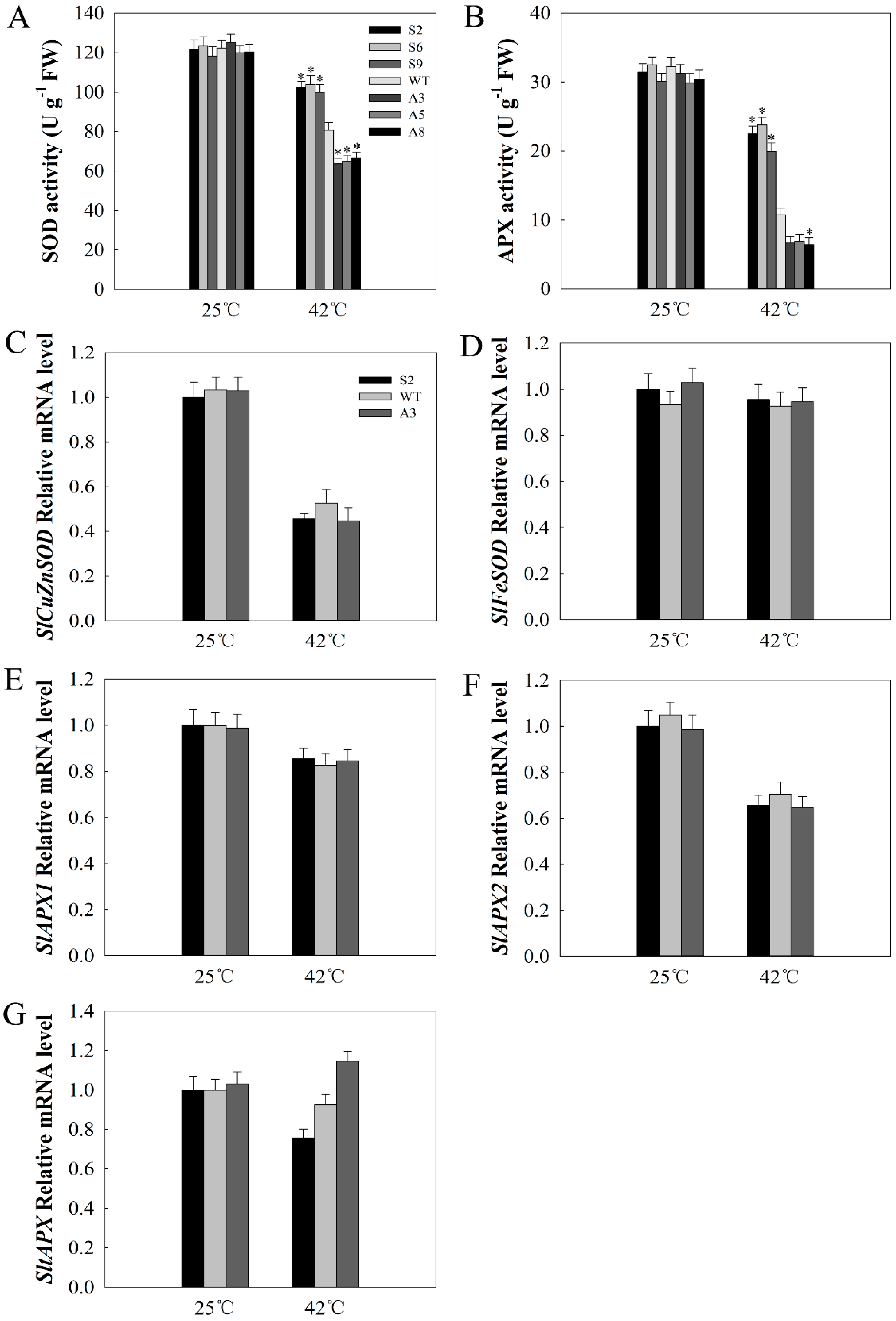
2.6. SlDnaJ20 Overexpression Alleviates ROS Accumulation by Maintaining High Levels of SOD and APX Activities
2.7. SlDnaJ20 Overexpression Alleviates Photoinhibition of Photosystem II (PSII) under Heat Stress
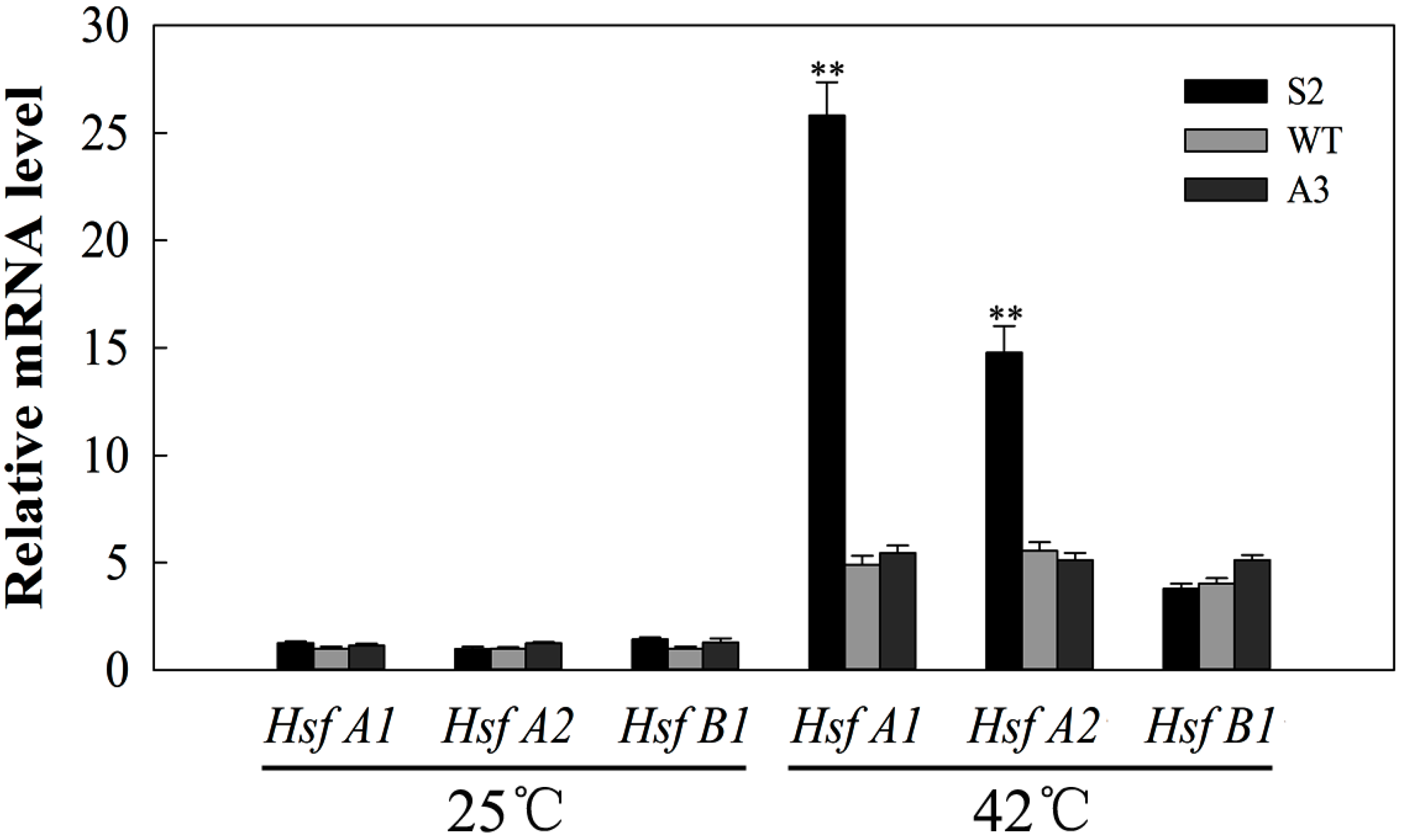
2.8. SlDnaJ20 Overexpression Promotes Expression of HSFs under Heat Stress
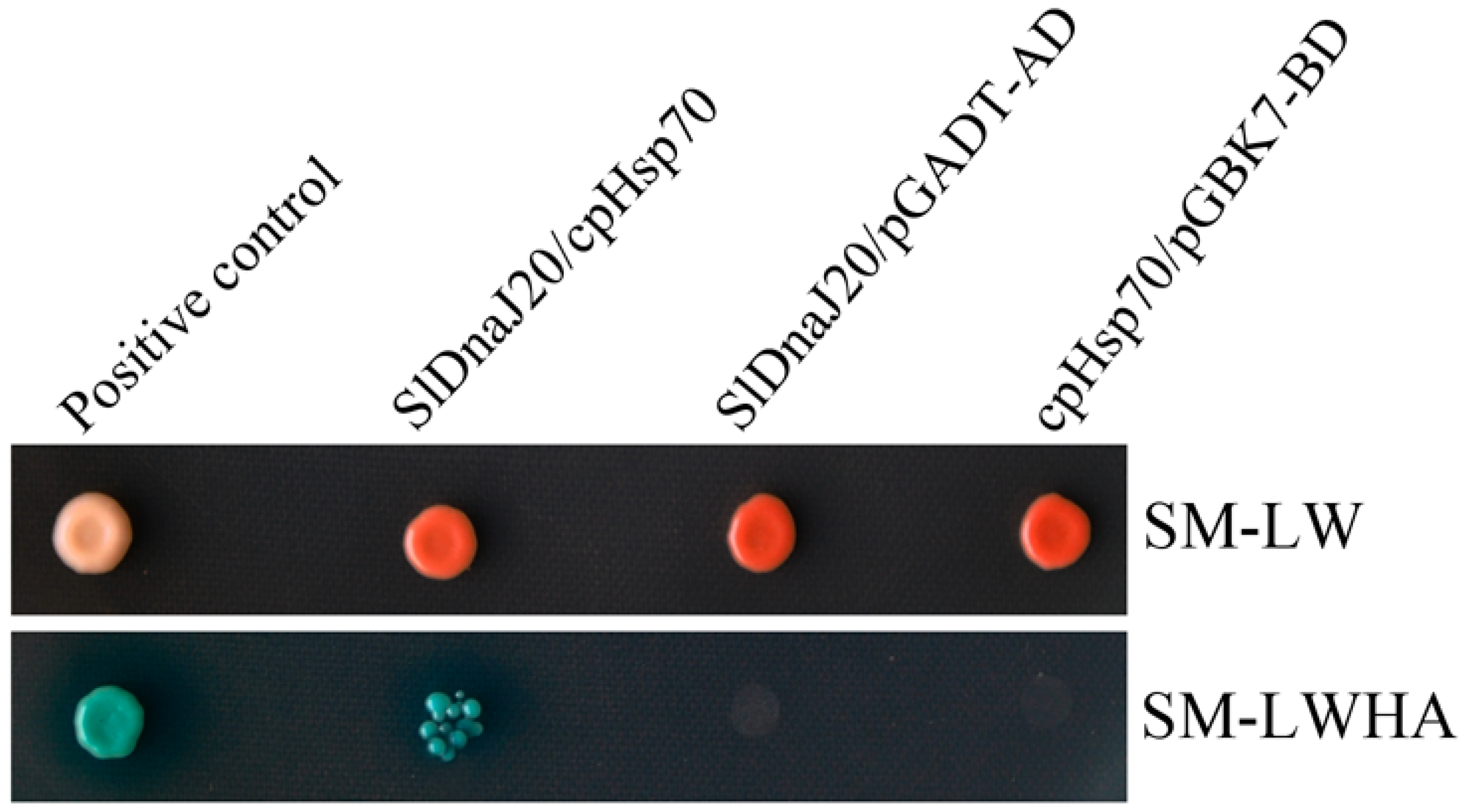
2.9. Interaction between SlDnaJ20 and Chloroplast Hsp70 (cpHsp70)
3. Discussion
4. Materials and Methods
4.1. Plant Materials, Growth Conditions, and Stress Treatments
4.2. Isolating and Sequencing of SlDnaJ20
4.3. Subcellular Localization of SlDnaJ20
4.4. Tomato Genetic Transformation and Identification
4.5. Real-Time Quantitative PCR (qPCR) Analysis
4.6. Antibody Production, Protein Acquisition, and Protein Level Analysis
4.7. Measurement of Net Photosynthetic Rate (Pn) and Chlorophyll Fluorescence
4.8. Histochemical Staining and Measurements of H2O2 and O2•−
4.9. Measurements of Chlorophyll Content and Antioxidative Enzyme Activities
4.10. Yeast Two-Hybrid Assays
4.11. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| APX | ascorbate peroxidase |
| CaMV35 S | cauliflower mosaic virus 35 S |
| DAB | 3,3′-diaminobenzidine |
| EGFP | enhanced green fluorescent protein |
| H2O2 | hydrogen peroxide |
| HSF | heat-shock transcription factor |
| HSP | heat-shock protein |
| NBT | nitroblue tetrazolium |
| O2•− | superoxide anion radical |
| PSII | photosystem II |
| qPCR | quantitative real-time polymerase chain reaction |
| ROS | reactive oxygen species |
| sHSP | small heat-shock protein |
| SlDnaJ20 | Solanum lycopersicum DnaJ protein 20 |
| SOD | superoxide dismutase |
References
- Xue, Y.; Xiong, A.; Li, X.; Zha, D.; Yao, Q. Overexpression of heat shock protein gene Hsp26 in Arabidopsis thaliana enhances heat tolerance. Biol. Plant 2010, 54, 105–111. [Google Scholar] [CrossRef]
- Sun, L.P.; Liu, Y.; Kong, X.P.; Zhang, D.; Pan, J.W.; Zhou, Y.; Wang, L.D.; Li, Q.; Yang, X.H. ZmHSP16.9, a cytosolic class I small heat shock protein in maize (Zea mays), confers heat tolerance in transgenic tobacco. Plant Cell Rep. 2012, 31, 1473–1484. [Google Scholar] [CrossRef] [PubMed]
- Lindquist, S.; Craig, E.A. The heat-shock proteins. Annu. Rev. Genet. 1988, 22, 631–677. [Google Scholar] [CrossRef]
- Hendrick, J.P.; Hartl, F.U. Molecular chaperone functions of heat-shock proteins. Annu. Rev. Biochem. 1993, 62, 349–384. [Google Scholar] [CrossRef] [PubMed]
- Hennessy, F.; Nicoll, W.S.; Zimmermann, R.; Cheetham, M.E.; Blatch, G.L. Not all J domains are created equal: Implications for the specificity of Hsp40-Hsp70 interactions. Protein Sci. 2005, 14, 1697–1709. [Google Scholar] [CrossRef] [PubMed]
- Craig, E.A.; Huang, P.; Aron, R.; Andrew, A. The diverse roles of J proteins, the obligate Hsp70 co-chaperone. Rev. Physiol. Biochem. Pharmacol. 2006, 156, 1–21. [Google Scholar] [PubMed]
- Caplan, A.J.; Cyr, D.M.; Douglas, M.G. Eukaryotic homologues of Escherichia coli dnaJ: A diverse protein family that functions with hsp70 stress proteins. Mol. Biol. Cell 1993, 4, 555–563. [Google Scholar] [CrossRef]
- Silver, P.A.; Way, J.C. Eukaryotic DnaJ homologs and the specificity of Hsp70 activity. Cell 1993, 74, 5–6. [Google Scholar] [CrossRef]
- Kelley, W.L. The J-domain family and the recruitment of chaperone power. Trends Biochem. Sci. 1998, 23, 222–227. [Google Scholar] [CrossRef]
- Martinez-Yamout, M.; Legge, G.B.; Zhang, O.; Wright, P.E.; Dyson, H.J. Solution structure of the cysteine-rich domain of the Escherichia coli chaperone protein DnaJ. J. Mol. Biol. 2000, 300, 805–818. [Google Scholar] [CrossRef]
- Bork, P.; Sander, C.; Valencia, A.; Bukau, B. A module of the DnaJ heat shock proteins found in malaria parasites. Trends Biochem. Sci. 1992, 17, 129. [Google Scholar] [CrossRef]
- Cheetham, M.E.; Caplan, A.J. Structure, function and evolution of DnaJ: Conservation and adaptation of chaperone function. Cell Stress Chaperon. 1998, 3, 28–36. [Google Scholar] [CrossRef]
- Li, G.L.; Chang, H.; Li, B.; Zhou, W.; Sun, D.Y.; Zhou, R.G. The roles of the atDjA2 and atDjA3 molecular chaperone proteins in improving thermotolerance of Arabidopsis thaliana seedlings. Plant Sci. 2007, 173, 408–416. [Google Scholar] [CrossRef]
- Yang, K.Z.; Xia, C.; Liu, X.L.; Dou, X.Y.; Wang, W.; Chen, L.Q.; Zhang, X.Q.; Xie, L.F.; He, L.Y.; Ma, X.; et al. A mutation in Thermosensitive Male Sterile 1, encoding a heat shock protein with DnaJ and PDI domains, leads to thermosensitive gametophytic male sterility in Arabidopsis. Plant J. 2009, 57, 870–882. [Google Scholar] [CrossRef] [PubMed]
- Zhou, W.; Zhou, T.; Li, M.X.; Zhao, C.L.; Jia, N.; Wang, X.X.; Sun, Y.Z.; Li, G.L.; Xu, M.; Zhou, R.G.; et al. The Arabidopsis J-protein AtDjB1 facilitates thermotolerance by protecting cells against heat-induced oxidative damage. New Phytol. 2012, 194, 364–378. [Google Scholar] [CrossRef]
- Kong, F.Y.; Deng, Y.S.; Wang, G.D.; Wang, J.R.; Liang, X.Q.; Meng, Q.W. LeCDJ1, a chloroplast DnaJ protein, facilitates heat tolerance in transgenic tomatoes. J. Integr. Plant Biol. 2014, 56, 63–74. [Google Scholar] [CrossRef]
- Wang, G.D.; Kong, F.Y.; Zhang, S.; Meng, X.; Wang, Y.; Meng, Q.W. A tomato chloroplast-targeted DnaJ protein protects Rubisco activity under heat stress. J. Exp. Bot. 2015, 66, 3027–3040. [Google Scholar] [CrossRef]
- Murata, N.; Takahashi, S.; Nishiyama, Y.; Allakhverdiev, S.I. Photoinhibition of photosystem II under environmental stress. Biochim. Biophys. Acta 2007, 1767, 414–421. [Google Scholar] [CrossRef]
- Wang, G.D.; Cai, G.H.; Kong, F.Y.; Deng, Y.S.; Ma, N.; Meng, Q.W. Overexpression of tomato chloroplast-targeted DnaJ protein enhances tolerance to drought stress and resistance to Pseudomonas solanacearum in transgenic tobacco. Plant Physiol. Bioch. 2014, 82, 95–104. [Google Scholar] [CrossRef]
- Scharf, K.D.; Rose, S.; Zott, W.; Schoff, F.; Nover, L. Three tomato genes code for heat stress transcription factors with a region of remarkable homology to the DNA binding domain of the yeast HSF. EMBO J. 1990, 9, 4495–4501. [Google Scholar] [CrossRef]
- Czarnecka-Verner, E.; Yuan, C.X.; Fox, P.C.; Gurley, W.B. Isolation and characterization of six heat shock transcription factor cDNA clones from soybean. Plant Mol. Biol. 1995, 29, 37–51. [Google Scholar] [CrossRef] [PubMed]
- Nover, L.; Bharti, K.; Doring, P.; Mishra, S.K.; Ganguli, A.; Scharf, K.D. Arabidopsis and the heat stress transcription factor world: How many heat stress transcription factors do we need. Cell Stress Chaperon. 2001, 6, 177–189. [Google Scholar] [CrossRef]
- Kotak, S.; Port, M.; Ganguli, A.; Bicker, F.; Koskull-Döring, P. Characterization of C-terminal domains of Arabidopsis heat stress transcription factors (Hsfs) and identification of a new signature combination of plant class A Hsfs with AHA and NES motifs essential for activator function and intracellular localization. Plant J. 2004, 39, 98–112. [Google Scholar] [CrossRef] [PubMed]
- Baniwal, S.K.; Bharti, K.; Chan, K.Y.; Fauth, M.; Ganguli, A.S.; Kotak, S.K.; Mishra, L.; Nover, M.; Port, K.D.; Scharf, K.D.; et al. Heat stress response in plants: A complex game with chaperones and more than twenty heat stress transcription factors. J. Biosci. 2004, 29, 471–487. [Google Scholar] [CrossRef] [PubMed]
- Chan-Schaminet, K.Y.; Baniwal, S.K.; Bublak, D.; Nover, L.; Scharf, K.D. Specific interaction between tomato HsfA1 and HsfA2 creates hetero-oligomeric superactivator complexes for synergistic activation of heat stress gene expression. J. Biol. Chem. 2009, 284, 20848–20857. [Google Scholar] [CrossRef] [PubMed]
- Hahn, A.; Bublak, D.; Schleiff, E.; Scharf, K.D. Crosstalk between Hsp90 and Hsp70 chaperones and heat stress transcription factors in tomato. Plant Cell 2011, 23, 741–755. [Google Scholar] [CrossRef] [PubMed]
- Miernyk, J.A. The J-domain proteins of Arabidopsis thaliana: An unexpectedly large and diverse family of chaperones. Cell Stress Chaperon. 2001, 6, 209–218. [Google Scholar] [CrossRef]
- Qiu, X.B.; Shao, Y.M.; Miao, S.; Wang, L. The diversity of the DnaJ/Hsp40 family, the crucial partners for Hsp70 chaperones. Cell. Mol. Life Sci. 2006, 63, 2560–2570. [Google Scholar] [CrossRef]
- Bekh-Ochir, D.; Shimada, S.; Yamagami, A.; Kanda, S.; Ogawa, K.; Nakazawa, M.; Matsui, M.; Sakuta, M.; Osada, H.; Asami, T.; et al. A novel mitochondrial DnaJ/Hsp40 family protein BIL2 promotes plant growth and resistance against environmental stress in brassinosteroid signaling. Planta 2013, 237, 1509–1525. [Google Scholar] [CrossRef]
- Fristedt, R.; Williams-Carrie, R.; Merchant, S.S.; Barkan, A. A thylakoid membrane protein harboring a DnaJ-type zinc finger domain is required for photosystem I accumulation in plants. J. Biol. Chem. 2014, 289, 30657–30667. [Google Scholar] [CrossRef]
- Kong, F.Y.; Deng, Y.S.; Zhou, B.; Wang, G.D.; Wang, Y.; Meng, Q.W. A chloroplast-targeted DnaJ protein contributes to maintenance of photosystem II under chilling stress. J. Exp. Bot. 2014, 65, 143–158. [Google Scholar] [CrossRef]
- Timperio, A.M.; Egidi, M.G.; Zolla, L. Proteomics applied on plant abiotic stresses: Role of heat shock proteins (HSP). J. Proteomics 2008, 71, 391–411. [Google Scholar] [CrossRef]
- Salvucci, M.E. Association of Rubisco activase with chaperonin-60b: A possible mechanism for protecting photosynthesis during heat stress. J. Exp. Bot. 2008, 59, 1923–1933. [Google Scholar] [CrossRef]
- Sharkey, T.D.; Schrader, S.M. High temperature stress. In Physiology and Molecular Biology of Stress Tolerance in Plants; Rao, K.V.M., Raghavendra, A.S., Reddy, K.J., Eds.; Springer: Dordrecht, The Netherlands, 2008; pp. 101–130. [Google Scholar]
- Zhang, R.; Sharkey, T.D. Photosynthetic electron transport and proton flux under moderate heat stress. Photosynth. Res. 2009, 100, 29–43. [Google Scholar] [CrossRef] [PubMed]
- Vandenabeele, J.; Dat, S.; Vranová, E.; Van Montagu, M.; Inzé, D.; Van Breusegem, F. Dual action of the active oxygen species during plant stress responses. CMLS Cell. Mol. Life Sci. 2000, 57, 779–795. [Google Scholar]
- Pospíšil, P. Production of reactive oxygen species by photosystem II. Biochim. Biophys. Acta 2009, 1787, 1151–1160. [Google Scholar] [CrossRef] [PubMed]
- Mishra, S.K.; Tripp, J.; Winkelhaus, S.; Tschiersch, B.; Theres, K.; Nover, L.; Scharf, K.D. In the complex family of heat stress transcription factors, HsfA1 has a unique role as master regulator of thermotolerance in tomato. Gene Dev. 2002, 16, 1555–1567. [Google Scholar] [CrossRef] [PubMed]
- Hermosa, M.R.; Cardoza, R.E.; Gutierrez, S.; Nicolas, C.; Monte, E. Transgenic expression of the trichoderma harzianum hsp70 gene increases Arabidopsis resistance to heat and other abiotic stresses. J. Plant Physiol. 2010, 167, 659–665. [Google Scholar]
- Chauhan, H.; Khurana, N.; Nijhavan, A.; Khurana, J.P.; Khurana, P. The wheat chloroplastic small heat shock protein (sHSP26) is involved in seed maturation and germination and imparts tolerance to heat stress. Plant Cell Environ. 2012, 35, 1912–1931. [Google Scholar] [CrossRef]
- Wang, J.; Wang, J.Z.; Lu, Y.Z.; Fang, Y.; Gao, X.; Wang, Z.H.; Zheng, W.J.; Xu, S.B. The heat responsive wheat TaRAD23 rescues developmental and thermotolerant defects of the rad23b mutant in Arabidopsis thaliana. Plant Sci. 2018, 274, 23–31. [Google Scholar] [CrossRef]
- Zhang, W.; Zhou, R.G.; Gao, Y.J.; Zheng, S.Z.; Xu, P.; Zhang, S.Q.; Sun, D.Y. Molecular and genetic evidence for the key role of AtCaM3 in heat-shock signal transduction in Arabidopsis. Plant Physiol. 2009, 149, 1773–1784. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.X.; Paakkarinen, V.; van Wijk, K.J.; Aro, E.M. Cotranslational assembly of the D1 protein into photosystem II. J. Biol. Chem. 1999, 274, 16062–16067. [Google Scholar] [CrossRef] [PubMed]
- Jiang, C.D.; Gao, H.Y.; Zou, Q.; Jiang, G.M.; Li, L.H. Leaf orientation, photorespiration and xanthophyll cycle protect young soybean leaves against high irradiance in field. Environ. Exp. Bot. 2006, 55, 87–96. [Google Scholar] [CrossRef]
- Pan, J.W.; Zhang, M.Y.; Kong, X.P.; Xing, X.; Liu, Y.; Zhou, Y.; Li, D.Q. ZmMPK17, a novel maize group D MAP kinase gene, is involved in multiple stress responses. Planta 2012, 235, 661–676. [Google Scholar] [CrossRef] [PubMed]
- Zong, X.J.; Li, D.P.; Gu, L.K.; Liu, L.X.; Hu, X.L.; Li, D.Q. Abscisic acid and hydrogen peroxide induce a novel maize group C MAP kinase gene, ZmMPK7, which is responsible for the removal of reactive oxygen species. Planta 2009, 229, 485–495. [Google Scholar] [CrossRef]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, G.; Cai, G.; Xu, N.; Zhang, L.; Sun, X.; Guan, J.; Meng, Q. Novel DnaJ Protein Facilitates Thermotolerance of Transgenic Tomatoes. Int. J. Mol. Sci. 2019, 20, 367. https://doi.org/10.3390/ijms20020367
Wang G, Cai G, Xu N, Zhang L, Sun X, Guan J, Meng Q. Novel DnaJ Protein Facilitates Thermotolerance of Transgenic Tomatoes. International Journal of Molecular Sciences. 2019; 20(2):367. https://doi.org/10.3390/ijms20020367
Chicago/Turabian StyleWang, Guodong, Guohua Cai, Na Xu, Litao Zhang, Xiuling Sun, Jing Guan, and Qingwei Meng. 2019. "Novel DnaJ Protein Facilitates Thermotolerance of Transgenic Tomatoes" International Journal of Molecular Sciences 20, no. 2: 367. https://doi.org/10.3390/ijms20020367
APA StyleWang, G., Cai, G., Xu, N., Zhang, L., Sun, X., Guan, J., & Meng, Q. (2019). Novel DnaJ Protein Facilitates Thermotolerance of Transgenic Tomatoes. International Journal of Molecular Sciences, 20(2), 367. https://doi.org/10.3390/ijms20020367



