miRNA Polymorphisms and Risk of Cardio-Cerebrovascular Diseases: A Systematic Review and Meta-Analysis
Abstract
1. Introduction
2. Results
2.1. Study Characteristics
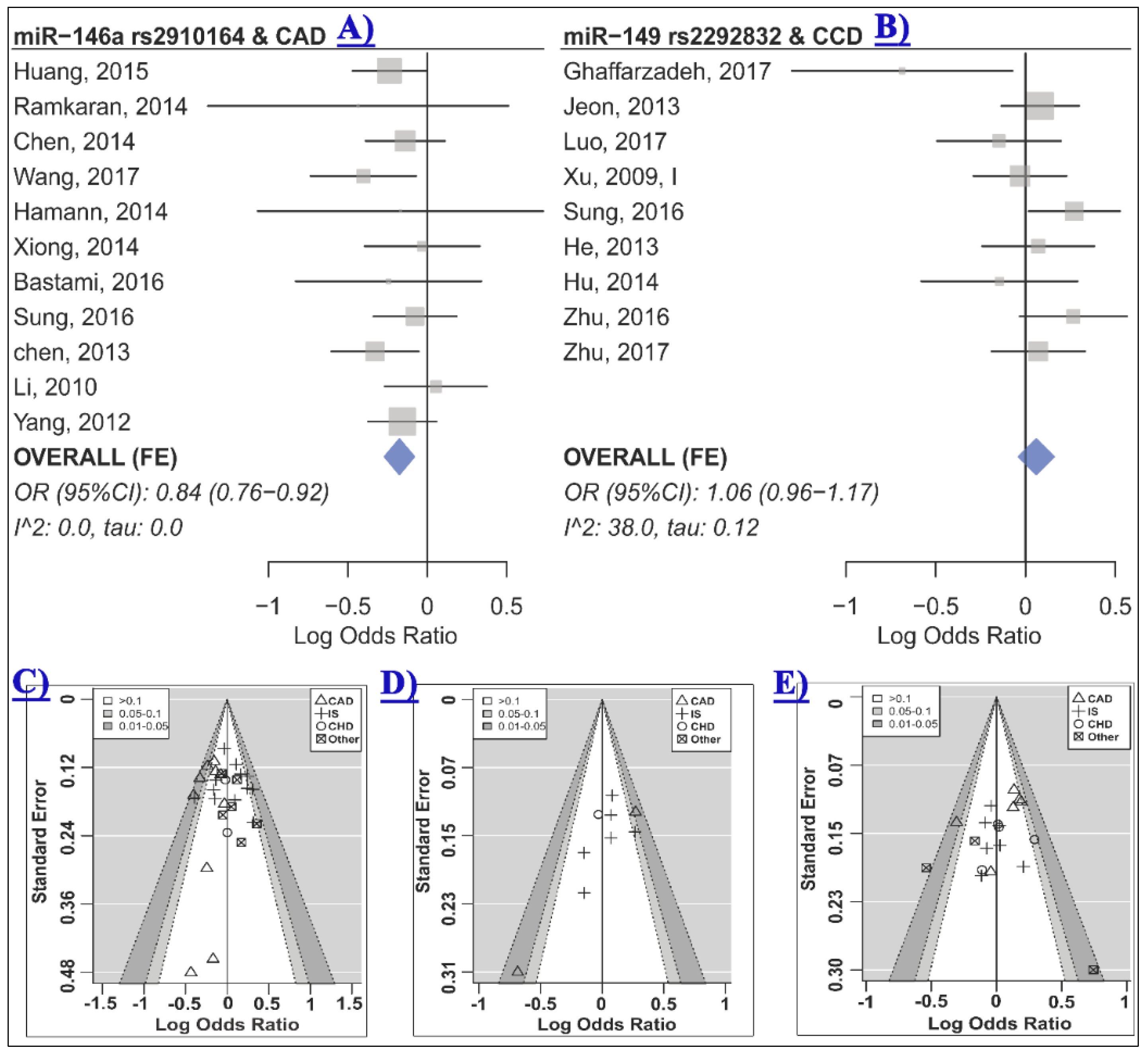
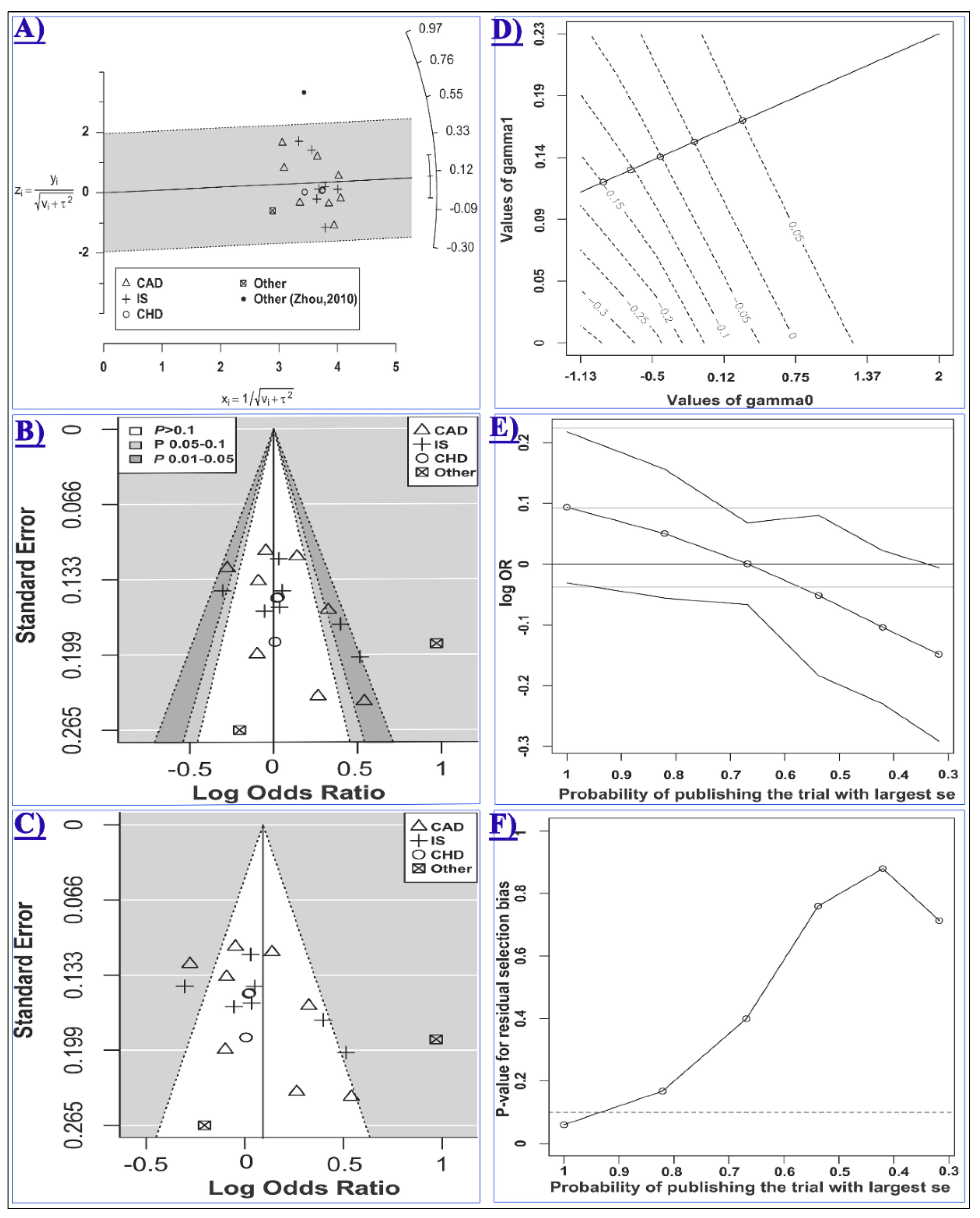
2.2. The Association of miR-146a rs2910164 and CCD Risk
2.3. The Association of miR-149 rs2292832 and CCD Risk
2.4. The Association of miR-149 rs71428439 and CCD Risk
2.5. The Association of miR-196a2 rs11614913 and CCD Risk
2.6. The Association of miR-218 rs11134527 and CCD Risk
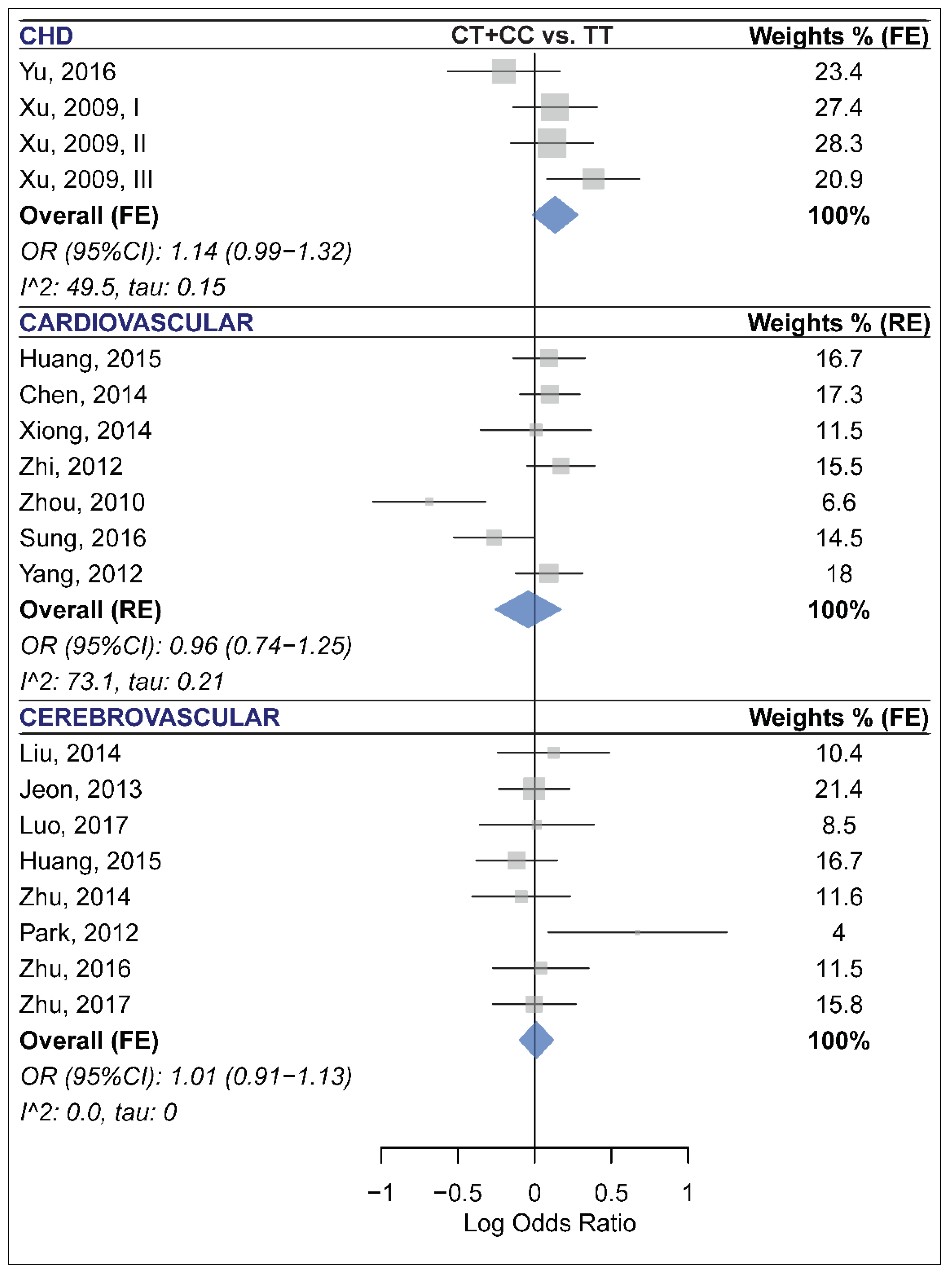
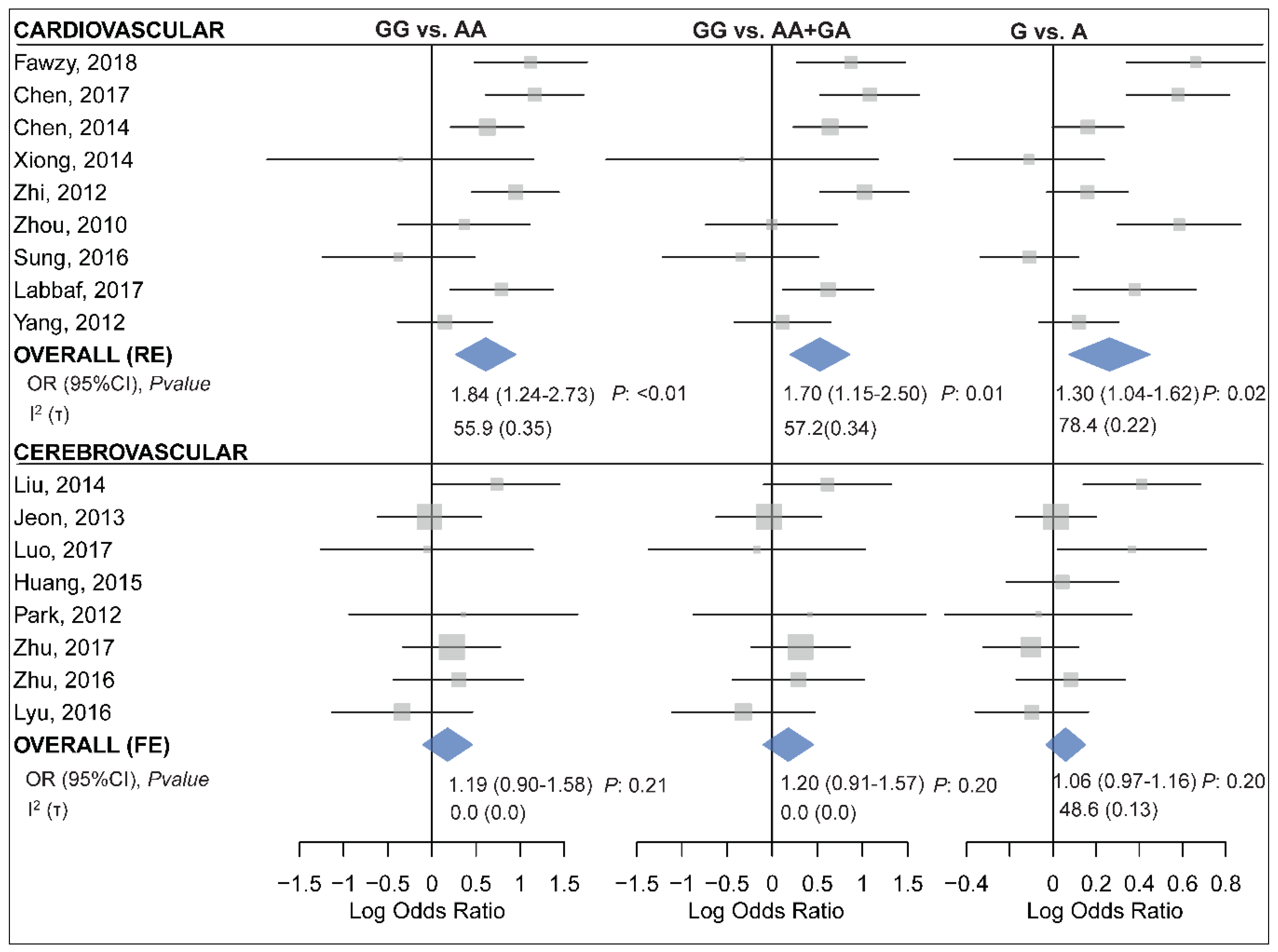
2.7. The Association of miR-499 rs3746444 and CCD Risk
3. Discussion
3.1. The Association of miRNA Polymorphisms with Risk of CCDs
3.2. Possible Pathogenetic Mechanisms and Effects of miR-146a rs2910164
3.3. Possible Pathogenetic Mechanisms and Effects of miR-499 rs3746444
4. Materials and Methods
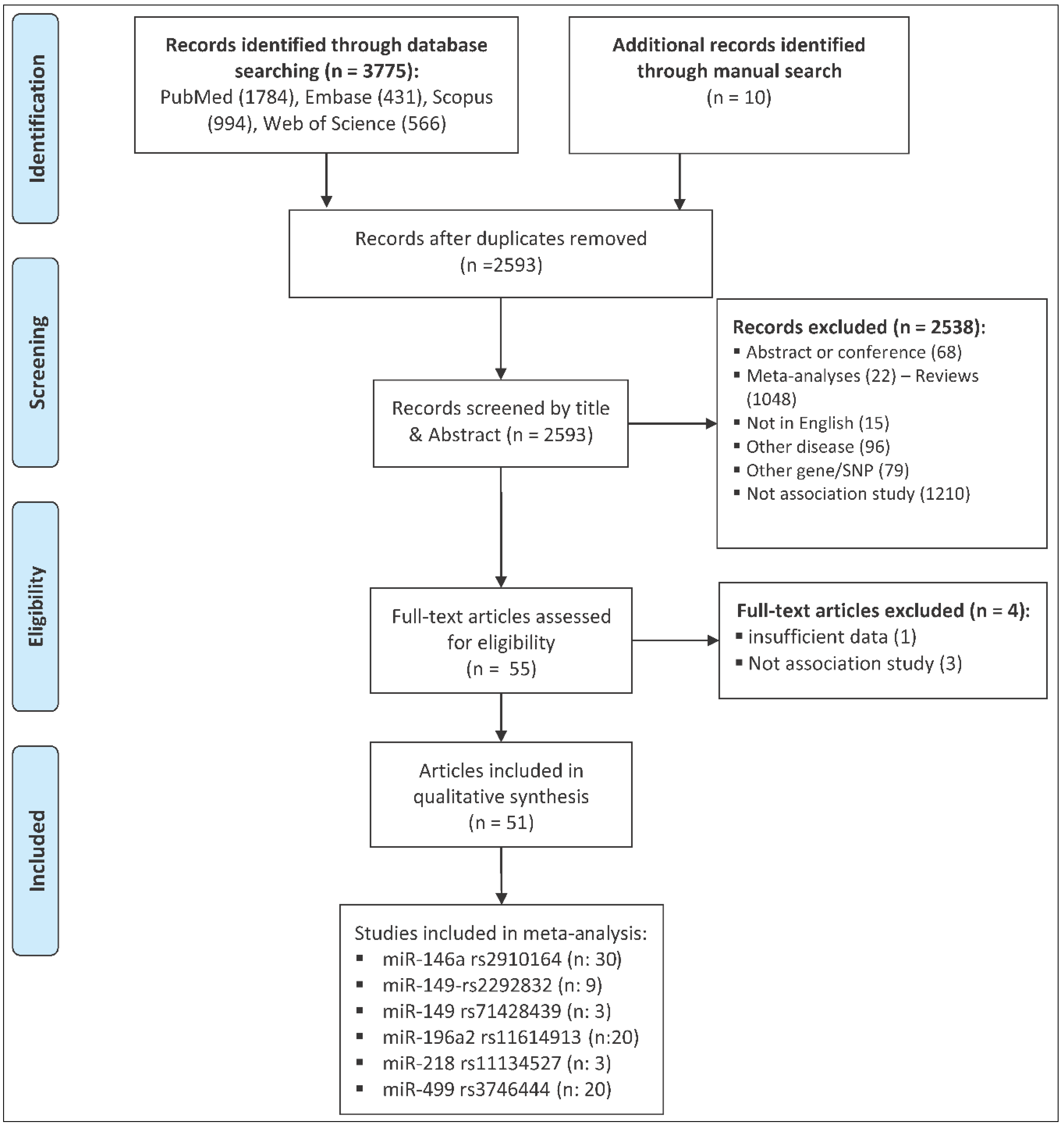
4.1. Publication Search
4.2. Inclusion and Exclusion Criteria
4.3. Data Extraction
4.4. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| CCD | Cardiocerebrovascular diseases |
| CVD | Cardiovascular diseases |
| CBVD | Cerebrovascular diseases |
| CHD | Congenital heart disease |
| CAD | Coronary artery disease |
| CI | Confidence interval |
| FE | Fixed-effects model |
| HWE | Hardy-Weinberg equilibrium |
| HWD | Deviation from HWE |
| IS | Ischemic stroke |
| OR | Odds ratio |
| RE | Random-effects model |
| SBI | Silent brain infarction |
| SNP | Single nucleotide polymorphism |
References
- Aho, K.; Harmsen, P.; Hatano, S.; Marquardsen, J.; Smirnov, V.E.; Strasser, T. Cerebrovascular disease in the community: Results of a WHO collaborative study. Bull. World Health Organ. 1980, 58, 113–130. [Google Scholar] [PubMed]
- Mozaffarian, D.; Benjamin, E.J.; Go, A.S.; Arnett, D.K.; Blaha, M.J.; Cushman, M.; Das, S.R.; de Ferranti, S.; Despres, J.P.; Fullerton, H.J.; et al. Heart Disease and Stroke Statistics-2016 Update: A Report from the American Heart Association. Circulation 2016, 133, e38–e360. [Google Scholar] [CrossRef] [PubMed]
- Oner, T.; Arslan, C.; Yenmis, G.; Arapi, B.; Tel, C.; Aydemir, B.; Sultuybek, G.K. Association of NFKB1A and microRNAs variations and the susceptibility to atherosclerosis. J. Genet. 2017, 96, 251–259. [Google Scholar] [CrossRef] [PubMed]
- Bastami, M.; Ghaderian, S.M.; Omrani, M.D.; Mirfakhraie, R.; Vakili, H.; Parsa, S.A.; Nariman-Saleh-Fam, Z.; Masotti, A. MiRNA-Related Polymorphisms in miR-146a and TCF21 Are Associated with Increased Susceptibility to Coronary Artery Disease in an Iranian Population. Genet. Test. Mol. Biomark. 2016, 20, 241–248. [Google Scholar] [CrossRef] [PubMed]
- Buraczynska, M.; Zukowski, P.; Wacinski, P.; Ksiazek, K.; Zaluska, W. Polymorphism in microRNA-196a2 contributes to the risk of cardiovascular disease in type 2 diabetes patients. J. Diabetes Complicat. 2014, 28, 617–620. [Google Scholar] [CrossRef] [PubMed]
- Cai, M.Y.; Cheng, J.; Zhou, M.Y.; Liang, L.L.; Lian, S.M.; Xie, X.S.; Xu, S.; Liu, X.; Xiong, X.D. The association between pre-miR-27a rs895819 polymorphism and myocardial infarction risk in a Chinese Han population. Lipids Health Dis. 2018, 17, 7. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Hong, H.; Chen, L.; Shi, X.; Chen, Y.; Weng, Q. Association of microRNA polymorphisms with the risk of myocardial infarction in a Chinese population. Tohoku J. Exp. Med. 2014, 233, 89–94. [Google Scholar] [CrossRef]
- Chen, L.; Wu, Y.T. Association of genetic polymorphisms in microRNAs precursor with the risk and prognosis of coronary heart disease. J. Xi’an Jiaotong Univ. (Med. Sci.) 2013, 34, 495–499. [Google Scholar]
- Chen, Q.Y.; Liu, N.; Ma, J.; Fang, Y.; Cao, Y.; Li, H.; Liu, Y.C. Effect of a pre-microRNA-149 (miR-149) genetic variation on the risk of ischemic stroke in a Chinese Han population. Genet. Mol. Res. 2015, 14, 2582–2589. [Google Scholar] [CrossRef]
- Chen, W.; Shao, D.; Gu, H.; Gong, J.; Zhang, J. Hsa-miR-499 rs3746444 T/C polymorphism is associated with increased risk of coronary artery disease in a Chinese population. Acta Cardiol. Sin. 2017, 33, 34–40. [Google Scholar] [CrossRef]
- Choi, G.H.; Ko, K.H.; Kim, J.O.; Kim, J.; Oh, S.H.; Han, I.B.; Cho, K.G.; Kim, O.J.; Bae, J.; Kim, N.K. Association of miR-34a, miR-130a, miR-150 and miR-155 polymorphisms with the risk of ischemic stroke. Int. J. Mol. Med. 2016, 38, 345–356. [Google Scholar] [CrossRef] [PubMed]
- Ding, S.L.; Wang, J.X.; Jiao, J.Q.; Tu, X.; Wang, Q.; Liu, F.; Li, Q.; Gao, J.; Zhou, Q.Y.; Gu, D.F.; et al. A pre-microRNA-149 (miR-149) genetic variation affects miR-149 maturation and its ability to regulate the Puma protein in apoptosis. J. Biol. Chem. 2013, 288, 26865–26877. [Google Scholar] [CrossRef] [PubMed]
- Fawzy, M.S.; Toraih, E.A.; Hamed, E.O.; Hussein, M.H.; Ismail, H.M. Association of miR-499a expression and seed region variant (rs3746444) with cardiovascular disease in Egyptian patients. Acta Cardiol. 2018, 73, 131–140. [Google Scholar] [CrossRef] [PubMed]
- Lyu, G.; Wang, S.; Wang, Q. Association of mir-146a rs2910164 and mir-499 rs3746444 polymorphisms with risk of ischemic stroke. New Med. 2016, 47, 257. [Google Scholar]
- Gao, X.; Yang, L.; Luo, H.; Tan, F.; Ma, X.; Lu, C. A Rare Rs139365823 Polymorphism in Pre-miR-138 Is Associated with Risk of Congenital Heart Disease in a Chinese Population. DNA Cell Biol. 2018, 37, 109–116. [Google Scholar] [CrossRef]
- Gao, X.; Yang, L.; Ma, Y.; Yang, J.; Zhang, G.; Huang, G.; Huang, Q.; Chen, L.; Fu, F.; Chen, Y.; et al. No association of functional variant in pri-miR-218 and risk of congenital heart disease in a Chinese population. Gene 2013, 523, 173–177. [Google Scholar] [CrossRef]
- Ghaffarzadeh, M.; Ghaedi, H.; Alipoor, B.; Omrani, M.D.; Kazerouni, F.; Shanaki, M.; Labbaf, A.; Pashaiefar, H.; Rahimipour, A. Association of MIR-149 (RS2292832) Variant with the Risk of Coronary Artery Disease. J. Med. Biochem. 2017, 36, 251–258. [Google Scholar] [CrossRef]
- Hamann, L.; Glaeser, C.; Schulz, S.; Gross, M.; Franke, A.; Nothlings, U.; Schumann, R.R. A micro RNA-146a polymorphism is associated with coronary restenosis. Int. J. Immunogenet. 2014, 41, 393–396. [Google Scholar] [CrossRef]
- Huang, S.; Lv, Z.; Deng, Q.; Li, L.; Yang, B.; Feng, J.; Wu, T.; Zhang, X.; Cheng, J. A genetic variant in Pre-miR-146a (rs2910164 C>G) Is associated with the decreased risk of acute coronary syndrome in a Chinese population. Tohoku J. Exp. Med. 2015, 237, 227–233. [Google Scholar] [CrossRef]
- Huang, S.L.; Zhou, S.Q.; Zhang, Y.W.; Lv, Z.Q.; Li, S.S.; Xie, C.H.; Ke, Y.B.; Deng, P.J.; Geng, Y.J.; Zhang, Q.; et al. Association of the Genetic Polymorphisms in Pre-MicroRNAs with Risk of Ischemic Stroke in a Chinese Population. PLoS ONE 2015, 10, e0117007. [Google Scholar] [CrossRef]
- Sun, J. Association of miRNA-146a and EPHX2 Polymorphisms with Risk of Ischemic Stroke in Changsha Han Population and the Mechanisms. Master’s Thesis, Central South University, Changsha, China, 2011. [Google Scholar]
- Jeon, Y.J.; Kim, O.J.; Kim, S.Y.; Oh, S.H.; Oh, D.; Kim, O.J.; Shin, B.S.; Kim, N.K. Association of the miR-146a, miR-149, miR-196a2, and miR-499 polymorphisms with ischemic stroke and silent brain infarction risk. Arterioscler. Thromb. Vasc. Biol. 2013, 33, 420–430. [Google Scholar] [CrossRef]
- Kim, J.; Choi, G.H.; Ko, K.H.; Kim, J.O.; Oh, S.H.; Park, Y.S.; Kim, O.J.; Kim, N.K. Association of the single nucleotide polymorphisms in microRNAs 130b, 200b, and 495 with Ischemic stroke susceptibility and post-stroke mortality. PLoS ONE 2016, 11, e0162519. [Google Scholar] [CrossRef] [PubMed]
- Li, L. Association of miRNA-146a Polymorphism with Risk of Cardiovascular Disease and Ischemia Stroke and the Mechanisms. Master’s Thesis, Central South University, Changsha, China, 2010. [Google Scholar]
- Labbaf, A.; Ghaedi, H.; Alipoor, B.; Omrani, M.D.; Kazerouni, F.; Shanaki, M.; Ghaffarzadeh, M.; Pashaiefar, H.; Rahimipour, A. The pre-mir-499 Variant rs3746444 May Contribute to Coronary Artery Disease Susceptibility: A Case-Control and Meta-Analysis Study. Clin. Lab. 2017, 63, 587–595. [Google Scholar] [CrossRef]
- Li, Y.; Zhu, J.; Ma, G.D.; Li, K.S.; Gui, L.L. Association between miR-146a gene polymorphism and lacunar infarction. Shandong Med. J. 2014, 54, 1–3. [Google Scholar]
- Liu, Y.; Ma, Y.; Zhang, B.; Wang, S.X.; Wang, X.M.; Yu, J.M. Genetic polymorphisms in pre-microRNAs and risk of ischemic stroke in a Chinese population. J. Mol. Neurosci. 2014, 52, 473–480. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.M.; Wang, Y.; Peng, W.; Wu, Z.; Wang, X.H.; Wang, M.L.; Wang, W.; Sun, J.; Zhang, Z.D.; Mo, X.M. Single-nucleotide polymorphism of the pri-miR-34b/c gene is not associated with susceptibility to congenital heart disease in the Han Chinese population. Genet. Mol. Res. 2013, 12, 2937–2944. [Google Scholar] [CrossRef] [PubMed]
- Luo, H.C.; Luo, Q.S.; Wang, C.F.; Lei, M.; Li, B.L.; Wei, Y.S. Association of miR-146a, miR-149, miR-196a2, miR-499 gene Polymorphisms with ischemic stroke in a Chinese people. Oncotarget 2017, 8, 81295–81304. [Google Scholar] [CrossRef]
- Hu, Y.M.; Li, S.J.; Jiang, X.F.; Li, G.; Zhang, M.L.; Zhang, Q.L.; Xiang, L. Study on the association of miR-146aC>G, miR-149T>C polymorphism with susceptibility to ischemic stroke. Prog. Mod. Biomed. 2014, 14, 5648–5651. [Google Scholar]
- Park, Y.S.; Jeon, Y.J.; Lee, B.E.; Kim, T.G.; Choi, J.U.; Kim, D.S.; Kim, N.K. Association of the miR-146aC>G, miR-196a2C>T, and miR-499A>G polymorphisms with moyamoya disease in the Korean population. Neurosci. Lett. 2012, 521, 71–75. [Google Scholar] [CrossRef]
- Qu, J.Y.; Xi, J.; Zhang, Y.H.; Zhang, C.N.; Song, L.; Song, Y.; Hui, R.T.; Chen, J.Z. Association of the microRNA-146a SNP rs2910164 with ischemic stroke incidence and prognosis in a Chinese population. Int. J. Mol. Sci. 2016, 17, 660. [Google Scholar] [CrossRef]
- Ramkaran, P.; Khan, S.; Phulukdaree, A.; Moodley, D.; Chuturgoon, A.A. miR-146a Polymorphism Influences Levels of miR-146a, IRAK-1, and TRAF-6 in Young Patients with Coronary Artery Disease. Cell Biochem. Biophys. 2014, 68, 259–266. [Google Scholar] [CrossRef] [PubMed]
- He, S.J.; Han, Y.F. Association between miR-149 polymorphism and ischemic stroke of Han population in Hanzhong of Shanxi. J. Mod. Lab. Med. 2013, 28, 32–34. [Google Scholar]
- Shen, J.; Zhang, M.; Sun, M.F.; Tang, K.; Zhou, B. The relationship of miR-146a gene polymorphism with carotid atherosclerosis in Chinese patients with type 2 diabetes mellitus. Thromb. Res. 2015, 136, 1149–1155. [Google Scholar] [CrossRef] [PubMed]
- Sima, X.; Sun, H.; Zhou, P.; You, C. A potential polymorphism in the promoter of let-7 is associated with an increased risk of Intracranial Aneurysm: A case-control study. Medicine 2015, 94. [Google Scholar] [CrossRef] [PubMed]
- Sima, X.T.; Xu, J.G.; Li, J.; You, C. Association between the hsa-miR-146a rs2910164 functional polymorphism with susceptibility to intracranial aneurysm. Genet. Mol. Res. 2015, 14, 7680–7686. [Google Scholar] [CrossRef] [PubMed]
- Sung, J.H.; Kim, S.H.; Yang, W.I.; Kim, W.J.; Moon, J.Y.; Kim, I.J.; Cha, D.H.; Cho, S.Y.; Kim, J.O.; Kim, K.A.; et al. miRNA polymorphisms (miR146a, miR149, miR196a2 and miR499) are associated with the risk of coronary artery disease. Mol. Med. Rep. 2016, 14, 2328–2342. [Google Scholar] [CrossRef]
- Wang, Y.Q.; Wang, X.T.; Li, Z.Y.; Chen, L.L.; Zhou, L.P.; Li, C.P.; Ouyang, D.S. Two Single Nucleotide Polymorphisms (rs2431697 and rs2910164) of miR-146a Are Associated with Risk of Coronary Artery Disease. Int. J. Environ. Res. Public Health 2017, 14, 514. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.H.; Liu, R.; Gu, H.Y. Association study of miR-146a rs2910164 CNG polymorphism and risk of congenital heart disease. J. Clin. Exp. Med. 2013, 12, 729–733. [Google Scholar]
- Wei, Y.S.; Xiang, Y.; Liao, P.H.; Wang, J.L.; Peng, Y.F. An rs4705342 T>C polymorphism in the promoter of miR-143/145 is associated with a decreased risk of ischemic stroke. Sci. Rep. 2016, 6. [Google Scholar] [CrossRef]
- Zhu, X. Association of Mirnas and Mthfr Gene Polymorphisms with Ischemic Stroke in the Chinese Han Population. Ph.D. Thesis, Qingdao University, Qingdao, China, 2016. [Google Scholar]
- Xiang, Y.; Guo, J.; Peng, Y.F.; Tan, T.; Huang, H.T.; Luo, H.C.; Wei, Y.S. Association of miR-21, miR-126 and miR-605 gene polymorphisms with ischemic stroke risk. Oncotarget 2017, 8, 95755–95763. [Google Scholar] [CrossRef]
- Xiong, X.D.; Cho, M.; Cai, X.P.; Cheng, J.; Jing, X.; Cen, J.M.; Liu, X.G.; Yang, X.L.; Suh, Y. A common variant in pre-miR-146 is associated with coronary artery disease risk and its mature miRNA expression. Mutat. Res. Fundam. Mol. Mech. Mutagen. 2014, 761, 15–20. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Hu, Z.B.; Xu, Z.F.; Gu, H.Y.; Yi, L.; Cao, H.L.; Chen, J.P.; Tian, T.; Liang, J.; Lin, Y.; et al. Functional Variant in microRNA-196a2 Contributes to the Susceptibility of Congenital Heart Disease in a Chinese Population. Hum. Mutat. 2009, 30, 1231–1236. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y. Common Genetic Variations in Pre-miRNAs and the Risk of Coronary Heart Disease in a Chinese Han Population. Ph.D. Thesis, Peking Union Medical College, Beijing, China, 2012. [Google Scholar]
- Yang, L.P.; Gao, X.B.; Luo, H.Y.; Huang, Q.Y.; Wei, Y.; Zhang, G.C.; Huang, G.Y.; Su, D.M.; Chen, L.W.; Lu, C.L.; et al. No Association of Pri-miR-143 rs41291957 Polymorphism with the Risk of Congenital Heart Disease in a Chinese Population. Pediatr. Cardiol. 2014, 35, 1057–1061. [Google Scholar] [CrossRef]
- Yu, K.; Ji, Y.; Wang, H.; Xuan, Q.K.; Li, B.B.; Xiao, J.J.; Sun, W.; Kong, X.Q. Association of miR-196a2, miR-27a, and miR-499 polymorphisms with isolated congenital heart disease in a Chinese population. Genet. Mol. Res. 2016, 15. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Yang, J.S.; Xue, Q.; Yang, D.; Lu, Y.B.; Guang, X.F.; Zhang, W.H.; Ba, R.Q.; Zhu, H.W.; Ma, X. An rs13293512 polymorphism in the promoter of let-7 is associated with a reduced risk of ischemic stroke. J. Thromb. Thrombolysis 2016, 42, 610–615. [Google Scholar] [CrossRef]
- Zhi, H.; Wang, L.N.; Ma, G.S.; Ye, X.Z.; Yu, X.J.; Zhu, Y.; Zhang, Y.; Zhang, J.J.; Wang, B. Polymorphisms of miRNAs genes are associated with the risk and prognosis of coronary artery disease. Clin. Res. Cardiol. 2012, 101, 289–296. [Google Scholar] [CrossRef] [PubMed]
- Zhong, H.D.; Cai, Y.J.; Cheng, J.F.; Cai, D.; Chen, L.; Su, C.B.; Li, K.Y.; Chen, P.H.; Xu, J.R.; Cui, L.L. Apolipoprotein E Epsilon 4 Enhances the Association between the rs2910164 Polymorphism of miR-146a and Risk of Atherosclerotic Cerebral Infarction. J. Atheroscler. Thromb. 2016, 23, 819–829. [Google Scholar] [CrossRef]
- Zhou, B.; Rao, L.; Peng, Y.; Wang, Y.Y.; Chen, Y.; Song, Y.P.; Zhang, L. Common genetic polymorphisms in pre-microRNAs were associated with increased risk of dilated cardiomyopathy. Clin. Chim. Acta 2010, 411, 1287–1290. [Google Scholar] [CrossRef]
- Zhu, H.; Zhang, H.; Bao, L.; Dai, M. Analysis of association of genetic polymorphisms of micrornas with ischemic stroke. Chin. J. Med. Genet. 2017, 34, 261–265. [Google Scholar] [CrossRef]
- Zhu, R.X.; Liu, X.; He, Z.Y.; Li, Q. miR-146a and miR-196a2 Polymorphisms in Patients with Ischemic Stroke in the Northern Chinese Han Population. Neurochem. Res. 2014, 39, 1709–1716. [Google Scholar] [CrossRef]
- Li, D.; Zhu, G.; Di, H.; Li, H.; Liu, X.; Zhao, M.; Zhang, Z.; Yang, Y. Associations between genetic variants located in mature microRNAs and risk of lung cancer. Oncotarget 2016, 7, 41715–41724. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.B.; Zheng, H.K.; Zhang, L.; An, Z.; Wang, X.P.; Shan, R.T.; Zhang, W.Q. A single nucleotide polymorphism located in microRNA-499a causes loss of function resulting in increased expression of osbpl1a and reduced serum HDL level. Oncol. Rep. 2017, 38, 3515–3521. [Google Scholar] [CrossRef] [PubMed]
- Liberati, A.; Altman, D.G.; Tetzlaff, J.; Mulrow, C.; Gøtzsche, P.C.; Ioannidis, J.P.A.; Clarke, M.; Devereaux, P.J.; Kleijnen, J.; Moher, D. The PRISMA statement for reporting systematic reviews and meta-analyses of studies that evaluate healthcare interventions: Explanation and elaboration. BMJ 2009, 339. [Google Scholar] [CrossRef] [PubMed]
- Schwarzer, G. Meta: An R package for meta-analysis. R News 2007, 7, 40–45. [Google Scholar]
- Sterne, J.A.C.; Sutton, A.J.; Ioannidis, J.P.A.; Terrin, N.; Jones, D.R.; Lau, J.; Carpenter, J.; Rücker, G.; Harbord, R.M.; Schmid, C.H.; et al. Recommendations for examining and interpreting funnel plot asymmetry in meta-analyses of randomised controlled trials. BMJ 2011, 343. [Google Scholar] [CrossRef] [PubMed]
- Peters, J.L.; Sutton, A.J.; Jones, D.R.; Abrams, K.R.; Rushton, L. Contour-enhanced meta-analysis funnel plots help distinguish publication bias from other causes of asymmetry. J. Clin. Epidemiol. 2008, 61, 991–996. [Google Scholar] [CrossRef] [PubMed]
- Ryan, B.M.; Robles, A.I.; Harris, C.C. Genetic variation in microRNA networks: The implications for cancer research. Nat. Rev. Cancer 2010, 10, 389–402. [Google Scholar] [CrossRef]
- Jazdzewski, K.; de la Chapelle, A. Genomic sequence matters: A SNP in microRNA-146a can turn anti-apoptotic. Cell Cycle 2009, 8, 1642–1643. [Google Scholar] [CrossRef]
- Alipoor, B.; Ghaedi, H.; Meshkani, R.; Omrani, M.D.; Sharifi, Z.; Golmohammadi, T. The rs2910164 variant is associated with reduced miR-146a expression but not cytokine levels in patients with type 2 diabetes. J. Endocrinol. Investig. 2018, 41, 557–566. [Google Scholar] [CrossRef]
- Vinci, S.; Gelmini, S.; Pratesi, N.; Conti, S.; Malentacchi, F.; Simi, L.; Pazzagli, M.; Orlando, C. Genetic variants in miR-146a, miR-149, miR-196a2, miR-499 and their influence on relative expression in lung cancers. Clin. Chem. Lab. Med. 2011, 49, 2073–2080. [Google Scholar] [CrossRef]
- Sluijter, J.P.; van Mil, A.; van Vliet, P.; Metz, C.H.; Liu, J.; Doevendans, P.A.; Goumans, M.J. MicroRNA-1 and -499 regulate differentiation and proliferation in human-derived cardiomyocyte progenitor cells. Arterioscler. Thromb. Vasc. Biol. 2010, 30, 859–868. [Google Scholar] [CrossRef] [PubMed]
- Wilson, K.D.; Hu, S.; Venkatasubrahmanyam, S.; Fu, J.D.; Sun, N.; Abilez, O.J.; Baugh, J.J.; Jia, F.; Ghosh, Z.; Li, R.A.; et al. Dynamic microRNA expression programs during cardiac differentiation of human embryonic stem cells: Role for miR-499. Circ. Cardiovasc. Genet. 2010, 3, 426–435. [Google Scholar] [CrossRef] [PubMed]
- Shieh, J.T.C.; Huang, Y.; Gilmore, J.; Srivastava, D. Elevated miR-499 Levels Blunt the Cardiac Stress Response. PLoS ONE 2011, 6, e19481. [Google Scholar] [CrossRef] [PubMed]
- Adachi, T.; Nakanishi, M.; Otsuka, Y.; Nishimura, K.; Hirokawa, G.; Goto, Y.; Nonogi, H.; Iwai, N. Plasma microRNA 499 as a biomarker of acute myocardial infarction. Clin. Chem. 2010, 56, 1183–1185. [Google Scholar] [CrossRef] [PubMed]
- Xin, Y.; Yang, C.; Han, Z. Circulating miR-499 as a potential biomarker for acute myocardial infarction. Ann. Transl. Med. 2016, 4, 135. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Jia, Z.; Zhang, C.; Sun, M.; Wang, W.; Chen, P.; Ma, K.; Zhang, Y.; Li, X.; Zhou, C. miR-499 protects cardiomyocytes from H2O2-induced apoptosis via its effects on Pdcd4 and Pacs2. RNA Biol. 2014, 11, 339–350. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.H.; He, K.; Shi, G. Effects of MicroRNA-499 on the Inflammatory Damage of Endothelial Cells during Coronary Artery Disease via the Targeting of PDCD4 through the NF-Κβ/TNF-α Signaling Pathway. Cell. Physiol. Biochem. 2017, 44, 110–124. [Google Scholar] [CrossRef]
- Bell, M.L.; Buvoli, M.; Leinwand, L.A. Uncoupling of expression of an intronic microRNA and its myosin host gene by exon skipping. Mol. Cell. Biol. 2010, 30, 1937–1945. [Google Scholar] [CrossRef]
- Huang, C.J.; Nguyen, P.N.; Choo, K.B.; Sugii, S.; Wee, K.; Cheong, S.K.; Kamarul, T. Frequent co-expression of miRNA-5p and -3p species and cross-targeting in induced pluripotent stem cells. Int. J. Med. Sci. 2014, 11, 824–833. [Google Scholar] [CrossRef]
- Choo, K.B.; Soon, Y.L.; Nguyen, P.N.N.; Hiew, M.S.Y.; Huang, C.-J. MicroRNA-5p and -3p co-expression and cross-targeting in colon cancer cells. J. Biomed. Sci. 2014, 21, 95. [Google Scholar] [CrossRef]
- Ro, S.; Park, C.; Young, D.; Sanders, K.M.; Yan, W. Tissue-dependent paired expression of miRNAs. Nucleic Acids Res. 2007, 35, 5944–5953. [Google Scholar] [CrossRef] [PubMed]
- Almeida, M.I.; Nicoloso, M.S.; Zeng, L.; Ivan, C.; Spizzo, R.; Gafa, R.; Xiao, L.; Zhang, X.; Vannini, I.; Fanini, F.; et al. Strand-specific miR-28-5p and miR-28-3p have distinct effects in colorectal cancer cells. Gastroenterology 2012, 142, 886–896.e889. [Google Scholar] [CrossRef] [PubMed]
- Xu, Z.; Han, Y.; Liu, J.; Jiang, F.; Hu, H.; Wang, Y.; Liu, Q.; Gong, Y.; Li, X. MiR-135b-5p and MiR-499a-3p Promote Cell Proliferation and Migration in Atherosclerosis by Directly Targeting MEF2C. Sci. Rep. 2015, 5, 12276. [Google Scholar] [CrossRef] [PubMed]
- Alshatwi, A.A.; Shafi, G.; Hasan, T.N.; Syed, N.A.; Al-Hazzani, A.A.; Alsaif, M.A.; Alsaif, A.A. Differential expression profile and genetic variants of microRNAs sequences in breast cancer patients. PLoS ONE 2012, 7, e30049. [Google Scholar] [CrossRef] [PubMed]
- Lau, J.; Ioannidis, J.P.; Schmid, C.H. Quantitative synthesis in systematic reviews. Ann. Intern. Med. 1997, 127, 820–826. [Google Scholar] [CrossRef] [PubMed]
- DerSimonian, R.; Laird, N. Meta-analysis in clinical trials. Control. Clin. Trials 1986, 7, 177–188. [Google Scholar] [CrossRef]
- Mantel, N.; Haenszel, W. Statistical aspects of the analysis of data from retrospective studies of disease. J. Natl. Cancer Inst. 1959, 22, 719–748. [Google Scholar]
- Galbraith, R.F. Graphical Display of Estimates Having Differing Standard Errors. Technometrics 1988, 30, 271–281. [Google Scholar] [CrossRef]
- Harbord, R.M.; Egger, M.; Sterne, J.A. A modified test for small-study effects in meta-analyses of controlled trials with binary endpoints. Stat. Med. 2006, 25, 3443–3457. [Google Scholar] [CrossRef]
- Rucker, G.; Schwarzer, G.; Carpenter, J. Arcsine test for publication bias in meta-analyses with binary outcomes. Stat. Med. 2008, 27, 746–763. [Google Scholar] [CrossRef]
- Copas, J.B.; Shi, J.Q. A sensitivity analysis for publication bias in systematic reviews. Stat. Methods Med. Res. 2001, 10, 251–265. [Google Scholar] [CrossRef] [PubMed]
- Copas, J. What Works?: Selectivity Models and Meta-Analysis. J. R. Stat. Soc. Ser. A 1999, 162, 95–109. [Google Scholar] [CrossRef]
- Duval, S.; Tweedie, R. Trim and fill: A simple funnel-plot-based method of testing and adjusting for publication bias in meta-analysis. Biometrics 2000, 56, 455–463. [Google Scholar] [CrossRef] [PubMed]
- Duval, S.; Tweedie, R. A Nonparametric “Trim and Fill” Method of Accounting for Publication Bias in Meta-Analysis. J. Am. Stat. Assoc. 2000, 95, 89–98. [Google Scholar] [CrossRef]
- Schwarzer, G.; Carpenter, J.; Rucker, G. Empirical evaluation suggests Copas selection model preferable to trim-and-fill method for selection bias in meta-analysis. J. Clin. Epidemiol. 2010, 63, 282–288. [Google Scholar] [CrossRef]
- Carpenter, J.R.; Schwarzer, G.; Rucker, G.; Kunstler, R. Empirical evaluation showed that the Copas selection model provided a useful summary in 80% of meta-analyses. J. Clin. Epidemiol. 2009, 62, 624–631. [Google Scholar] [CrossRef]
- Zintzaras, E.; Lau, J. Synthesis of genetic association studies for pertinent gene-disease associations requires appropriate methodological and statistical approaches. J. Clin. Epidemiol. 2008, 61, 634–645. [Google Scholar] [CrossRef]
- Zintzaras, E. Variance estimation of allele-based odds ratio in the absence of Hardy-Weinberg equilibrium. Eur. J. Epidemiol. 2008, 23, 323–326. [Google Scholar] [CrossRef]
- Zintzaras, E.; Koufakis, T.; Ziakas, P.D.; Rodopoulou, P.; Giannouli, S.; Voulgarelis, M. A meta-analysis of genotypes and haplotypes of methylenetetrahydrofolate reductase gene polymorphisms in acute lymphoblastic leukemia. Eur. J. Epidemiol. 2006, 21, 501–510. [Google Scholar] [CrossRef]
- Srivastava, K.; Srivastava, A. Comprehensive Review of Genetic Association Studies and Meta-Analyses on miRNA Polymorphisms and Cancer Risk. PLoS ONE 2012, 7, e50966. [Google Scholar] [CrossRef]
- Trikalinos, T.A.; Salanti, G.; Khoury, M.J.; Ioannidis, J.P. Impact of violations and deviations in Hardy-Weinberg equilibrium on postulated gene-disease associations. Am. J. Epidemiol. 2006, 163, 300–309. [Google Scholar] [CrossRef] [PubMed]
| Author | miRNA Polymorphism | Region | Genotyping | Source a | Cases b | Controls b | Condition | HWE c |
|---|---|---|---|---|---|---|---|---|
| Bastami, 2016 [4] | miR-146a-rs2910164 | Iran | TaqMan | HB | 34/155/111 | 22/128/150 | CAD | 0.52 |
| Chen, 2013 [8] | miR-146a-rs2910164 | China | TaqMan | NA | 172/305/181 | 134/330/194 | CAD | 0.81 |
| Chen, 2014 [7] | miR-146a-rs2910164 | China | PCR-LDR | NA | 187/463/269 | 153/435/301 | CAD | 0.89 |
| Hamann, 2014 [18] | miR-146a-rs2910164 | Germany | HRM | PB | 12/74/120 | 10/73/117 | CAD | 0.87 |
| Hu, 2014 [30] | miR-146a-rs2910164 | China | PCR-RFLP | NA | 75/87/34 | 97/82/26 | IS | 0.24 |
| Huang, 2015-a [19] | miR-146a-rs2910164 | China | TaqMan | HB | 266/308/143 | 237/348/132 | CAD | 0.87 |
| Huang, 2015-b [20] | miR-146a-rs2910164 | China | TaqMan | HB | 189/261/81 | 219/257/55 | IS | 0.12 |
| Jeon, 2013 [22] | miR-146a-rs2910164 | South Korea | PCR-RFLP | HB | 360/506/185 | 211/266/76 | IS | 0.64 |
| Li, 2010 [24] | miR-146a-rs2910164 | China | PCR-RFLP | HB | 149/184/82 | 345/455/210 | CVD | 0.01 |
| Li, 2014 [26] | miR-146a-rs2910164 | China | SNaPshot | HB | 73/85/15 | 111/136/51 | LI | 0.45 |
| Liu, 2014 [27] | miR-146a-rs2910164 | China | PCR-RFLP | HB | 85/159/52 | 116/198/77 | IS | 0.71 |
| Luo, 2017 [29] | miR-146a-rs2910164 | China | SNaPshot | HB | 129/130/39 | 119/139/45 | IS | 0.74 |
| Lyu, 2016 [14] | miR-146a-rs2910164 | China | TaqMan | HB | 119/198/61 | 153/187/38 | IS | 0.10 |
| Park, 2012 [31] | miR-146a-rs2910164 | South Korea | PCR-RFLP | HB | 38/56/13 | 91/113/36 | MD | 0.97 |
| Qu, 2016 [32] | miR-146a-rs2910164 | China | PCR-LDR | NA | 355/618/166 | 483/869/233 | IS | 0.00 |
| Ramkaran, 2014 [33] | miR-146a-rs2910164 | South Africa | PCR-RFLP | NA | 13/43/50 | 9/46/45 | CAD | 0.69 |
| Shen, 2015 [35] | miR-146a-rs2910164 | China | TaqMan | HB | 217/283/96 | 153/177/49 | CA | 0.91 |
| Sima, 2015 [37] | miR-146a-rs2910164 | China | PCR-RFLP | HB | 37/100/27 | 134/254/90 | IA | 0.13 |
| Sun, 2011 [21] | miR-146a-rs2910164 | China | PCR-RFLP | HB | 146/170/65 | 228/304/118 | IS | 0.38 |
| Sung, 2016 [38] | miR-146a-rs2910164 | South Korea | PCR-RFLP | HB | 203/242/77 | 202/260/73 | CAD | 0.50 |
| Wang, 2013 [40] | miR-146a-rs2910164 | China | PCR-RFLP | HB | 84/121/45 | 48/69/17 | CHD | 0.38 |
| Wang, 2017 [39] | miR-146a-rs2910164 | China | MassARRAY | HB | 136/155/62 | 105/179/84 | CAD | 0.70 |
| Xiong, 2014 [44] | miR-146a-rs2910164 | China | PCR-RFLP | HB | 113/141/41 | 97/125/61 | CAD | 0.10 |
| Xu, 2009, I [45] | miR-146a-rs2910164 | China | PCR-RFLP | HB | 161/245/95 | 164/255/86 | CHD | 0.48 |
| Yang, 2012 [46] | miR-146a-rs2910164 | China | TaqMan | NA | 272/392/165 | 271/457/189 | CAD | 0.92 |
| Zhong, 2016 [51] | miR-146a-rs2910164 | China | CE | HB | 141/128/28 | 113/152/35 | IS | 0.16 |
| Zhou, 2010 [52] | miR-146a-rs2910164 | China | PCR-RFLP | HB | 78/113/30 | 120/165/36 | DM | 0.08 |
| Zhu, 2014 [54] | miR-146a-rs2910164 | China | PCR-LDR | HB | 145/173/50 | 132/185/64 | IS | 0.97 |
| Zhu, 2017 [53] | miR-146a-rs2910164 | China | TaqMan | HB | 170/267/86 | 204/251/55 | IS | 0.58 |
| Zhu, 2016 [42] | miR-146a-rs2910164 | China | PCR-RFLP | HB | 131/194/71 | 154/179/45 | IS | 0.10 |
| Ghaffarzadeh, 2017 [17] | miR-149-rs2292832 | Iran | PCR-RFLP | HB | 53/124/95 | 17/79/53 | CAD | 0.16 |
| He, 2013 [34] | miR-149-rs2292832 | China | PCR-RFLP | NA | 138/162/57 | 160/175/38 | IS | 0.37 |
| Hu, 2014 [30] | miR-149-rs2292832 | China | PCR-RFLP | NA | 79/76/41 | 80/89/36 | IS | 0.24 |
| Jeon, 2013 [22] | miR-149-rs2292832 | South Korea | PCR-RFLP | HB | 479/472/100 | 262/238/53 | IS | 0.97 |
| Luo, 2017 [29] | miR-149-rs2292832 | China | SNaPshot | HB | 131/127/40 | 121/136/46 | IS | 0.50 |
| Sung, 2016 [38] | miR-149-rs2292832 | South Korea | PCR-RFLP | HB | 227/248/47 | 263/219/53 | CAD | 0.51 |
| Xu, 2009, I [45] | miR-149-rs2292832 | China | PCR-RFLP | HB | 224/233/44 | 220/236/49 | CHD | 0.24 |
| Zhu, 2017 [53] | miR-149-rs2292832 | China | TaqMan | HB | 232/221/70 | 240/213/57 | IS | 0.79 |
| Zhu, 2016 [42] | miR-149-rs2292832 | China | PCR-RFLP | HB | 165/179/52 | 190/158/30 | IS | 0.39 |
| Chen, 2014 [7] | miR-149-rs71428439 | China | PCR-LDR | NA | 375/389/155 | 384/381/124 | CAD | 0.07 |
| Chen, 2015 [9] | miR-149-rs71428439 | China | Sequencing | HB | 162/146/40 | 149/172/61 | IS | 0.38 |
| Ding, 2013 [12] | miR-149-rs71428439 | China | Sequencing | NA | 95/130/64 | 132/126/38 | CAD | 0.42 |
| Buraczynska, 2014 [5] | miR-196a2-rs11614913 | Poland | PCR-RFLP | PB | 85/240/209 | 125/417/292 | CVD | 0.25 |
| Chen, 2014 [7] | miR-196a2-rs11614913 | China | PCR-LDR | NA | 312/450/157 | 322/406/161 | CAD | 0.11 |
| Huang, 2015-a [19] | miR-196a2-rs11614913 | China | TaqMan | HB | 190/381/147 | 204/360/156 | CAD | 0.95 |
| Huang, 2015-b [20] | miR-196a2-rs11614913 | China | TaqMan | HB | 166/265/100 | 153/266/112 | IS | 0.91 |
| Jeon, 2013 [22] | miR-196a2-rs11614913 | South Korea | PCR-RFLP | HB | 297/533/221 | 156/292/105 | IS | 0.14 |
| Liu, 2014 [27] | miR-196a2-rs11614913 | China | PCR-RFLP | HB | 64/181/51 | 93/214/84 | IS | 0.07 |
| Luo, 2017 [29] | miR-196a2-rs11614913 | China | SNaPshot | HB | 73/138/87 | 75/159/69 | IS | 0.43 |
| Park, 2012 [31] | miR-196a2-rs11614913 | South Korea | PCR-RFLP | HB | 18/64/25 | 68/115/57 | MD | 0.60 |
| Sung, 2016 [38] | miR-196a2-rs11614913 | South Korea | PCR-RFLP | HB | 179/236/107 | 153/274/108 | CAD | 0.51 |
| Xiong, 2014 [44] | miR-196a2-rs11614913 | China | PCR-RFLP | HB | 86/131/78 | 83/132/68 | CAD | 0.32 |
| Xu, 2009, I [45] | miR-196a2-rs11614913 | China | PCR-RFLP | HB | 140/241/120 | 155/262/88 | CHD | 0.23 |
| Xu, 2009, II [45] | miR-196a2-rs11614913 | China | PCR-RFLP | HB | 143/245/114 | 167/283/91 | CHD | 0.13 |
| Xu, 2009, III [45] | miR-196a2-rs11614913 | China | PCR-RFLP | HB | 77/168/76 | 233/380/124 | CHD | 0.16 |
| Yang, 2012 [46] | miR-196a2-rs11614913 | China | TaqMan | NA | 202/463/163 | 241/463/217 | CAD | 0.89 |
| Yu, 2016 [48] | miR-196a2-rs11614913 | China | TaqMan | PB | 93/152/52 | 75/137/64 | CHD | 0.98 |
| Zhi, 2012 [50] | miR-196a2-rs11614913 | China | PCR-RFLP | PB | 291/470/155 | 208/278/98 | CAD | 0.80 |
| Zhou, 2010 [52] | miR-196a2-rs11614913 | China | PCR-RFLP | HB | 93/111/17 | 86/176/59 | DM | 0.07 |
| Zhu, 2014 [54] | miR-196a2-rs11614913 | China | PCR-LDR | HB | 108/189/71 | 105/198/78 | IS | 0.43 |
| Zhu, 2017 [53] | miR-196a2-rs11614913 | China | TaqMan | HB | 150/273/100 | 146/260/104 | IS | 0.40 |
| Zhu, 2016 [42] | miR-196a2-rs11614913 | China | PCR-RFLP | HB | 112/205/79 | 110/196/72 | IS | 0.59 |
| Chen, 2014 [7] | miR-499-rs3746444 | China | PCR-LDR | NA | 612/237/70 | 606/246/37 | CAD | 0.08 |
| Chen, 2017 [10] | miR-499-rs3746444 | China | MassArray | HB | 264/110/47 | 342/103/19 | CAD | 0.00 |
| Fawzy, 2018 [13] | miR-499-rs3746444 | Egypt | TaqMan | PB | 103/116/74 | 64/42/15 | CAD | 0.09 |
| Huang, 2015-b [20] | miR-499-rs3746444 | China | TaqMan | HB | 398/133/0 | 403/128/0 | IS | 0.00 |
| Jeon, 2013 [22] | miR-499-rs3746444 | South Korea | PCR-RFLP | HB | 688/330/33 | 365/170/18 | IS | 0.83 |
| Labbaf, 2017 | miR-499-rs3746444 | Iran | PCR-RFLP | HB | 68/142/78 | 48/77/25 | CAD | 0.61 |
| Liu, 2014 [27] | miR-499-rs3746444 | China | PCR-RFLP | HB | 181/96/19 | 278/99/14 | IS | 0.23 |
| Luo, 2017 [29] | miR-499-rs3746444 | China | SNaPshot | HB | 215/78/5 | 244/53/6 | IS | 0.22 |
| Lyu, 2016 [14] | miR-499-rs3746444 | China | TaqMan | HB | 257/110/11 | 250/113/15 | IS | 0.72 |
| Park, 2012 [31] | miR-499-rs3746444 | South Korea | PCR-RFLP | HB | 76/27/4 | 163/71/6 | MD | 0.73 |
| Sung, 2016 [38] | miR-499-rs3746444 | South Korea | PCR-RFLP | HB | 358/155/9 | 354/168/13 | CAD | 0.23 |
| Xiong, 2014 [44] | miR-499-rs3746444 | China | PCR-RFLP | HB | 227/65/3 | 212/67/4 | CAD | 0.78 |
| Xu, 2009, I [45] | miR-499-rs3746444 | China | PCR-RFLP | HB | 373/123/5 | 367/118/20 | CHD | 0.02 |
| Xu, 2009, II [45] | miR-499-rs3746444 | China | PCR-RFLP | HB | 373/113/16 | 407/121/13 | CHD | 0.35 |
| Yang, 2012 [46] | miR-499-rs3746444 | China | TaqMan | NA | 589/210/28 | 683/212/28 | CAD | 0.03 |
| Yu, 2016 [48] | miR-499-rs3746444 | China | TaqMan | PB | 209/82/6 | 195/76/5 | CHD | 0.56 |
| Zhi, 2012 [50] | miR-499-rs3746444 | China | PCR-RFLP | PB | 629/201/86 | 396/167/21 | CAD | 0.60 |
| Zhou, 2010 [52] | miR-499-rs3746444 | China | PCR-RFLP | HB | 104/104/13 | 219/83/19 | DM | 0.01 |
| Zhu, 2017 [53] | miR-499-rs3746444 | China | TaqMan | HB | 349/124/32 | 328/158/24 | IS | 0.96 |
| Zhu, 2016 [42] | miR-499-rs3746444 | China | PCR-RFLP | HB | 255/123/18 | 249/116/13 | IS | 0.44 |
| Genetic Models | na | Samples | OR b (95% CI) | Pc | PHetd | I2 | τ | Pbiase |
|---|---|---|---|---|---|---|---|---|
| miR-146a rs2910164 | ||||||||
| Homozygote (GG vs. CC) | 30 | 13186/14497 | 0.99 (0.85–1.15) | 0.86 | <0.01 | 67.2 | 0.29 | 0.94 |
| Heterozygote (GC vs. CC) | 30 | 13186/14497 | 0.97 (0.90–1.05) | 0.44 | 0.03 | 35.7 | 0.12 | 0.83 |
| Dominant (GG+GC vs. CC) | 30 | 13186/14497 | 0.98 (0.89–1.07) | 0.58 | <0.01 | 57.5 | 0.17 | 0.89 |
| Recessive (GG vs. GC+CC) | 30 | 13186/14497 | 1.00 (0.90–1.13) | 0.94 | <0.01 | 58.9 | 0.21 | 0.61 |
| Allelic (G vs. C) | 30 | 13186/14497 | 0.99 (0.92–1.06) | 0.72 | <0.01 | 68.7 | 0.14 | 0.94 |
| miR-149 rs2292832 | ||||||||
| Homozygote (CC vs. TT) | 9 | 4116/3511 | 1.11 (0.84–1.46) | 0.40 | 0.04 | 51.1 | 0.24 | - |
| Heterozygote (CT vs. TT) | 9 | 4116/3511 | 1.06 (0.96–1.17) | 0.23 | 0.12 | 38.0 | 0.12 | - |
| Dominant (CT+CC vs. TT) | 9 | 4116/3511 | 1.08 (0.99–1.19) | 0.10 | 0.07 | 45.0 | 0.13 | - |
| Recessive (CC vs. TT+CT) | 9 | 4116/3511 | 1.11 (0.97–1.28) | 0.13 | 0.19 | 29.2 | 0.14 | - |
| Allelic (C vs. T) | 9 | 4116/3511 | 1.07 (1.00–1.15) | 0.05 | 0.07 | 44.9 | 0.09 | - |
| miR-149 rs71428439 | ||||||||
| Homozygote (GG vs. AA) | 3 | 1556/1567 | 1.21 (0.23–6.36) | 0.66 | <0.01 | 87.7 | 0.54 | - |
| Heterozygote (GA vs. AA) | 3 | 1556/1567 | 1.04 (0.51–2.12) | 0.82 | 0.043 | 68.0 | 0.21 | - |
| Dominant (GA+GG vs. AA) | 3 | 1556/1567 | 1.09(0.41–2.89) | 0.73 | <0.01 | 84.2 | 0.31 | - |
| Recessive (GG vs. AA+GA) | 3 | 1556/1567 | 1.18 (0.33–4.17) | 0.62 | <0.01 | 82.2 | 0.40 | - |
| Allelic (G vs. A) | 3 | 1556/1567 | 1.10 (0.46–2.60) | 0.68 | <0.01 | 89.4 | 0.29 | - |
| miR-196a2 rs11614913 | ||||||||
| Homozygote (CC vs. TT) | 20 | 10144/10433 | 1.02 (0.87–1.20) | 0.76 | <0.01 | 60.2 | 0.23 | 0.59 |
| Heterozygote (CT vs. TT) | 20 | 10144/10433 | 1.02 (0.92–1.12) | 0.73 | 0.03 | 41.2 | 0.13 | 0.49 |
| Dominant (CT+CC vs. TT) | 20 | 10144/10433 | 1.02 (0.92–1.13) | 0.70 | 0.01 | 49.5 | 0.14 | 0.55 |
| Recessive (CC vs. TT+CT) | 20 | 10144/10433 | 1.01 (0.89–1.16) | 0.82 | <0.01 | 60.3 | 0.19 | 0.46 |
| Allelic (C vs. T) | 20 | 10144/10433 | 1.01 (0.94–1.09) | 0.70 | <0.01 | 59.6 | 0.11 | 0.57 |
| miR-218 rs11134527 | ||||||||
| Homozygote (GG vs. AA) | 3 | 2322/2754 | 0.96 (0.81–1.13) | 0.68 | 0.39 | 0 | 0 | - |
| Heterozygote (GA vs. AA) | 3 | 2322/2754 | 0.95 (0.84–1.08) | 0.51 | 0.43 | 0 | 0 | - |
| Dominant (GA+GG vs. AA) | 3 | 2322/2754 | 0.96 (0.85–1.08) | 0.51 | 0.79 | 0 | 0 | - |
| Recessive (GG vs. AA+GA) | 3 | 2322/2754 | 0.98 (0.85–1.14) | 0.86 | 0.10 | 54.7 | 0.15 | - |
| Allelic (G vs. A) | 3 | 2322/2754 | 0.97 (0.90–1.05) | 0.59 | 0.67 | 0 | 0 | - |
| miR-499 rs3746444 | ||||||||
| Homozygote (GG vs. AA) | 19 | 9033/8345 | 1.41 (1.06–1.87) | 0.02 | <0.01 | 59.7 | 0.42 | 0.81 |
| Heterozygote (GA vs. AA) | 20 | 9564/8876 | 1.10 (0.95–1.26) | 0.18 | <0.01 | 67.7 | 0.22 | 0.07 |
| Heterozygote-Trim&fill f | - | - | 1.10 (0.95–1.26) | 0.18 | - | - | - | - |
| Heterozygote-Copas f | - | - | 1.05 (0.94–1.17) | 0.35 | - | - | - | - |
| Dominant (GA+GG vs. AA) | 20 | 9564/8876 | 1.15 (0.99–1.32) | 0.05 | <0.01 | 69.0 | 0.22 | 0.18 |
| Recessive (GG vs. AA+GA) | 19 | 9033/8345 | 1.35 (1.03–1.77) | 0.03 | <0.01 | 57.7 | 0.40 | 0.44 |
| Allelic (G vs. A) | 20 | 9564/8876 | 1.16 (1.03–1.30) | 0.02 | <0.01 | 71.3 | 0.20 | 0.91 |
| Genetic Models | na | Samples | OR b (95% CI) | Pc | PHetd | I2 | τ | M e |
|---|---|---|---|---|---|---|---|---|
| Disease category: CVD | ||||||||
| Homozygote (GG vs. CC) | 12 | 5394/6298 | 0.84 (0.68–1.05) | 0.12 | <0.01 | 62.3 | 0.25 | RE |
| Homozygote HWE | 11 | 5126/5288 | 0.79 (0.66–0.94) | <0.01 | 0.14 | 32.1 | 0.13 | FE |
| Homozygote HWD-adj | 12 | 5394/6298 | 0.84 (0.67–1.06) | 0.13 | <0.01 | 64.0 | 0.26 | RE |
| Heterozygote (GC vs. CC) | 12 | 5394/6298 | 0.85 (0.78–0.93) | <0.01 | 0.70 | 0.0 | 0.00 | FE |
| Dominant (GG+GC vs. CC) | 12 | 5394/6298 | 0.85 (0.79–0.93) | <0.01 | 0.16 | 29.1 | 0.10 | FE |
| Recessive (GG vs. GC+CC) | 12 | 5394/6298 | 0.93 (0.78–1.12) | 0.42 | <0.01 | 65.6 | 0.22 | RE |
| Allelic (G vs. C) | 12 | 5394/6298 | 0.91 (0.82–1.02) | 0.10 | <0.01 | 64.0 | 0.13 | RE |
| Disease category: CBVD | ||||||||
| Homozygote (GG vs. CC) | 17 | 7041/8570 | 1.04 (0.76–1.44) | 0.78 | <0.01 | 77.7 | 0.42 | RE |
| Heterozygote (GC vs. CC) | 17 | 7041/8570 | 1.05 (0.98–1.13) | 0.19 | 0.06 | 37.5 | 0.12 | FE |
| Dominant (GG+GC vs. CC) | 17 | 7041/8570 | 1.05 (0.91–1.21) | 0.51 | <0.01 | 67.6 | 0.21 | RE |
| Recessive (GG vs. GC+CC) | 17 | 7041/8570 | 1.02 (0.78–1.34) | 0.87 | <0.01 | 72.6 | 0.33 | RE |
| Allelic (G vs. C) | 17 | 7041/8570 | 1.01 (0.89–1.16) | 0.83 | <0.01 | 80.3 | 0.21 | RE |
| Disease type: CAD | ||||||||
| Homozygote (GG vs. CC) | 11 | 5173/5977 | 0.82 (0.65–1.03) | 0.08 | <0.01 | 63.0 | 0.25 | RE |
| Homozygote HWE | 10 | 4905/4967 | 0.78 (0.69–0.88) | <0.01 | 0.22 | 24.2 | 0.11 | FE |
| Homozygote HWD-adj | 11 | 5173/5977 | 0.82 (0.65–1.04) | 0.09 | <0.01 | 64.9 | 0.26 | RE |
| Heterozygote (GC vs. CC) | 11 | 5173/5977 | 0.84 (0.76–0.92) | <0.01 | 0.74 | 0.0 | 0.00 | FE |
| Dominant (GG+GC vs. CC) | 11 | 5173/5977 | 0.84 (0.77–0.92) | <0.01 | 0.19 | 26.2 | 0.09 | FE |
| Recessive (GG vs. GC+CC) | 11 | 5173/5977 | 0.92 (0.75–1.11) | 0.34 | <0.01 | 67.5 | 0.22 | RE |
| Allelic (G vs. C) | 11 | 5173/5977 | 0.90 (0.80–1.01) | 0.07 | <0.01 | 64.7 | 0.13 | RE |
| Disease type: IS | ||||||||
| Homozygote (GG vs. CC) | 13 | 5628/7175 | 1.09 (0.72–1.65) | 0.66 | <0.01 | 81.3 | 0.45 | RE |
| Heterozygote (GC vs. CC) | 13 | 5628/7175 | 1.04 (0.91–1.18) | 0.57 | 0.03 | 48.3 | 0.14 | FE |
| Dominant (GG+GC vs. CC) | 13 | 5628/7175 | 1.04 (0.86–1.25) | 0.66 | <0.01 | 74.3 | 0.24 | RE |
| Recessive (GG vs. GC+CC) | 13 | 5628/7175 | 1.08 (0.78–1.52) | 0.61 | <0.01 | 75.5 | 0.34 | FE |
| Allelic (G vs. C) | 13 | 5628/7175 | 1.02 (0.86–1.22) | 0.77 | <0.01 | 84.3 | 0.23 | RE |
| Genetic Models | na | Samples | OR b (95% CI) | Pc | PHetd | I2 | τ | M e |
|---|---|---|---|---|---|---|---|---|
| Disease category: CBVD (IS, SBI) | ||||||||
| Homozygote (CC vs. TT) | 6 | 2821/2322 | 1.25 (1.04–1.50) | 0.02 | 0.09 | 47.9 | 0.22 | FE |
| Heterozygote (CT vs. TT) | 6 | 2821/2322 | 1.06 (0.95–1.20) | 0.30 | 0.53 | 0.0 | 0.00 | FE |
| Dominant (CT+CC vs. TT) | 6 | 2821/2322 | 1.10 (0.99–1.24) | 0.08 | 0.27 | 21.6 | 0.07 | FE |
| Recessive (CC vs. TT+CT) | 6 | 2821/2322 | 1.22 (1.03–1.45) | 0.02 | 0.17 | 35.5 | 0.16 | FE |
| Allelic (C vs. T) | 6 | 2821/2322 | 1.11 (1.02–1.20) | 0.02 | 0.10 | 46.3 | 0.10 | FE |
| Disease type: IS | ||||||||
| Homozygote (CC vs. TT) | 6 | 2448/2322 | 1.31 (1.09–1.58) | <0.01 | 0.14 | 39.6 | 0.19 | FE |
| Heterozygote (CT vs. TT) | 6 | 2448/2322 | 1.07 (0.95–1.21) | 0.26 | 0.52 | 0.0 | 0.00 | FE |
| Dominant (CT+CC vs. TT) | 6 | 2448/2322 | 1.12 (1.00–1.26) | 0.047 | 0.28 | 20.7 | 0.07 | FE |
| Recessive (CC vs. TT+CT) | 6 | 2448/2322 | 1.28 (1.08–1.52) | 0.01 | 0.29 | 18.6 | 0.10 | FE |
| Allelic (C vs. T) | 6 | 2448/2322 | 1.13 (1.04–1.23) | <0.01 | 0.13 | 41.1 | 0.09 | FE |
| Genetic Models | na | Samples | OR b (95% CI) | Pc | PHetd | I2 | τ | M e |
|---|---|---|---|---|---|---|---|---|
| Disease category: CHD | ||||||||
| Homozygote (CC vs. TT) | 4 | 1621/2059 | 1.31 (0.65–2.64) | 0.31 | 0.01 | 75.1 | 0.34 | RE |
| Heterozygote (CT vs. TT) | 4 | 1621/2059 | 1.06 (0.91–1.24) | 0.43 | 0.39 | 1.1 | 0.02 | FE |
| Dominant (CT+CC vs. TT) | 4 | 1621/2059 | 1.14 (0.99–1.32) | 0.07 | 0.11 | 49.5 | 0.15 | FE |
| Recessive (CC vs. TT+CT) | 4 | 1621/2059 | 1.26 (0.71–2.25) | 0.28 | 0.01 | 72.5 | 0.28 | RE |
| Allelic (C vs. T) | 4 | 1621/2059 | 1.13 (0.82–1.57) | 0.32 | 0.01 | 72.8 | 0.16 | RE |
| Disease category: CVD | ||||||||
| Homozygote (CC vs. TT) | 7 | 4419/4253 | 0.89 (0.61–1.29) | 0.47 | <0.01 | 68.5 | 0.25 | RE |
| Heterozygote (CT vs. TT) | 7 | 4419/4253 | 0.99 (0.77–1.27) | 0.94 | <0.01 | 69.2 | 0.20 | RE |
| Dominant (CT+CC vs. TT) | 7 | 4419/4253 | 0.96 (0.74–1.25) | 0.71 | <0.01 | 73.1 | 0.21 | RE |
| Recessive (CC vs. TT+CT) | 7 | 4419/4253 | 0.89 (0.69–1.15) | 0.33 | 0.04 | 55.3 | 0.16 | RE |
| Allelic (C vs. T) | 7 | 4419/4253 | 0.95 (0.79–1.13) | 0.48 | <0.01 | 71.1 | 0.13 | RE |
| Disease category: CBVD | ||||||||
| Homozygote(CC vs. TT) | 8 | 3570/3287 | 1.01 (0.88–1.16) | 0.90 | 0.57 | 0.0 | 0.00 | FE |
| Heterozygote (CT vs. TT) | 8 | 3570/3287 | 1.01 (0.90–1.13) | 0.82 | 0.33 | 12.7 | 0.06 | FE |
| Dominant(CT+CC vs. TT) | 8 | 3570/3287 | 1.01 (0.91–1.13) | 0.83 | 0.46 | 0.0 | 0.00 | FE |
| Recessive(CC vs. TT+CT) | 8 | 3570/3287 | 1.00 (0.89–1.13) | 0.99 | 0.38 | 6.0 | 0.04 | FE |
| Allelic (C vs. T) | 8 | 3570/3287 | 1.00 (0.94–1.08) | 0.89 | 0.62 | 0.0 | 0.00 | FE |
| Disease type: CAD | ||||||||
| Homozygote(CC vs. TT) | 6 | 4198/3932 | 0.99 (0.87–1.12) | 0.84 | 0.81 | 0.0 | 0.00 | FE |
| Heterozygote (CT vs. TT) | 6 | 4198/3932 | 1.08 (0.98–1.20) | 0.11 | 0.08 | 49.0 | 0.13 | FE |
| Dominant (CT+CC vs. TT) | 6 | 4198/3932 | 1.06 (0.96–1.16) | 0.25 | 0.19 | 32.4 | 0.08 | FE |
| Recessive (CC vs. TT+CT) | 6 | 4198/3932 | 0.94 (0.84–1.04) | 0.24 | 0.61 | 0.0 | 0.00 | FE |
| Allelic (C vs. T) | 6 | 4198/3932 | 1.00 (0.94–1.07) | 0.93 | 0.63 | 0.0 | 0.00 | FE |
| Disease type: IS | ||||||||
| Homozygote (CC vs. TT) | 7 | 3090/3047 | 0.98 (0.85–1.14) | 0.82 | 0.73 | 0.0 | 0.00 | FE |
| Heterozygote (CT vs. TT) | 7 | 3090/3047 | 0.99 (0.88–1.12) | 0.92 | 0.91 | 0.0 | 0.00 | FE |
| Dominant (CT+CC vs. TT) | 7 | 3090/3047 | 0.99 (0.89–1.11) | 0.89 | 0.95 | 0.0 | 0.00 | FE |
| Recessive (CC vs. TT+CT) | 7 | 3090/3047 | 0.99 (0.87–1.12) | 0.87 | 0.33 | 13.3 | 0.07 | FE |
| Allelic (C vs. T) | 7 | 3090/3047 | 0.99 (0.93–1.07) | 0.85 | 0.75 | 0.0 | 0.00 | FE |
| Genetic Models | na | Samples | OR b (95% CI) | Pc | PHetd | I2 | τ | M e |
|---|---|---|---|---|---|---|---|---|
| Disease category: CHD | ||||||||
| Homozygote (GG vs. AA) | 3 | 1300/1322 | 0.73 (0.07–7.44) | 0.61 | 0.02 | 74.0 | 0.83 | RE |
| Heterozygote | 3 | 1300/1322 | 1.02 (0.85–1.22) | 0.84 | 1.00 | 0.0 | 0.00 | FE |
| Dominant | 3 | 1300/1322 | 0.99 (0.83–1.17) | 0.88 | 0.77 | 0.0 | 0.00 | FE |
| Recessive | 3 | 1300/1322 | 0.72 (0.07–7.43) | 0.60 | 0.02 | 74.3 | 0.83 | RE |
| Allelic (G vs. A) | 3 | 1300/1322 | 0.96 (0.82–1.12) | 0.59 | 0.31 | 14.3 | 0.06 | FE |
| Disease category: CVD | ||||||||
| Homozygote (GG vs. AA) | 9 | 4702/4270 | 1.84 (1.24–2.73) | <0.01 | 0.02 | 55.9 | 0.35 | RE |
| Heterozygote | 9 | 4702/4270 | 1.18 (0.88–1.58) | 0.22 | 0.00 | 80.6 | 0.31 | RE |
| Dominant | 9 | 4702/4270 | 1.29 (0.97–1.70) | 0.07 | 0.00 | 79.9 | 0.28 | RE |
| Recessive | 9 | 4702/4270 | 1.70 (1.15–2.50) | 0.01 | 0.02 | 57.2 | 0.34 | RE |
| Allelic (G vs. A) | 9 | 4702/4270 | 1.30 (1.04–1.62) | 0.02 | 0.00 | 78.4 | 0.22 | RE |
| Disease category: CBVD | ||||||||
| Homozygote (GG vs. AA) | 8 | 3562/3284 | 1.19 (0.90–1.58) | 0.21 | 0.57 | 0.0 | 0.00 | FE |
| Heterozygote | 8 | 3562/3284 | 1.05 (0.85–1.31) | 0.59 | 0.02 | 58.6 | 0.19 | RE |
| Dominant | 8 | 3562/3284 | 1.07 (0.87–1.31) | 0.45 | 0.03 | 56.0 | 0.17 | RE |
| Recessive | 8 | 3562/3284 | 1.20 (0.91–1.57) | 0.20 | 0.64 | 0.0 | 0.00 | FE |
| Allelic (G vs. A) | 8 | 3562/3284 | 1.06 (0.97–1.16) | 0.20 | 0.06 | 48.6 | 0.13 | FE |
| Disease type: CAD | ||||||||
| Homozygote (GG vs. AA) | 8 | 4405/3949 | 1.91 (1.19–3.07) | 0.01 | 0.01 | 62.8 | 0.39 | RE |
| Heterozygote | 8 | 4405/3949 | 1.06 (0.85–1.34) | 0.54 | 0.01 | 63.6 | 0.19 | RE |
| Dominant | 8 | 4405/3949 | 1.20 (0.92–1.57) | 0.16 | 0.00 | 72.8 | 0.23 | RE |
| Recessive | 8 | 4405/3949 | 1.82 (1.19–2.77) | 0.01 | 0.02 | 57.3 | 0.34 | RE |
| Allelic (G vs. A) | 8 | 4405/3949 | 1.26 (0.99–1.62) | 0.06 | 0.00 | 79.0 | 0.22 | RE |
| Disease type: IS | ||||||||
| Homozygote (GG vs. AA) | 7 | 3082/3044 | 1.20 (0.90–1.60) | 0.21 | 0.48 | 0.0 | 0.00 | FE |
| Heterozygote | 7 | 3082/3044 | 1.06 (0.82–1.36) | 0.61 | 0.01 | 64.9 | 0.21 | RE |
| Dominant | 7 | 3082/3044 | 1.07 (0.85–1.36) | 0.49 | 0.01 | 63.5 | 0.20 | RE |
| Recessive | 7 | 3082/3044 | 1.21 (0.91–1.61) | 0.19 | 0.58 | 0.0 | 0.00 | FE |
| Allelic (G vs. A) | 7 | 3082/3044 | 1.08 (0.89–1.30) | 0.38 | 0.03 | 58.0 | 0.15 | RE |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bastami, M.; Choupani, J.; Saadatian, Z.; Zununi Vahed, S.; Mansoori, Y.; Daraei, A.; Samadi Kafil, H.; Masotti, A.; Nariman-saleh-fam, Z. miRNA Polymorphisms and Risk of Cardio-Cerebrovascular Diseases: A Systematic Review and Meta-Analysis. Int. J. Mol. Sci. 2019, 20, 293. https://doi.org/10.3390/ijms20020293
Bastami M, Choupani J, Saadatian Z, Zununi Vahed S, Mansoori Y, Daraei A, Samadi Kafil H, Masotti A, Nariman-saleh-fam Z. miRNA Polymorphisms and Risk of Cardio-Cerebrovascular Diseases: A Systematic Review and Meta-Analysis. International Journal of Molecular Sciences. 2019; 20(2):293. https://doi.org/10.3390/ijms20020293
Chicago/Turabian StyleBastami, Milad, Jalal Choupani, Zahra Saadatian, Sepideh Zununi Vahed, Yaser Mansoori, Abdolreza Daraei, Hossein Samadi Kafil, Andrea Masotti, and Ziba Nariman-saleh-fam. 2019. "miRNA Polymorphisms and Risk of Cardio-Cerebrovascular Diseases: A Systematic Review and Meta-Analysis" International Journal of Molecular Sciences 20, no. 2: 293. https://doi.org/10.3390/ijms20020293
APA StyleBastami, M., Choupani, J., Saadatian, Z., Zununi Vahed, S., Mansoori, Y., Daraei, A., Samadi Kafil, H., Masotti, A., & Nariman-saleh-fam, Z. (2019). miRNA Polymorphisms and Risk of Cardio-Cerebrovascular Diseases: A Systematic Review and Meta-Analysis. International Journal of Molecular Sciences, 20(2), 293. https://doi.org/10.3390/ijms20020293








