Preferential Localization of MUC1 Glycoprotein in Exosomes Secreted by Non-Small Cell Lung Carcinoma Cells
Abstract
1. Introduction
2. Results
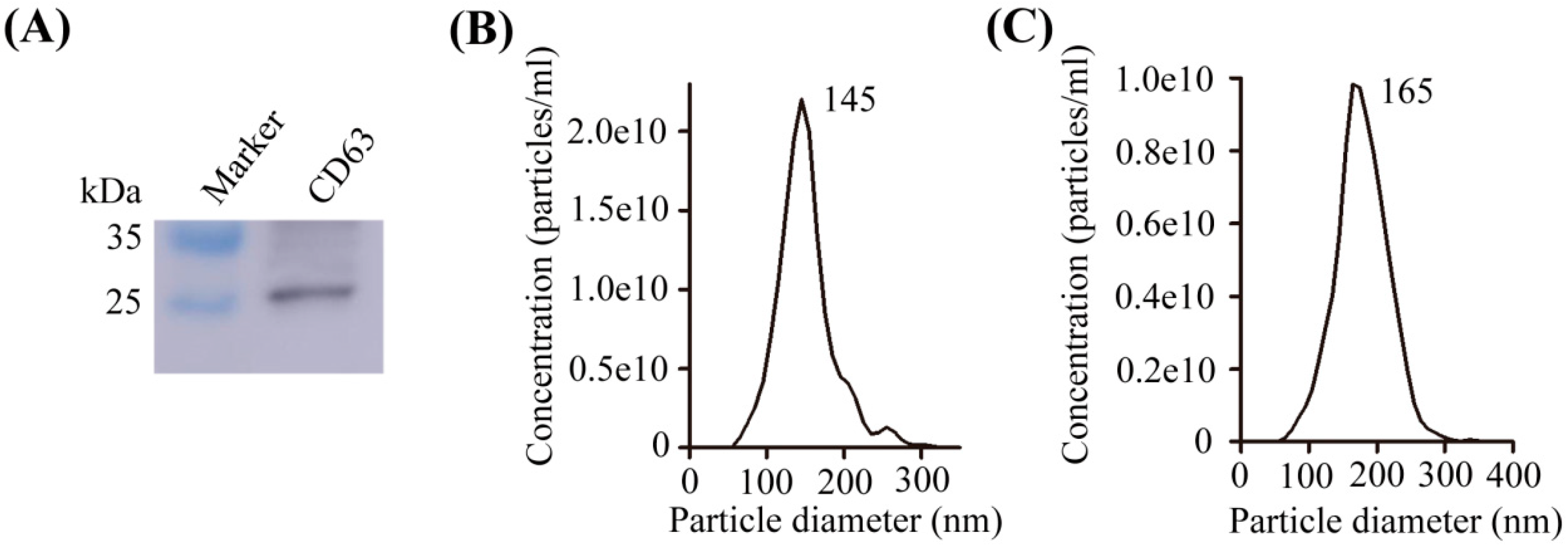
2.1. Characterization of Exosomes
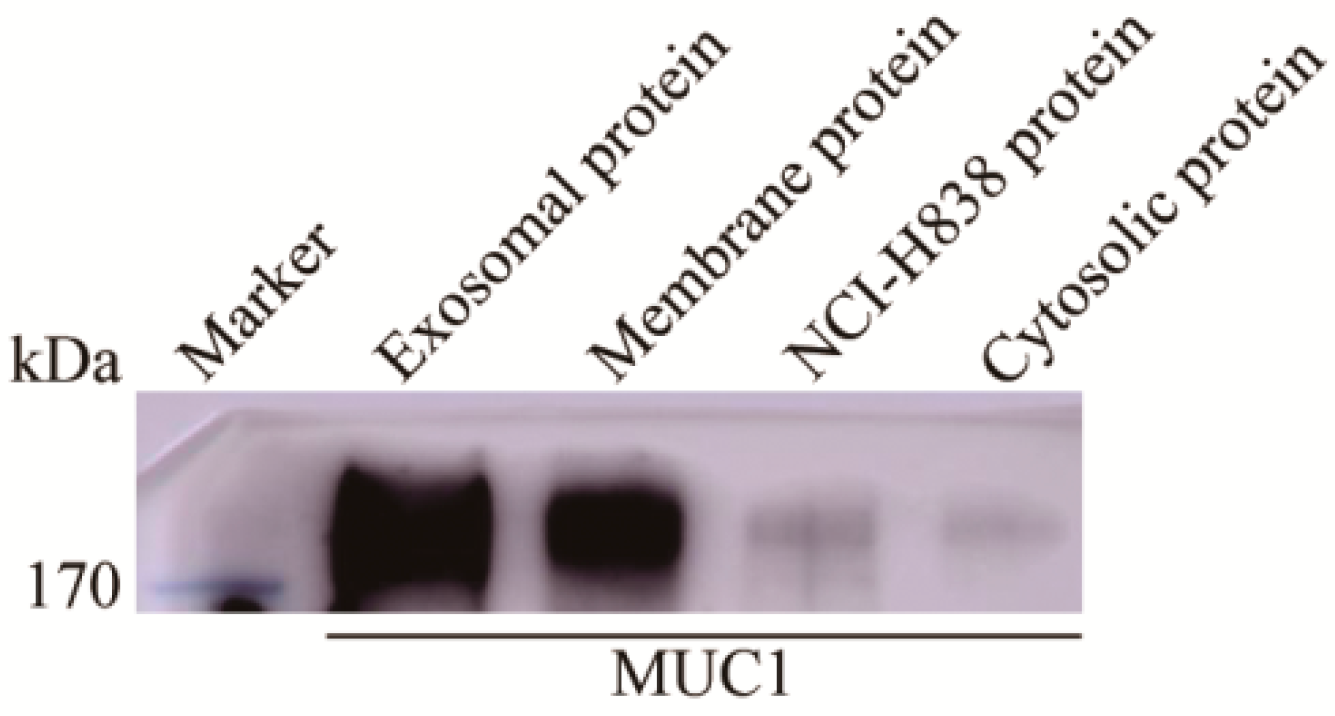
2.2. Proteins Preferentially Localized in the Exosomes of NCI-H838 Cells
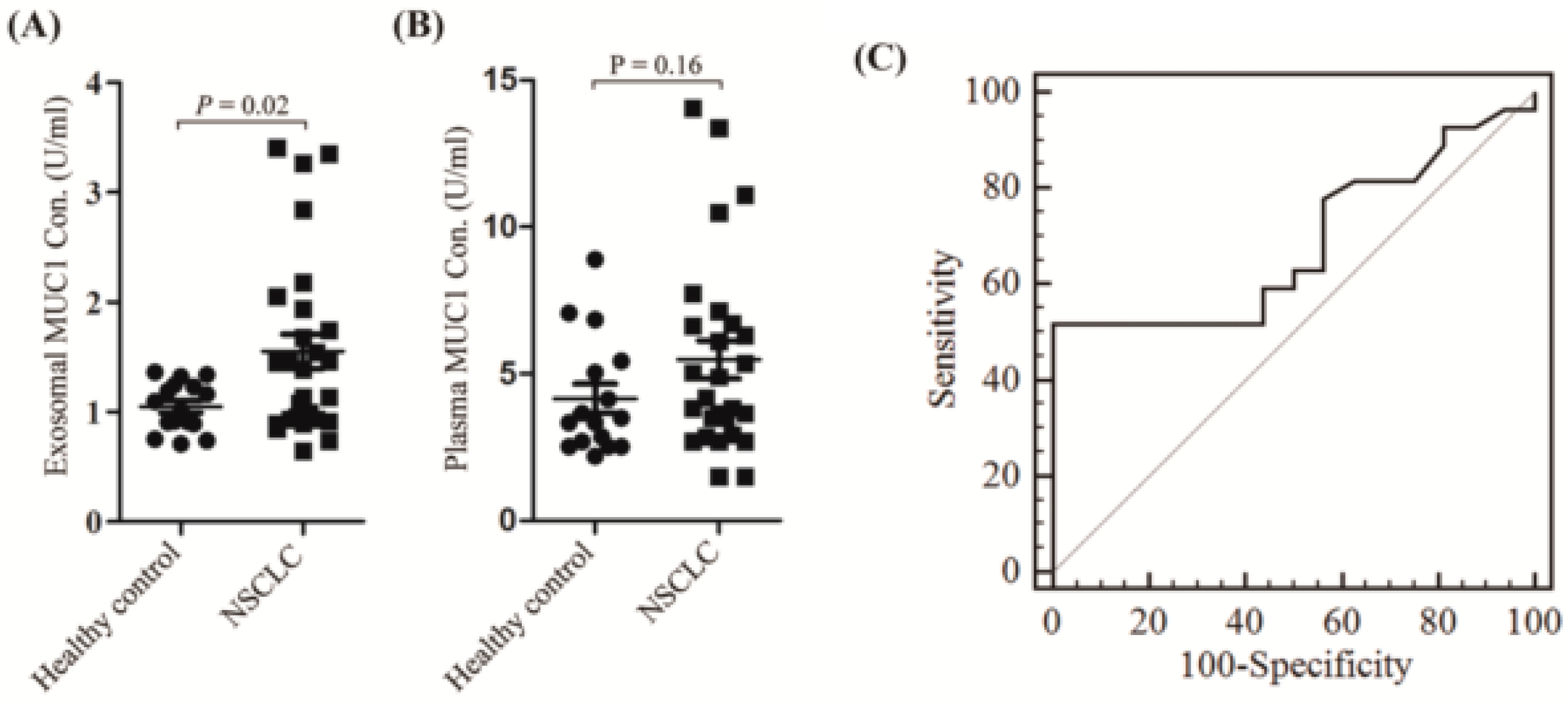
2.3. Exosomal Expressed MUC1 Is Significantly Different between NSCLC Patients and Healthy Controls
3. Discussion
4. Materials and Methods
4.1. Patients and Cell Line
4.2. Western Blot Analysis
4.3. Nanoparticle Tracking Analysis
4.4. Isolation of Exosomes
4.5. Mass Spectrometry Method and Data Analysis
4.6. Electrochemiluminescence Immunoassay Analysis (EIA)
4.7. Statistical Analysis
5. Conclusions
Supplementary Materials
Authors Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| NSCLC | Non-small cell lung cancer |
| SCLC | Small cell lung cancer |
| LC | Liquid chromatography |
| MS | Mass spectrometry |
| NTA | Nanoparticle tracking analysis |
| EIA | Electrochemiluminescence immunoassay analysis |
| AUC | Area under the curve |
| ADC | Adenocarcinoma |
| SCC | Squamous cell carcinoma |
References
- Ferlay, J.; Soerjomataram, I.; Dikshit, R.; Eser, S.; Mathers, C.; Rebelo, M.; Parkin, D.M.; Forman, D.; Bray, F. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int. J. Cancer 2015, 136, E359–E386. [Google Scholar] [CrossRef] [PubMed]
- Torre, L.A.; Bray, F.; Siegel, R.L.; Ferlay, J.; Lortet-Tieulent, J.; Jemal, A. Global cancer statistics, 2012. CA Cancer J. Clin. 2015, 65, 87–108. [Google Scholar] [CrossRef]
- Travis, W.D.; Brambilla, E.; Noguchi, M.; Nicholson, A.G.; Geisinger, K.R.; Yatabe, Y.; Beer, D.G.; Powell, C.A.; Riely, G.J.; Van Schil, P.E.; et al. International association for the study of lung cancer/american thoracic society/european respiratory society international multidisciplinary classification of lung adenocarcinoma. J. Thorac. Oncol. 2011, 6, 244–285. [Google Scholar] [CrossRef]
- Borghaei, H.; Paz-Ares, L.; Horn, L.; Spigel, D.R.; Steins, M.; Ready, N.E.; Chow, L.Q.; Vokes, E.E.; Felip, E.; Holgado, E.; et al. Nivolumab versus Docetaxel in Advanced Nonsquamous Non-Small-Cell Lung Cancer. N. Engl. J. Med. 2015, 373, 1627–1639. [Google Scholar] [CrossRef] [PubMed]
- Vansteenkiste, J.; Crino, L.; Dooms, C.; Douillard, J.Y.; Faivre-Finn, C.; Lim, E.; Rocco, G.; Senan, S.; Van Schil, P.; Veronesi, G.; et al. 2nd ESMO Consensus Conference on Lung Cancer: early-stage non-small-cell lung cancer consensus on diagnosis, treatment and follow-up. Ann. Oncol. 2014, 25, 1462–1474. [Google Scholar] [CrossRef]
- Goldstraw, P.; Chansky, K.; Crowley, J.; Rami-Porta, R.; Asamura, H.; Eberhardt, W.E.; Nicholson, A.G.; Groome, P.; Mitchell, A.; Bolejack, V. The IASLC Lung Cancer Staging Project: Proposals for Revision of the TNM Stage Groupings in the Forthcoming (Eighth) Edition of the TNM Classification for Lung Cancer. J. Thorac. Oncol. 2016, 11, 39–51. [Google Scholar] [CrossRef]
- Chae, Y.K.; Pan, A.; Davis, A.A.; Mohindra, N.; Matsangou, M.; Villaflor, V.; Giles, F. Recent Advances and Future Strategies for Immune-Checkpoint Inhibition in Small-Cell Lung Cancer. Clin. Lung Cancer 2017, 18, 132–140. [Google Scholar] [CrossRef] [PubMed]
- Ada, G. The coming of age of tumour immunotherapy. Immunol. Cell Biol. 1999, 77, 180–185. [Google Scholar] [CrossRef]
- De Mello, R.A.; Veloso, A.F.; Esrom Catarina, P.; Nadine, S.; Antoniou, G. Potential role of immunotherapy in advanced non-small-cell lung cancer. Onco Targets Ther. 2017, 10, 21–30. [Google Scholar] [CrossRef]
- Hirsch, F.R.; Franklin, W.A.; Gazdar, A.F.; Bunn, P.A., Jr. Early detection of lung cancer: clinical perspectives of recent advances in biology and radiology. Clin. Cancer. Res. 2001, 7, 5–22. [Google Scholar]
- Hou, J.; Meng, F.; Chan, L.W.; Cho, W.C.; Wong, S.C. Circulating Plasma MicroRNAs As Diagnostic Markers for NSCLC. Front Genet 2016, 7, 193. [Google Scholar] [CrossRef] [PubMed]
- Aberle, D.R.; Adams, A.M.; Berg, C.D.; Black, W.C.; Clapp, J.D.; Fagerstrom, R.M.; Gareen, I.F.; Gatsonis, C.; Marcus, P.M.; Sicks, J.D. Reduced lung-cancer mortality with low-dose computed tomographic screening. N. Engl. J. Med. 2011, 365, 395–409. [Google Scholar] [PubMed]
- Pan, B.T.; Teng, K.; Wu, C.; Adam, M.; Johnstone, R.M. Electron microscopic evidence for externalization of the transferrin receptor in vesicular form in sheep reticulocytes. J. Cell Biol. 1985, 101, 942–948. [Google Scholar] [CrossRef] [PubMed]
- Mathivanan, S.; Simpson, R.J. ExoCarta: A compendium of exosomal proteins and RNA. Proteomics 2009, 9, 4997–5000. [Google Scholar] [CrossRef] [PubMed]
- Valadi, H.; Ekstrom, K.; Bossios, A.; Sjostrand, M.; Lee, J.J.; Lotvall, J.O. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat. Cell Biol. 2007, 9, 654–659. [Google Scholar] [CrossRef]
- Taylor, D.D.; Gercel-Taylor, C. Exosomes/microvesicles: mediators of cancer-associated immunosuppressive microenvironments. Semin. Immunopathol. 2011, 33, 441–454. [Google Scholar] [CrossRef] [PubMed]
- Khan, S.; Bennit, H.F.; Turay, D.; Perez, M.; Mirshahidi, S.; Yuan, Y.; Wall, N.R. Early diagnostic value of survivin and its alternative splice variants in breast cancer. BMC Cancer 2014, 14, 176. [Google Scholar] [CrossRef]
- Peinado, H.; Aleckovic, M.; Lavotshkin, S.; Matei, I.; Costa-Silva, B.; Moreno-Bueno, G.; Hergueta-Redondo, M.; Williams, C.; Garcia-Santos, G.; Ghajar, C.; et al. Melanoma exosomes educate bone marrow progenitor cells toward a pro-metastatic phenotype through MET. Nat. Med. 2012, 18, 883–891. [Google Scholar] [CrossRef]
- Taylor, D.D.; Gercel-Taylor, C. MicroRNA signatures of tumor-derived exosomes as diagnostic biomarkers of ovarian cancer. Gynecol. Oncol. 2008, 110, 13–21. [Google Scholar] [CrossRef]
- Skog, J.; Wurdinger, T.; van Rijn, S.; Meijer, D.H.; Gainche, L.; Sena-Esteves, M.; Curry, W.T., Jr.; Carter, B.S.; Krichevsky, A.M.; Breakefield, X.O. Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat. Cell Biol. 2008, 10, 1470–1476. [Google Scholar] [CrossRef]
- Rabinowits, G.; Gercel-Taylor, C.; Day, J.M.; Taylor, D.D.; Kloecker, G.H. Exosomal microRNA: a diagnostic marker for lung cancer. Clin. Lung Cancer 2009, 10, 42–46. [Google Scholar] [CrossRef]
- Yamashita, T.; Kamada, H.; Kanasaki, S.; Maeda, Y.; Nagano, K.; Abe, Y.; Inoue, M.; Yoshioka, Y.; Tsutsumi, Y.; Katayama, S.; et al. Epidermal growth factor receptor localized to exosome membranes as a possible biomarker for lung cancer diagnosis. Pharmazie 2013, 68, 969–973. [Google Scholar] [PubMed]
- Sandfeld-Paulsen, B.; Jakobsen, K.R.; Baek, R.; Folkersen, B.H.; Rasmussen, T.R.; Meldgaard, P.; Varming, K.; Jorgensen, M.M.; Sorensen, B.S. Exosomal Proteins as Diagnostic Biomarkers in Lung Cancer. J. Thorac. Oncol. 2016, 11, 1701–1710. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.J.; Sertamo, K.; Pierrard, M.A.; Mesmin, C.; Kim, S.Y.; Schlesser, M.; Berchem, G.; Domon, B. Verification of the biomarker candidates for non-small-cell lung cancer using a targeted proteomics approach. J. Proteome Res. 2015, 14, 1412–1419. [Google Scholar] [CrossRef]
- Yang, S.; Chen, L.; Chan, D.W.; Li, Q.K.; Zhang, H. Protein signatures of molecular pathways in non-small cell lung carcinoma (NSCLC): Comparison of glycoproteomics and global proteomics. Clin. Proteom. 2017, 14, 31. [Google Scholar] [CrossRef] [PubMed]
- Vykoukal, J.; Sun, N.; Aguilar-Bonavides, C.; Katayama, H.; Tanaka, I.; Fahrmann, J.F.; Capello, M.; Fujimoto, J.; Aguilar, M.; Wistuba, I.I.; et al. Plasma-derived extracellular vesicle proteins as a source of biomarkers for lung adenocarcinoma. Oncotarget 2017, 8, 95466–95480. [Google Scholar] [CrossRef] [PubMed]
- Bigbee, W.L.; Gopalakrishnan, V.; Weissfeld, J.L.; Wilson, D.O.; Dacic, S.; Lokshin, A.E.; Siegfried, J.M. A multiplexed serum biomarker immunoassay panel discriminates clinical lung cancer patients from high-risk individuals found to be cancer-free by CT screening. J. Thorac. Oncol. 2012, 7, 698–708. [Google Scholar] [CrossRef] [PubMed]
- Wang, N.; Song, X.; Liu, L.; Niu, L.; Wang, X.; Song, X.; Xie, L. Circulating exosomes contain protein biomarkers of metastatic non-small-cell lung cancer. Cancer Sci. 2018, 109, 1701–1709. [Google Scholar] [CrossRef]
- Birse, C.E.; Lagier, R.J.; FitzHugh, W.; Pass, H.I.; Rom, W.N.; Edell, E.S.; Bungum, A.O.; Maldonado, F.; Jett, J.R.; Mesri, M.; et al. Blood-based lung cancer biomarkers identified through proteomic discovery in cancer tissues, cell lines and conditioned medium. Clin. Proteom. 2015, 12, 18. [Google Scholar] [CrossRef]
- Dai, L.; Qu, Y.; Li, J.; Wang, X.; Wang, K.; Wang, P.; Jiang, B.H.; Zhang, J. Serological proteome analysis approach-based identification of ENO1 as a tumor-associated antigen and its autoantibody could enhance the sensitivity of CEA and CYFRA 21-1 in the detection of non-small cell lung cancer. Oncotarget 2017, 8, 36664–36673. [Google Scholar] [CrossRef]
- Clark, D.J.; Fondrie, W.E.; Yang, A.; Mao, L. Triple SILAC quantitative proteomic analysis reveals differential abundance of cell signaling proteins between normal and lung cancer-derived exosomes. J. Proteom. 2016, 133, 161–169. [Google Scholar] [CrossRef] [PubMed]
- Ring, B.Z.; Seitz, R.S.; Beck, R.A.; Shasteen, W.J.; Soltermann, A.; Arbogast, S.; Robert, F.; Schreeder, M.T.; Ross, D.T. A novel five-antibody immunohistochemical test for subclassification of lung carcinoma. Mod. Pathol. 2009, 22, 1032–1043. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Xia, Z.; Shang, Z.; Sun, K.; Niu, X.; Qian, L.; Fan, L.Y.; Cao, C.X.; Xiao, H. Facile preparation of salivary extracellular vesicles for cancer proteomics. Sci. Rep. 2016, 6, 24669. [Google Scholar] [CrossRef]
- Gao, W.M.; Kuick, R.; Orchekowski, R.P.; Misek, D.E.; Qiu, J.; Greenberg, A.K.; Rom, W.N.; Brenner, D.E.; Omenn, G.S.; Haab, B.B.; et al. Distinctive serum protein profiles involving abundant proteins in lung cancer patients based upon antibody microarray analysis. BMC Cancer 2005, 5, 110. [Google Scholar] [CrossRef] [PubMed]
- Ostroff, R.M.; Bigbee, W.L.; Franklin, W.; Gold, L.; Mehan, M.; Miller, Y.E.; Pass, H.I.; Rom, W.N.; Siegfried, J.M.; Stewart, A.; et al. Unlocking biomarker discovery: large scale application of aptamer proteomic technology for early detection of lung cancer. PLoS ONE 2010, 5, e15003. [Google Scholar] [CrossRef] [PubMed]
- Li, X.J.; Hayward, C.; Fong, P.Y.; Dominguez, M.; Hunsucker, S.W.; Lee, L.W.; McLean, M.; Law, S.; Butler, H.; Schirm, M.; et al. A blood-based proteomic classifier for the molecular characterization of pulmonary nodules. Sci. Transl. Med. 2013, 5, 207ra142. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Zhong, W.; Chen, C.; Meng, Q.; Wei, J. Circulating Antibodies to Linear Peptide Antigens Derived from ANXA1 and FOXP3 in Lung Cancer. Anticancer Res. 2017, 37, 3151–3155. [Google Scholar]
- Zhong, L.; Coe, S.P.; Stromberg, A.J.; Khattar, N.H.; Jett, J.R.; Hirschowitz, E.A. Profiling tumor-associated antibodies for early detection of non-small cell lung cancer. J. Thorac. Oncol. 2006, 1, 513–519. [Google Scholar] [CrossRef]
- Dai, L.; Tsay, J.C.; Li, J.; Yie, T.A.; Munger, J.S.; Pass, H.; Rom, W.N.; Zhang, Y.; Tan, E.M.; Zhang, J.Y. Autoantibodies against tumor-associated antigens in the early detection of lung cancer. Lung Cancer 2016, 99, 172–179. [Google Scholar] [CrossRef]
- Dai, L.; Li, J.; Tsay, J.J.; Yie, T.A.; Munger, J.S.; Pass, H.; Rom, W.N.; Tan, E.M.; Zhang, J.Y. Identification of autoantibodies to ECH1 and HNRNPA2B1 as potential biomarkers in the early detection of lung cancer. Oncoimmunology 2017, 6, e1310359. [Google Scholar] [CrossRef]
- Song, W.; Delyria, E.S.; Chen, J.; Huang, W.; Lee, J.S.; Mittendorf, E.A.; Ibrahim, N.; Radvanyi, L.G.; Li, Y.; Lu, H.; et al. MUC1 glycopeptide epitopes predicted by computational glycomics. Int. J. Oncol. 2012, 41, 1977–1984. [Google Scholar] [CrossRef]
- Li, W.; Li, C.; Zhou, T.; Liu, X.; Liu, X.; Li, X.; Chen, D. Role of exosomal proteins in cancer diagnosis. Mol. Cancer 2017, 16, 145. [Google Scholar] [CrossRef] [PubMed]
- Sandfeld-Paulsen, B.; Aggerholm-Pedersen, N.; Baek, R.; Jakobsen, K.R.; Meldgaard, P.; Folkersen, B.H.; Rasmussen, T.R.; Varming, K.; Jorgensen, M.M.; Sorensen, B.S. Exosomal proteins as prognostic biomarkers in non-small cell lung cancer. Mol. Oncol. 2016, 10, 1595–1602. [Google Scholar] [CrossRef] [PubMed]
- Apostolopoulos, V.; Stojanovska, L.; Gargosky, S.E. MUC1 (CD227): a multi-tasked molecule. Cell. Mol. Life Sci. 2015, 72, 4475–4500. [Google Scholar] [CrossRef] [PubMed]
- Movahedin, M.; Brooks, T.M.; Supekar, N.T.; Gokanapudi, N.; Boons, G.J.; Brooks, C.L. Glycosylation of MUC1 influences the binding of a therapeutic antibody by altering the conformational equilibrium of the antigen. Glycobiology 2017, 27, 677–687. [Google Scholar] [CrossRef] [PubMed]
- Buisine, M.P.; Devisme, L.; Copin, M.C.; Durand-Reville, M.; Gosselin, B.; Aubert, J.P.; Porchet, N. Developmental mucin gene expression in the human respiratory tract. Am. J. Respir. Cell Mol. Biol. 1999, 20, 209–218. [Google Scholar] [CrossRef]
- Awaya, H.; Takeshima, Y.; Yamasaki, M.; Inai, K. Expression of MUC1, MUC2, MUC5AC, and MUC6 in atypical adenomatous hyperplasia, bronchioloalveolar carcinoma, adenocarcinoma with mixed subtypes, and mucinous bronchioloalveolar carcinoma of the lung. Am. J. Clin. Pathol. 2004, 121, 644–653. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Zhang, S.; Ni, W.; Zhai, X.; Xie, F.; Yuan, H.; Gao, S.; Tai, G. Development and application of a double- antibody sandwich ELISA kit for the detection of serum MUC1 in lung cancer patients. Cancer Biomark. 2016, 17, 369–376. [Google Scholar] [CrossRef]
- Kikuchi, T.; Hassanein, M.; Amann, J.M.; Liu, Q.; Slebos, R.J.; Rahman, S.M.; Kaufman, J.M.; Zhang, X.; Hoeksema, M.D.; Harris, B.K.; et al. In-depth proteomic analysis of nonsmall cell lung cancer to discover molecular targets and candidate biomarkers. Mol. Cell. Proteom. 2012, 11, 916–932. [Google Scholar] [CrossRef] [PubMed]
- Schneider, J. Detection of lung cancer in silicosis patients using a tumor-marker panel. Cancer Biomark. 2010, 6, 137–148. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, R.; Alam, M.; Rajabi, H.; Kufe, D. The MUC1-C oncoprotein binds to the BH3 domain of the pro-apoptotic BAX protein and blocks BAX function. J. Biol. Chem. 2012, 287, 20866–20875. [Google Scholar] [CrossRef] [PubMed]
- Zhang, K.; Sikut, R.; Hansson, G.C. A MUC1 mucin secreted from a colon carcinoma cell line inhibits target cell lysis by natural killer cells. Cell. Immunol. 1997, 176, 158–165. [Google Scholar] [CrossRef] [PubMed]
- Moreno, M.; Bontkes, H.J.; Scheper, R.J.; Kenemans, P.; Verheijen, R.H.; von Mensdorff-Pouilly, S. High level of MUC1 in serum of ovarian and breast cancer patients inhibits huHMFG-1 dependent cell-mediated cytotoxicity (ADCC). Cancer Lett. 2007, 257, 47–55. [Google Scholar] [CrossRef] [PubMed]
- Wisniewski, J.R.; Zougman, A.; Nagaraj, N.; Mann, M. Universal sample preparation method for proteome analysis. Nat. Methods 2009, 6, 359–362. [Google Scholar] [CrossRef]
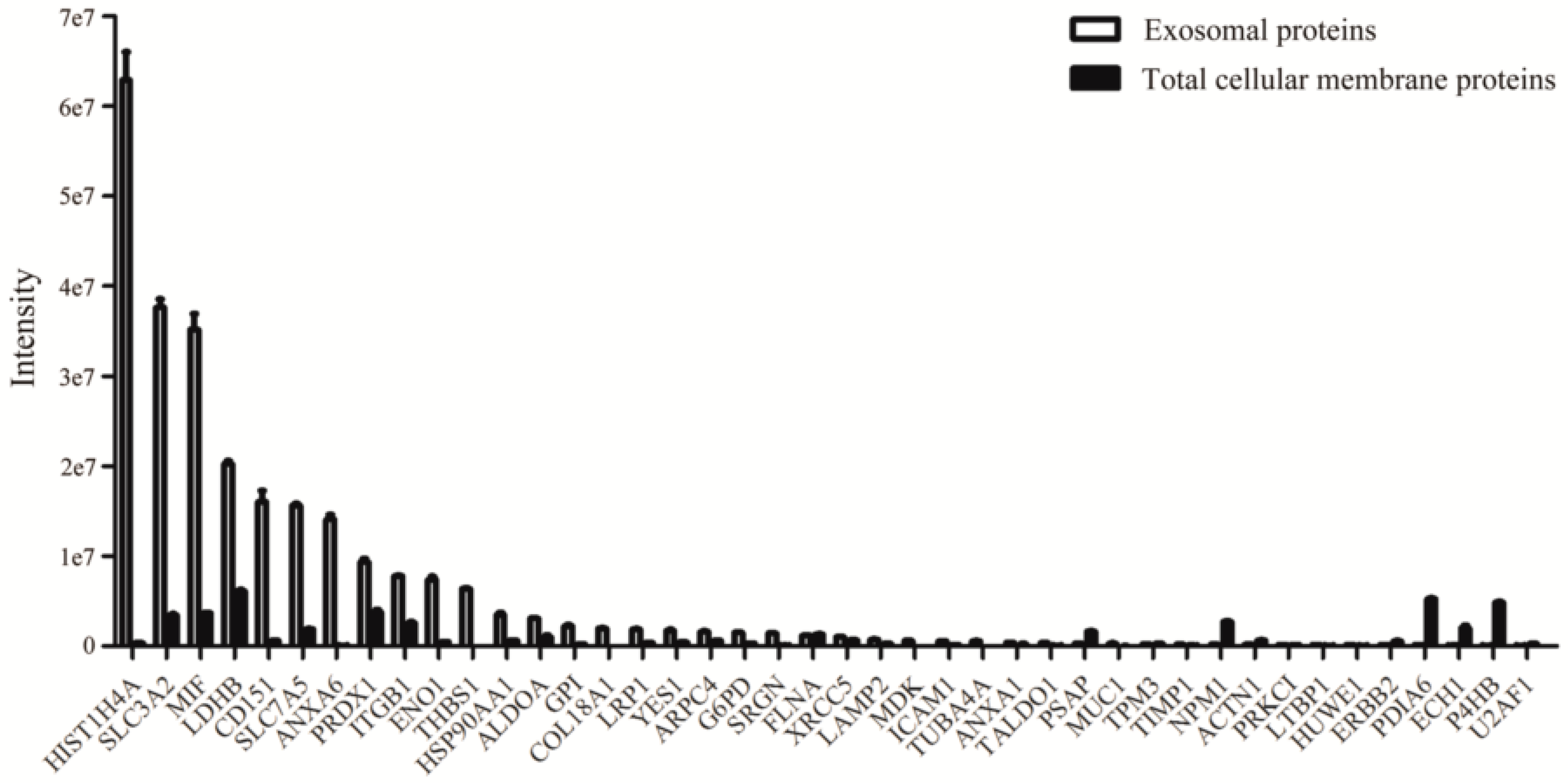
| Protein Accessions | Protein Descriptions | Ratio of Mass Spectrometry Signal Intensity in Exosomal to Membrane Proteins | References |
|---|---|---|---|
| P48509 | CD151 antigen (CD151) | 27.092 | [23] |
| P07996 | Thrombospondin-1 (THBS1) | 259.860 | [24,25,26,27] |
| P06744 | Glucose-6-phosphate isomerase (GPI) | 10.700 | [24] |
| P37837 | Transaldolase (TALDO1) | 8.424 | [24] |
| Q5NKV8 | Intercellular adhesion molecule 1 (ICAM1) | 6.001 | [24] |
| Q5U077 | L-lactate dehydrogenase (LDHB) | 3.307 | [24] |
| Q60FE6 | Filamin A (FLNA) | 0.900 | [24] |
| P12814 | Alpha-actinin-1 (ACTN1) | 0.251 | [24] |
| P11413 | Glucose-6-phosphate 1-dehydrogenase (G6PD) | 5.411 | [24] |
| Q14766 | Latent-transforming growth factor beta-binding protein 1 (LTBP1) | 2.894 | [25] |
| Q59EN5 | Prosaposin variant (PSAP) | 0.176 | [25] |
| Q01081 | Splicing factor U2AF 35 kDa subunit (U2AF1) | 0.182 | [25] |
| Q5HYB6 | Epididymis luminal protein 189 (TPM3) | 0.838 | [26] |
| Q7Z6Z7 | E3 ubiquitin-protein ligase HUWE1 (HUWE1) | 1.730 | [26] |
| P10124 | Serglycin (SRGN) | 18.260 | [26] |
| P14174 | Macrophage migration inhibitory factor (MIF) | 9.645 | [27] |
| X5DNK3 | Receptor protein-tyrosine kinase (Fragment) (ERBB2) | 0.179 | [27] |
| P62805 | Histone H4 (HIST1H4A) | 186.179 | [28] |
| P39060 | Collagen alpha-1(XVIII) chain (COL18A1) | 133.480 | [28] |
| P08133 | Annexin A6 (ANXA6) | 242.502 | [28] |
| P68366 | Tubulin alpha-4A chain (TUBA4A) | 13.347 | [28] |
| P59998 | Actin-related protein 2/3 complex subunit 4 (ARPC4) | 2.852 | [28] |
| Q15084 | Protein disulfide-isomerase A6 (PDIA6) | 0.016 | [28] |
| P07237 | Protein disulfide-isomerase (P4HB) | 0.011 | [28] |
| Q6FGX5 | TIMP metallopeptidase inhibitor 1, isoform CRA_a (TIMP1) | 2.671 | [29] |
| P21741 | Midkine (MDK) | 34.132 | [29] |
| P06733 | Alpha-enolase (ENO1) | 16.102 | [24,30] |
| P08195 | 4F2 cell-surface antigen heavy chain (SLC3A2) | 10.947 | [31] |
| P05556 | Integrin beta-1 (ITGB1) | 3.037 | [31] |
| P13473 | Lysosome-associated membrane glycoprotein 2 (LAMP2) | 2.789 | [31] |
| Q01650 | Large neutral amino acids transporter small subunit 1 (SLC7A5) | 8.272 | [32] |
| P15941 | Mucin-1 (MUC1) | 8.977 | [32,33,34] |
| P07900 | Heat shock protein HSP 90-alpha (HSP90AA1) | 5.526 | [35] |
| P07947 | Tyrosine-protein kinase Yes (YES1) | 4.321 | [35] |
| P41743 | Protein kinase C iota type (PRKCI) | 1.545 | [35] |
| Q07954 | Prolow-density lipoprotein receptor-related protein 1 (LRP1) | 5.011 | [36] |
| V9HWN7 | Fructose-bisphosphate aldolase (ALDOA) | 3.090 | [24,36] |
| Q06830 | Peroxiredoxin-1 (PRDX1) | 2.424 | [36] |
| Q5TZZ9 | Annexin (ANXA1) | 2.113 | [37] |
| P13010 | X-ray repair cross-complementing protein 5 (XRCC5) | 1.456 | [38] |
| P06748 | Nucleophosmin (NPM1) | 0.063 | [39] |
| Q13011 | Delta(3,5)-Delta(2,4)-dienoyl-CoA isomerase, mitochondrial (ECH1) | 0.031 | [40] |
| Number | Exosomal MUC1 Con. (U/mL) | Plasma MUC1 Con. (U/mL) | Exosome-Plasma MUC1 Ratio (%) | |
|---|---|---|---|---|
| NSCLC patients | 1 | 1.49 | 6.11 | 24.38 |
| 2 | 2.05 | 6.63 | 30.97 | |
| 3 | 1.13 | 2.85 | 39.76 | |
| 4 | 0.94 | 4.90 | 19.10 | |
| 5 | 1.13 | 3.50 | 32.44 | |
| 6 | 1.94 | 7.14 | 27.15 | |
| 7 | 1.00 | 1.49 | 66.63 | |
| 8 | 0.74 | 1.49 | 49.43 | |
| 9 | 3.27 | 5.05 | 64.74 | |
| 10 | 3.35 | 14.06 | 23.86 | |
| 11 | 3.41 | 10.49 | 32.47 | |
| 12 | 0.94 | 2.69 | 34.77 | |
| 13 | 0.95 | 4.18 | 22.80 | |
| 14 | 1.54 | 11.09 | 13.89 | |
| 15 | 1.39 | 13.38 | 10.38 | |
| 16 | 0.89 | 3.50 | 25.53 | |
| 17 | 1.47 | 2.93 | 49.95 | |
| 18 | 0.85 | 2.69 | 31.55 | |
| 19 | 0.91 | 2.69 | 33.99 | |
| 20 | 1.45 | 6.31 | 23.00 | |
| 21 | 2.85 | 3.81 | 74.65 | |
| 22 | 1.74 | 7.73 | 22.51 | |
| 23 | 2.18 | 5.35 | 40.77 | |
| 24 | 0.64 | 3.66 | 17.64 | |
| 25 | 1.68 | 6.70 | 25.05 | |
| 26 | 0.89 | 3.81 | 23.42 | |
| 27 | 1.08 | 3.66 | 29.59 | |
| Healthy controls | 1 | 1.19 | 5.43 | 21.93 |
| 2 | 0.90 | 3.34 | 27.09 | |
| 3 | 0.91 | 2.52 | 36.17 | |
| 4 | 1.16 | 2.69 | 43.23 | |
| 5 | 1.34 | 4.13 | 32.41 | |
| 6 | 1.23 | 7.07 | 17.39 | |
| 7 | 1.36 | 8.90 | 15.29 | |
| 8 | 1.24 | 5.05 | 24.65 | |
| 9 | 1.09 | 3.34 | 32.79 | |
| 10 | 0.74 | 2.85 | 25.88 | |
| 11 | 1.32 | 6.85 | 19.31 | |
| 12 | 1.01 | 3.50 | 28.91 | |
| 13 | 0.89 | 2.52 | 35.37 | |
| 14 | 0.71 | 3.66 | 19.42 | |
| 15 | 0.75 | 2.19 | 34.27 | |
| 16 | 0.91 | 2.52 | 36.21 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pan, D.; Chen, J.; Feng, C.; Wu, W.; Wang, Y.; Tong, J.; Zhou, D. Preferential Localization of MUC1 Glycoprotein in Exosomes Secreted by Non-Small Cell Lung Carcinoma Cells. Int. J. Mol. Sci. 2019, 20, 323. https://doi.org/10.3390/ijms20020323
Pan D, Chen J, Feng C, Wu W, Wang Y, Tong J, Zhou D. Preferential Localization of MUC1 Glycoprotein in Exosomes Secreted by Non-Small Cell Lung Carcinoma Cells. International Journal of Molecular Sciences. 2019; 20(2):323. https://doi.org/10.3390/ijms20020323
Chicago/Turabian StylePan, Deng, Jiaxi Chen, Chunchao Feng, Weibo Wu, Yanjin Wang, Jiao Tong, and Dapeng Zhou. 2019. "Preferential Localization of MUC1 Glycoprotein in Exosomes Secreted by Non-Small Cell Lung Carcinoma Cells" International Journal of Molecular Sciences 20, no. 2: 323. https://doi.org/10.3390/ijms20020323
APA StylePan, D., Chen, J., Feng, C., Wu, W., Wang, Y., Tong, J., & Zhou, D. (2019). Preferential Localization of MUC1 Glycoprotein in Exosomes Secreted by Non-Small Cell Lung Carcinoma Cells. International Journal of Molecular Sciences, 20(2), 323. https://doi.org/10.3390/ijms20020323





