Regulated Assembly of LPS, Its Structural Alterations and Cellular Response to LPS Defects
Abstract
1. Introduction
2. Regulatory Steps in LPS Biosynthesis
3. Essentiality of LPS and the Minimal LPS Structure
4. Regulation of Synthesis of UDP-GlcNAc-Precursor for LPS Biosynthesis
5. Coupled Regulation of LPS and Phospholipids–Regulation of Amounts of Kdo2-Lipid A Synthesis
6. Assembly of LPS Requires LapB
7. Transport of LPS
7.1. MsbA-Mediated Transport of LPS Across the Inner Membrane
7.2. LPS Translocation and Assembly in the Outer Membrane
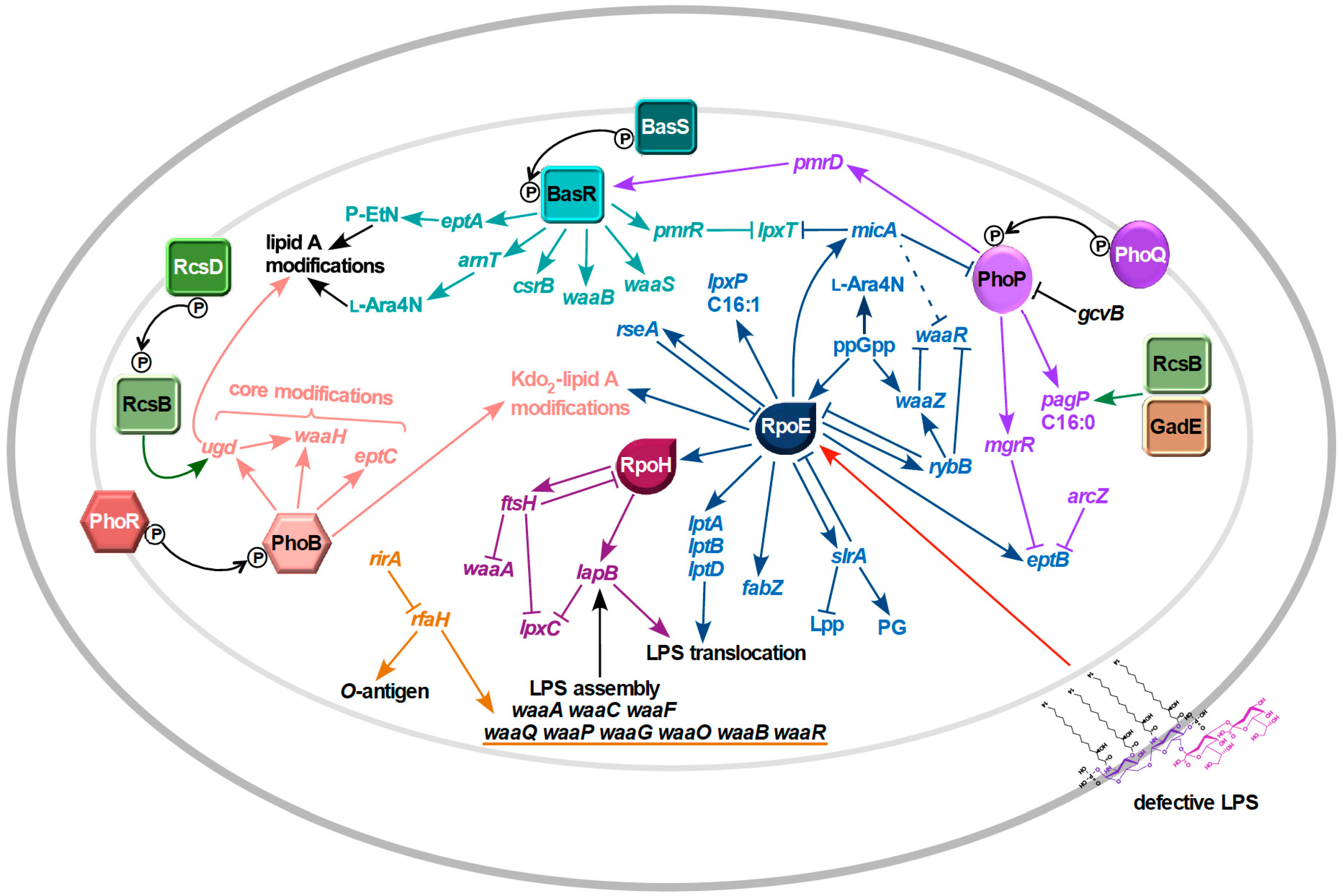
8. Regulated Structural Alterations in LPS
8.1. Regulation of Lipid A Modifications
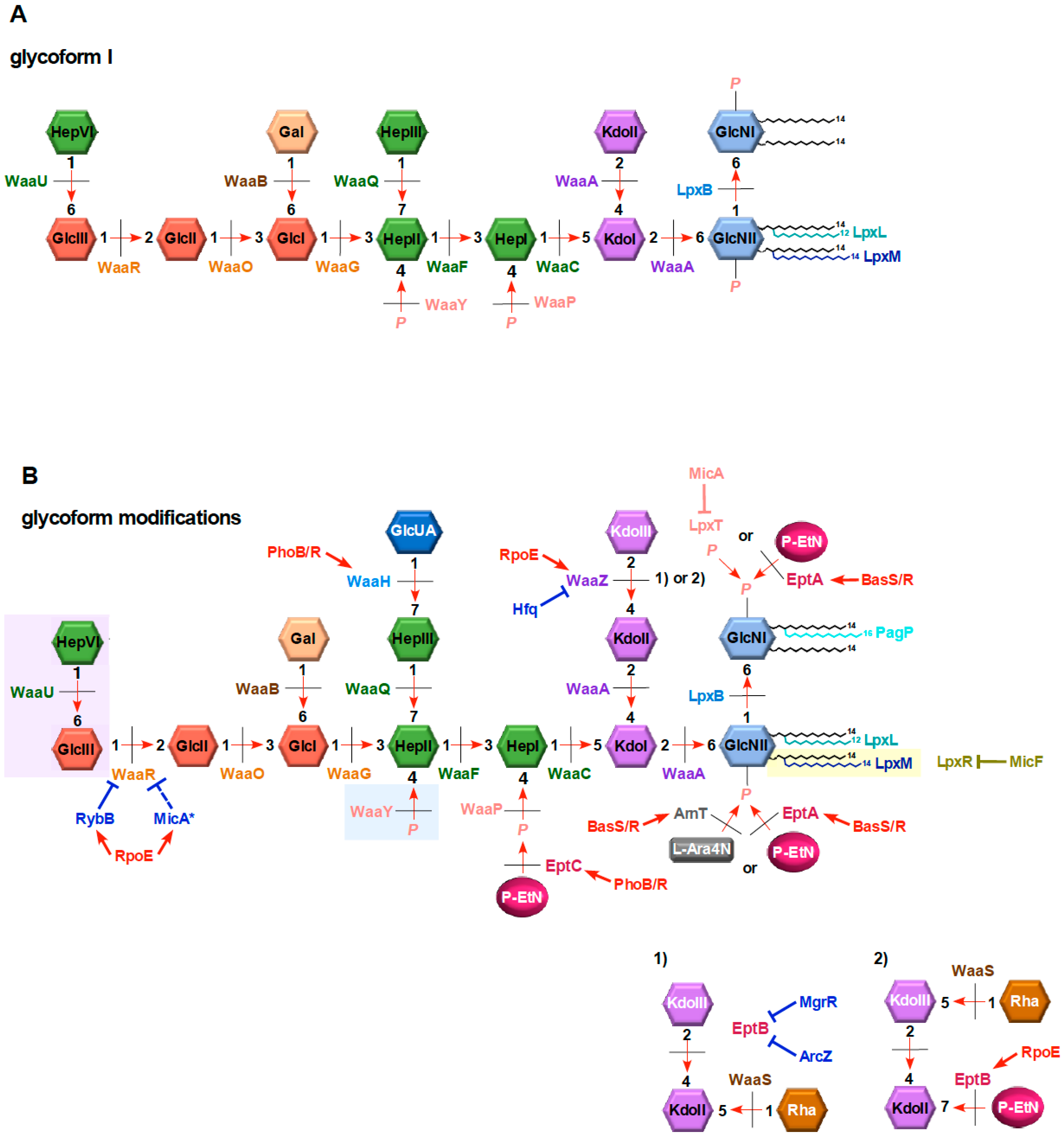
8.2. Regulation of Inner Core Modifications and Switches Between Different Glycoforms
8.3. Modifications in the Heptose Region of the LPS Inner Core
8.4. Glycoform Switches
8.5. Transcriptional Regulation of Major LPS Biosynthetic Operons by RfaH
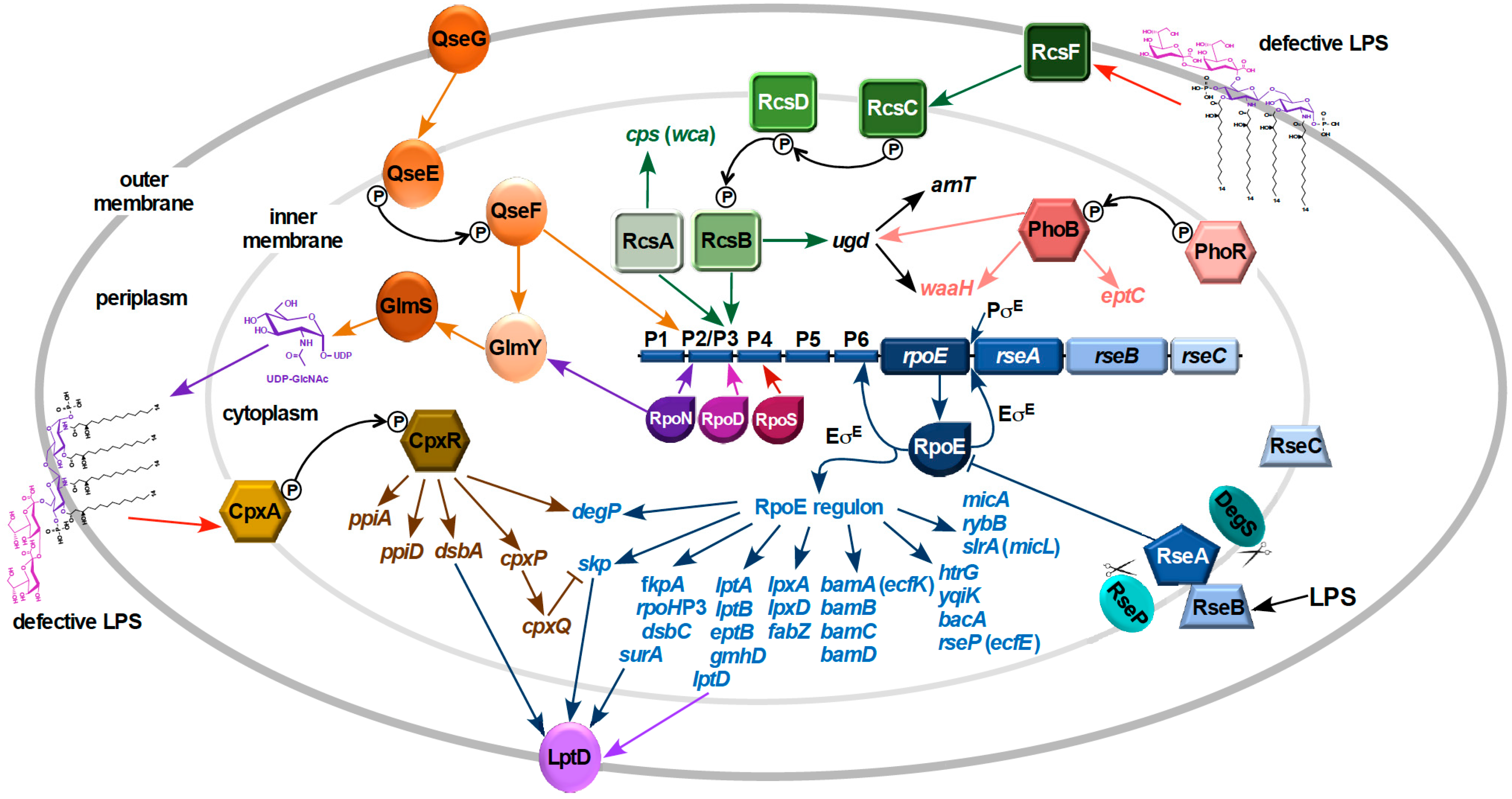
9. Signal Transduction in Response to LPS Defects
10. Mechanism of Signal Transduction
11. Conclusions
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| AMV | ammonium metavanadate |
| GlcUA | glucuronic acid |
| IM | inner membrane |
| Kdo | 3-deoxy-α-d-manno-oct-2-ulsonic acid |
| l-Ara4N | 4-amino-4-deoxy-l-arabinose |
| LPS | lipopolysaccharide |
| OM | outer membrane |
| OMP | outer membrane protein |
| ops | operon polarity suppressor |
| P-EtN | phosphoethanolamine |
| sRNA | noncoding small regulatory RNA |
| TCS | two-component system |
| TPR | tetratricopeptide repeats |
| UTR | untranslated region |
References
- Nikaido, H. Outer membrane. In Escherichia coli and Salmonella: Cellular and Molecular Biology, 2nd ed.; Neidhardt, F.C., Curtiss, R., Ingraham, J.L., Eds.; American Society for Microbiology Press: Washington, DC, USA, 1996; Volume 1, pp. 29–47. [Google Scholar]
- Anderson, M.S.; Bulawa, C.E.; Raetz, C.R.H. The biosynthesis of Gram-negative endotoxin. Formation of lipid A precursors from UDP-GlcNAc in extracts of Escherichia coli. J. Biol. Chem. 1985, 260, 15536–15541. [Google Scholar]
- Babinski, K.J.; Kanjilal, S.J.; Raetz, C.R.H. Accumulation of the lipid A precursor UDP-2,3-diacylglucosamine in an Escherichia coli mutant lacking the lpxH gene. J. Biol. Chem. 2002, 277, 25947–25956. [Google Scholar] [CrossRef] [PubMed]
- Raetz, C.R.H.; Whitfield, C. Lipopolysaccharide endotoxins. Annu. Rev. Biochem. 2002, 71, 635–700. [Google Scholar] [CrossRef]
- Vorachek-Warren, M.K.; Ramirez, S.; Cotter, R.J.; Raetz, C.R.H. A triple mutant of Escherichia coli lacking secondary acyl chains on lipid A. J. Biol. Chem. 2002, 277, 14194–14205. [Google Scholar] [CrossRef] [PubMed]
- Carty, S.M.; Sreekumar, K.R.; Raetz, C.R.H. Effect of cold shock on lipid A biosynthesis in Escherichia coli. Induction at 12 °C of an acyltransferase specific for palmitoleoyl-acyl carrier protein. J. Biol. Chem. 1999, 274, 9677–9685. [Google Scholar] [CrossRef] [PubMed]
- Klein, G.; Lindner, B.; Brabetz, W.; Brade, H.; Raina, S. Escherichia coli K-12 suppressor-free mutants lacking early glycosyltransferases and late acyltransferases. Minimal lipopolysaccharide structure and induction of envelope stress response. J. Biol. Chem. 2009, 284, 15369–15389. [Google Scholar] [CrossRef]
- Holst, O. The structures of core regions from enterobacterial lipopolysaccharides—An update. FEMS Microbiol. Lett. 2007, 271, 3–11. [Google Scholar] [CrossRef]
- Klein, G.; Kobylak, N.; Lindner, B.; Stupak, A.; Raina, S. Assembly of lipopolysaccharide in Escherichia coli requires the essential LapB heat shock protein. J. Biol. Chem. 2014, 289, 14829–14853. [Google Scholar] [CrossRef]
- Klein, G.; Raina, S. Small regulatory bacterial RNAs regulating the envelope stress response. Biochem. Soc. Trans. 2017, 45, 417–425. [Google Scholar] [CrossRef] [PubMed]
- Klein, G.; Stupak, A.; Biernacka, D.; Wojtkiewicz, P.; Lindner, B.; Raina, S. Multiple transcriptional factors regulate transcription of the rpoE gene in Escherichia coli under different growth conditions and when the lipopolysaccharide biosynthesis is defective. J. Biol. Chem. 2016, 291, 22999–23019. [Google Scholar] [CrossRef]
- Bailey, M.J.; Hughes, C.; Koronakis, V. RfaH and the ops element, components of a novel system controlling bacterial transcription elongation. Mol. Microbiol. 1997, 26, 845–851. [Google Scholar] [CrossRef]
- Klein, G.; Raina, S. Regulated control of the assembly and diversity of LPS by noncoding sRNAs. Biomed. Res. Int. 2015, 2015, 153561. [Google Scholar] [CrossRef]
- Dartigalongue, C.; Missiakas, D.; Raina, S. Characterization of the Escherichia coli σE regulon. J. Biol. Chem. 2001, 276, 20866–20875. [Google Scholar] [CrossRef] [PubMed]
- Henry, R.; Vithanage, N.; Harrison, P.; Seemann, T.; Coutts, S.; Moffatt, J.H.; Nation, R.L.; Li, J.; Harper, M.; Adler, B.; et al. Colistin-resistant, lipopolysaccharide-deficient Acinetobacter baumannii responds to lipopolysaccharide loss through increased expression of genes involved in the synthesis and transport of lipoproteins, phospholipids, and poly-β-1,6-N-acetylglucosamine. Antimicrob. Agents Chemother. 2012, 56, 59–69. [Google Scholar] [CrossRef] [PubMed]
- Steeghs, L.; den Hartog, R.; den Boer, A.; Zomer, B.; Roholl, P.; van der Ley, P. Meningitis bacterium is viable without endotoxin. Nature 1998, 392, 449–450. [Google Scholar] [CrossRef] [PubMed]
- Missiakas, D.; Betton, J.M.; Raina, S. New components of protein folding in extracytoplasmic compartments of Escherichia coli SurA, FkpA and Skp/OmpH. Mol. Microbiol. 1996, 21, 871–884. [Google Scholar] [CrossRef] [PubMed]
- Crhanova, M.; Malcova, M.; Mazgajova, M.; Karasova, D.; Sebkova, A.; Fucikova, A.; Bortlicek, Z.; Pilousova, L.; Kyrova, K.; Dekanova, M.; et al. LPS structure influences protein secretion in Salmonella enterica. Vet. Microbiol. 2011, 152, 131–137. [Google Scholar] [CrossRef]
- Ebbensgaard, A.; Mordhorst, H.; Aarestrup, F.M.; Hansen, E.B. The role of outer membrane proteins and lipopolysaccharides for the sensitivity of Escherichia coli to antimicrobial peptides. Front. Microbiol. 2018. [Google Scholar] [CrossRef]
- Vivijs, B.; Aertsen, A.; Michiels, C.W. Identification of genes required for growth of Escherichia coli MG1655 at moderately low pH. Front. Microbiol. 2016, 7, 1672. [Google Scholar] [CrossRef]
- Wang, Z.; Wang, J.; Ren, G.; Li, Y.; Wang, X. Deletion of the genes waaC, waaF, or waaG in Escherichia coli W3110 disables the flagella biosynthesis. J. Basic Microbiol. 2016, 56, 1021–1035. [Google Scholar] [CrossRef]
- Schwalm, J.; Mahoney, T.F.; Soltes, G.R.; Silhavy, T.J. Role for Skp in LptD assembly in Escherichia coli. J. Bacteriol. 2013, 195, 3734–3742. [Google Scholar] [CrossRef] [PubMed]
- Murata, M.; Fujimoto, H.; Nishimura, K.; Charoensuk, K.; Nagamitsu, H.; Raina, S.; Kosaka, T.; Oshima, T.; Ogasawara, N.; Yamada, M. Molecular strategy for survival at a critical high temperature in Escherichia coli. PLoS ONE 2011, 6, e20063. [Google Scholar] [CrossRef] [PubMed]
- Yethon, J.A.; Heinrichs, D.E.; Monteiro, M.A.; Perry, M.B.; Whitfield, C. Involvement of waaY, waaQ, and waaP in the modification of Escherichia coli lipopolysaccharide and their role in the formation of a stable outer membrane. J. Biol. Chem. 1998, 273, 26310–26316. [Google Scholar] [CrossRef]
- Reynolds, C.M.; Raetz, C.R.H. Replacement of lipopolysaccharide with free lipid A molecules in Escherichia coli mutants lacking all core sugars. Biochemistry 2009, 48, 9627–9640. [Google Scholar] [CrossRef]
- Urban, J.H.; Vogel, J. Two seemingly homologous noncoding RNAs act hierarchically to activate glmS mRNA translation. PLoS Biol. 2008, 6, e64. [Google Scholar] [CrossRef] [PubMed]
- Göpel, Y.; Papenfort, K.; Reichenbach, B.; Vogel, J.; Görke, B. Targeted decay of a regulatory small RNA by an adaptor protein for RNase E and counteraction by an anti-adaptor RNA. Genes Dev. 2013, 27, 552–564. [Google Scholar] [CrossRef]
- Reichenbach, B.; Göpel, Y.; Görke, B. Dual control by perfectly overlapping σ54- and σ70-promoters adjusts small RNA GlmY expression to different environmental signals. Mol. Microbiol. 2009, 74, 1054–1070. [Google Scholar] [CrossRef] [PubMed]
- Ogura, T.; Inoue, K.; Tatsuta, T.; Suzaki, T.; Karata, K.; Young, K.; Su, L.H.; Fierke, C.A.; Jackman, J.E.; Raetz, C.R.H.; et al. Balanced biosynthesis of major membrane components through regulated degradation of the committed enzyme of lipid A biosynthesis by the protease FtsH (HflB) in Escherichia coli. Mol. Microbiol. 1999, 31, 833–844. [Google Scholar] [CrossRef]
- Führer, F.; Langklotz, S.; Narberhaus, F. The C-terminal end of LpxC is required for degradation by the FtsH protease. Mol. Microbiol. 2006, 59, 1025–1036. [Google Scholar] [CrossRef] [PubMed]
- Mahalakshmi, S.; Sunayana, M.R.; Saisree, L.; Reddy, M. yciM is an essential gene required for regulation of lipopolysaccharide synthesis in Escherichia coli. Mol. Microbiol. 2014, 91, 145–157. [Google Scholar] [CrossRef] [PubMed]
- Zeng, D.; Zhao, J.; Chung, H.S.; Guan, Z.; Raetz, C.R.H.; Zhou, P. Mutants resistant to LpxC inhibitors by rebalancing cellular homeostasis. J. Biol. Chem. 2013, 288, 5475–5486. [Google Scholar] [CrossRef] [PubMed]
- Guo, M.S.; Updegrove, T.B.; Gogol, E.B.; Shabalina, S.A.; Gross, C.A.; Storz, G. MicL, a new σE-dependent sRNA, combats envelope stress by repressing synthesis of Lpp, the major outer membrane lipoprotein. Genes Dev. 2014, 28, 1620–1634. [Google Scholar] [CrossRef] [PubMed]
- Metzger IV, L.E.; Raetz, C.R.H. Purification and characterization of the lipid A disaccharide synthase (LpxB) from Escherichia coli, a peripheral membrane protein. Biochemistry 2009, 48, 11559–11571. [Google Scholar] [CrossRef] [PubMed]
- Emiola, A.; Andrews, S.S.; Heller, C.; George, J. Crosstalk between the lipopolysaccharide and phospholipid pathways during outer membrane biogenesis in Escherichia coli. Proc. Natl. Acad. Sci. USA 2016, 113, 3108–3113. [Google Scholar] [CrossRef]
- Katz, C.; Ron, E.Z. Dual role of FtsH in regulating lipopolysaccharide biosynthesis in Escherichia coli. J. Bacteriol. 2008, 190, 7117–7122. [Google Scholar] [CrossRef] [PubMed]
- Prince, C.; Jia, Z. An unexpected duo: Rubredoxin binds nine TPR motifs to form LapB, an essential regulator of lipopolysaccharide synthesis. Structure 2015, 23, 1500–1506. [Google Scholar] [CrossRef]
- Putker, F.; Grutsch, A.; Tommassen, J.; Bos, M.P. Ght protein of Neisseria meningitidis is involved in the regulation of lipopolysaccharide biosynthesis. J. Bacteriol. 2014, 196, 780–789. [Google Scholar] [CrossRef] [PubMed]
- Moreau, P.L.; Loiseau, L. Characterization of acetic acid-detoxifying Escherichia coli evolved under phosphate starvation conditions. Microb. Cell Fact. 2016, 15, 42. [Google Scholar] [CrossRef]
- Sutterlin, H.A.; Shi, H.; May, K.L.; Miguel, A.; Khare, S.; Huang, K.C.; Silhavy, T.J. Disruption of lipid homeostasis in the Gram-negative cell envelope activates a novel cell death pathway. Proc. Natl. Acad. Sci. USA 2016, 113, E1565–E1574. [Google Scholar] [CrossRef]
- Ward, A.; Reyes, C.L.; Yu, J.; Roth, C.B.; Chang, G. Flexibility in the ABC transporter MsbA: Alternating access with a twist. Proc. Natl. Acad. Sci. USA 2007, 104, 19005–19010. [Google Scholar] [CrossRef]
- Mi, W.; Li, Y.; Yoon, S.H.; Ernst, R.K.; Walz, T.; Liao, M. Structural basis of MsbA-mediated lipopolysaccharide transport. Nature 2017, 549, 233–237. [Google Scholar] [CrossRef] [PubMed]
- Ho, H.; Miu, A.; Alexander, M.K.; Garcia, N.K.; Oh, A.; Zilberleyb, I.; Reichelt, M.; Austin, C.D.; Tam, C.; Shriver, S.; et al. Structural basis for dual-mode inhibition of the ABC transporter MsbA. Nature 2018, 557, 196–201. [Google Scholar] [CrossRef]
- Hicks, G.; Jia, Z. Structural basis for the lipopolysaccharide export activity of the bacterial lipopolysaccharide transport system. Int. J. Mol. Sci. 2018, 19, 2680. [Google Scholar] [CrossRef] [PubMed]
- Sperandeo, P.; Martorana, A.M.; Polissi, A. The lipopolysaccharide transport (Lpt) machinery: A nonconventional transporter for lipopolysaccharide assembly at the outer membrane of Gram-negative bacteria. J. Biol. Chem. 2017, 292, 17981–17990. [Google Scholar] [CrossRef] [PubMed]
- Bohl, T.E.; Aihara, H. Current progress in the structural and biochemical characterization of proteins involved in the assembly of lipopolysaccharide. Int. J. Microbiol. 2018, 2018, 5319146. [Google Scholar] [CrossRef] [PubMed]
- Okuda, S.; Sherman, D.J.; Silhavy, T.J.; Ruiz, N.; Kahne, D. Lipopolysaccharide transport and assembly at the outer membrane: The PEZ model. Nat. Rev. Microbiol. 2016, 14, 337–345. [Google Scholar] [CrossRef] [PubMed]
- Luo, Q.; Yang, X.; Yu, S.; Shi, H.; Wang, K.; Xiao, L.; Zhu, G.; Sun, C.; Li, T.; Li, D.; et al. Structural basis for lipopolysaccharide extraction by ABC transporter LptB2FG. Nat. Struct. Mol. Biol. 2017, 24, 469–474. [Google Scholar] [CrossRef]
- Sherman, D.J.; Xie, R.; Taylor, R.J.; George, A.H.; Okuda, S.; Foster, P.J.; Needleman, D.J.; Kahne, D. Lipopolysaccharide is transported to the cell surface by a membrane-to-membrane protein bridge. Science 2018, 359, 798–801. [Google Scholar] [CrossRef]
- Qiao, S.; Luo, Q.; Zhao, Y.; Zhang, X.C.; Huang, Y. Structural basis for lipopolysaccharide insertion in the bacterial outer membrane. Nature 2014, 511, 108–111. [Google Scholar] [CrossRef]
- Dong, H.; Xiang, Q.; Gu, Y.; Wang, Z.; Paterson, N.G.; Stansfeld, P.J.; He, C.; Zhang, Y.; Wang, W.; Dong, C. Structural basis for outer membrane lipopolysaccharide insertion. Nature 2014, 511, 52–56. [Google Scholar] [CrossRef]
- Missiakas, D.; Raina, S. Protein folding in the bacterial periplasm. J. Bacteriol. 1997, 179, 2465–2471. [Google Scholar] [CrossRef] [PubMed]
- Chng, S.S.; Xue, M.; Garner, R.A.; Kadokura, H.; Boyd, D.; Beckwith, J.; Kahne, D. Disulfide rearrangement triggered by translocon assembly controls lipopolysaccharide export. Science 2012, 337, 1665–1668. [Google Scholar] [CrossRef] [PubMed]
- Narita, S.; Masui, C.; Suzuki, T.; Dohmae, N.; Akiyama, Y. Protease homolog BepA (YfgC) promotes assembly and degradation of β-barrel membrane proteins in Escherichia coli. Proc. Natl. Acad. Sci. USA 2013, 110, E3612–E3621. [Google Scholar] [CrossRef] [PubMed]
- Klein, G.; Müller-Loennies, S.; Lindner, B.; Kobylak, N.; Brade, H.; Raina, S. Molecular and structural basis of inner core lipopolysaccharide alterations in Escherichia coli: Incorporation of glucuronic acid and phosphoethanolamine in the heptose region. J. Biol. Chem. 2013, 288, 8111–8127. [Google Scholar] [CrossRef] [PubMed]
- Szczesny, M.; Beloin, C.; Ghigo, J.M. Increased osmolarity in biofilm triggers RcsB-dependent lipid A palmitoylation in Escherichia coli. mBio 2018, 9, e01415–e01418. [Google Scholar] [CrossRef] [PubMed]
- Raetz, C.R.; Reynolds, C.M.; Trent, M.S.; Bishop, R.E. Lipid A modification systems in Gram-negative bacteria. Annu. Rev. Biochem. 2007, 76, 295–329. [Google Scholar] [CrossRef]
- Rubin, E.J.; Herrera, C.M.; Crofts, A.A.; Trent, M.S. PmrD is required for modifications to Escherichia coli endotoxin that promote antimicrobial resistance. Antimicrob. Agents Chemother. 2015, 59, 2051–2061. [Google Scholar] [CrossRef]
- Herrera, C.M.; Hankins, J.V.; Trent, M.S. Activation of PmrA inhibits LpxT-dependent phosphorylation of lipid A promoting resistance to antimicrobial peptides. Mol. Microbiol. 2010, 76, 1444–1460. [Google Scholar] [CrossRef]
- Kato, A.; Chen, H.D.; Latifi, T.; Groisman, E.A. Reciprocal control between a bacterium’s regulatory system and the modification status of its lipopolysaccharide. Mol. Cell 2012, 47, 897–908. [Google Scholar] [CrossRef]
- Hong, X.; Chen, H.D.; Groisman, E.A. Gene expression kinetics governs stimulus-specific decoration of the Salmonella outer membrane. Sci. Signal. 2018, 11, eaar7921. [Google Scholar] [CrossRef]
- Klein, G.; Lindner, B.; Brade, H.; Raina, S. Molecular basis of lipopolysaccharide heterogeneity in Escherichia coli: Envelope stress-responsive regulators control the incorporation of glycoforms with a third 3-deoxy-α-d-manno-oct-2-ulosonic acid and rhamnose. J. Biol. Chem. 2011, 286, 42787–42807. [Google Scholar] [CrossRef] [PubMed]
- Coornaert, A.; Lu, A.; Mandin, P.; Springer, M.; Gottesman, S.; Guillier, M. MicA sRNA links the PhoP regulon to cell envelope stress. Mol. Microbiol. 2010, 76, 467–479. [Google Scholar] [CrossRef] [PubMed]
- Gogol, E.B.; Rhodius, V.A.; Papenfort, K.; Vogel, J.; Gross, C.A. Small RNAs endow a transcriptional activator with essential repressor functions for single-tier control of a global stress regulon. Proc. Natl. Acad. Sci. USA 2012, 108, 12875–12880. [Google Scholar] [CrossRef] [PubMed]
- Coornaert, A.; Chiaruttini, C.; Springer, M.; Guillier, M. Post-transcriptional control of the Escherichia coli PhoQ-PhoP two-component system by multiple sRNAs involves a novel pairing region of GcvB. PLoS Genet. 2013, 9, e1003156. [Google Scholar] [CrossRef]
- Bishop, R.E.; Gibbons, H.S.; Guina, T.; Trent, M.S.; Miller, S.I.; Raetz, C.R.H. Transfer of palmitate from phospholipids to lipid A in outer membranes of Gram-negative bacteria. EMBO J. 2000, 19, 5071–5080. [Google Scholar] [CrossRef]
- Trent, M.S.; Pabich, W.; Raetz, C.R.H.; Miller, S.I. A PhoP/PhoQ-induced lipase (PagL) that catalyzes 3-O-deacylation of lipid A precursors in membranes of Salmonella typhimurium. J. Biol. Chem. 2001, 276, 9083–9092. [Google Scholar] [CrossRef]
- Han, K.; Tjaden, B.; Lory, S. GRIL-seq provides a method for identifying direct targets of bacterial small regulatory RNA by in vivo proximity ligation. Nat. Microbiol. 2016, 2, 16239. [Google Scholar] [CrossRef] [PubMed]
- Kawano, M.; Manabe, T.; Kawasaki, K. Salmonella enterica serovar Typhimurium lipopolysaccharide deacylation enhances its intracellular growth within macrophages. FEBS Lett. 2010, 584, 207–212. [Google Scholar] [CrossRef] [PubMed]
- Reynolds, C.M.; Ribeiro, A.A.; McGrath, S.C.; Cotter, R.J.; Raetz, C.R.H.; Trent, M.S. An outer membrane enzyme encoded by Salmonella typhimurium lpxR that removes the 3′-acyloxyacyl moiety of lipid A. J. Biol. Chem. 2006, 281, 21974–21987. [Google Scholar] [CrossRef]
- Corcoran, C.P.; Podkaminski, D.; Papenfort, K.; Urban, J.H.; Hinton, J.C.D.; Vogel, J. Superfolder GFP reporters validate diverse new mRNA targets of the classic porin regulator, MicF RNA. Mol. Microbiol. 2012, 84, 428–445. [Google Scholar] [CrossRef]
- Moon, K.; Six, D.A.; Lee, H.J.; Lee, C.R.; Raetz, C.R.H.; Gottesman, S. Complex transcriptional and post-transcriptional regulation of an enzyme for lipopolysaccharide modification. Mol. Microbiol. 2013, 89, 52–64. [Google Scholar] [CrossRef]
- Moon, K.; Gottesman, S. A PhoQ/P-regulated small RNA regulates sensitivity of Escherichia coli to antimicrobial peptides. Mol. Microbiol. 2009, 74, 1314–1330. [Google Scholar] [CrossRef]
- Müller-Loennies, S.; Holst, O.; Lindner, B.; Brade, H. Isolation and structural analysis of phosphorylated oligosaccharides obtained from Escherichia coli J-5 lipopolysaccharides. Eur. J. Biochem. 1999, 260, 235–249. [Google Scholar] [CrossRef] [PubMed]
- Fresno, S.; Jiménez, N.; Canals, R.; Merino, S.; Corsaro, M.M.; Lanzetta, R.; Parrilli, M.; Pieretti, G.; Regué, M.; Tomás, J.M. A second galacturonic acid transferase is required for core lipopolysaccharide biosynthesis and complete capsule association with the cell surface in Klebsiella pneumoniae. J. Bacteriol. 2007, 189, 1128–1137. [Google Scholar] [CrossRef]
- Müller-Loennies, S.; Lindner, B.; Brade, H. Structural analysis of oligosaccharides from lipopolysaccharide (LPS) of Escherichia coli K12 strain W3100 reveals a link between inner and outer core LPS biosynthesis. J. Biol. Chem. 2003, 278, 34090–34101. [Google Scholar] [CrossRef] [PubMed]
- Reynolds, C.M.; Kalb, S.R.; Cotter, R.J.; Raetz, C.R.H. A phosphoethanolamine transferase specific for the outer 3-deoxy-d-manno-octulosonic acid residue of Escherichia coli lipopolysaccharide. Identification of the eptB gene and Ca2+ hypersensitivity of an eptB deletion mutant. J. Biol. Chem. 2005, 280, 21202–21211. [Google Scholar] [CrossRef]
- Amar, A.; Pezzoni, M.; Pizarro, R.A.; Costa, C.S. New envelope stress factors involved in σE activation and conditional lethality of rpoE mutations in Salmonella enterica. Microbiology 2018, 164, 1293–1307. [Google Scholar] [CrossRef]
- Burmann, B.M.; Knauer, S.H.; Sevostyanova, A.; Schweimer, K.; Mooney, R.A.; Landick, R.; Artsimovitch, I.; Rösch, P. An α helix to β barrel domain switch transforms the transcription factor RfaH into a translation factor. Cell 2012, 150, 291–303. [Google Scholar] [CrossRef] [PubMed]
- Majdalani, N.; Heck, M.; Stout, V.; Gottesman, S. Role of RcsF in signaling to the Rcs phosphorelay pathway in Escherichia coli. J. Bacteriol. 2005, 187, 6770–6778. [Google Scholar] [CrossRef] [PubMed]
- Raina, S.; Missiakas, D.; Georgopoulos, C. The rpoE gene encoding the σE (σ24) heat shock sigma factor of Escherichia coli. EMBO J. 1995, 14, 1043–1055. [Google Scholar] [CrossRef] [PubMed]
- Rouvière, P.E.; De Las Peñas, A.; Mecsas, J.; Lu, C.Z.; Rudd, K.E.; Gross, C.A. rpoE, the gene encoding the second heat-shock sigma factor, σE, in Escherichia coli. EMBO J. 1995, 14, 1032–1042. [Google Scholar] [CrossRef] [PubMed]
- Missiakas, D.; Mayer, M.P.; Lemaire, M.; Georgopoulos, C.; Raina, S. Modulation of the Escherichia coli σE (RpoE) heat-shock transcription-factor activity by the RseA, RseB and RseC proteins. Mol. Microbiol. 1997, 24, 355–371. [Google Scholar] [CrossRef] [PubMed]
- Lima, S.; Guo, M.S.; Chaba, R.; Gross, C.A.; Sauer, R.T. Dual molecular signals mediate the bacterial response to outer-membrane stress. Science 2013, 340, 837–841. [Google Scholar] [CrossRef] [PubMed]
- Sperandeo, P.; Lau, F.K.; Carpentieri, A.; De Castro, C.; Molinaro, A.; Dehò, G.; Silhavy, T.J.; Polissi, A. Functional analysis of the protein machinery required for transport of lipopolysaccharide to the outer membrane of Escherichia coli. J. Bacteriol. 2008, 190, 4460–4469. [Google Scholar] [CrossRef]
- Konovalova, A.; Mitchell, A.M.; Silhavy, T.J. A lipoprotein/β-barrel complex monitors lipopolysaccharide integrity transducing information across the outer membrane. eLife 2016, 5, e15276. [Google Scholar] [CrossRef] [PubMed]
- Noor, R.; Murata, M.; Nagamitsu, H.; Klein, G.; Raina, S.; Yamada, M. Dissection of σE-dependent cell lysis in Escherichia coli: Roles of RpoE regulators RseA, RseB and periplasmic folding catalysts PpiD. Genes Cells 2009, 14, 885–899. [Google Scholar] [CrossRef]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Klein, G.; Raina, S. Regulated Assembly of LPS, Its Structural Alterations and Cellular Response to LPS Defects. Int. J. Mol. Sci. 2019, 20, 356. https://doi.org/10.3390/ijms20020356
Klein G, Raina S. Regulated Assembly of LPS, Its Structural Alterations and Cellular Response to LPS Defects. International Journal of Molecular Sciences. 2019; 20(2):356. https://doi.org/10.3390/ijms20020356
Chicago/Turabian StyleKlein, Gracjana, and Satish Raina. 2019. "Regulated Assembly of LPS, Its Structural Alterations and Cellular Response to LPS Defects" International Journal of Molecular Sciences 20, no. 2: 356. https://doi.org/10.3390/ijms20020356
APA StyleKlein, G., & Raina, S. (2019). Regulated Assembly of LPS, Its Structural Alterations and Cellular Response to LPS Defects. International Journal of Molecular Sciences, 20(2), 356. https://doi.org/10.3390/ijms20020356





