Proteomics of PTI and Two ETI Immune Reactions in Potato Leaves
Abstract
1. Introduction
2. Results and Discussion
2.1. General Characteristics of the Protein Dataset
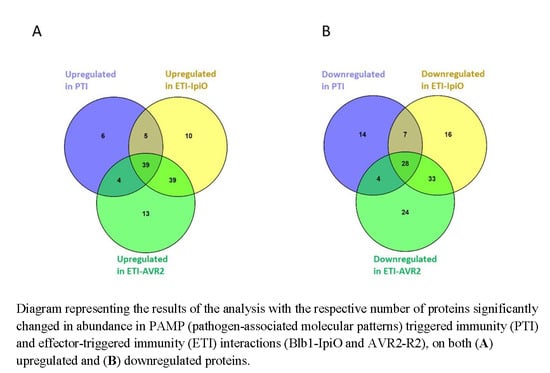
2.2. Proteins with Increased Abundance in the PTI Interaction
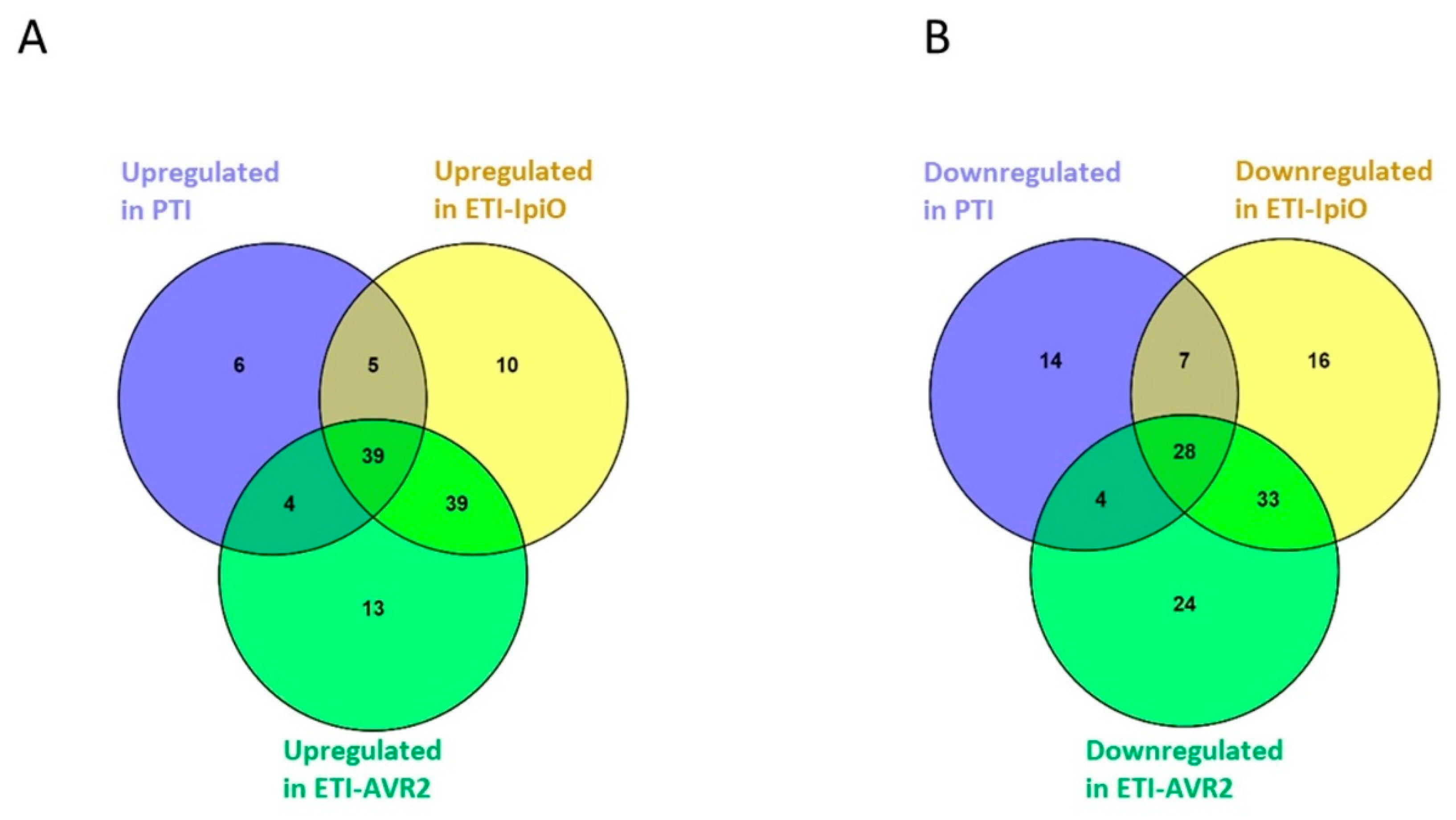
2.3. Proteins with Decreased Abundance in the PTI Interaction
2.4. Proteins with Increased Abundance in ETI Interactions
2.5. Proteins with Decreased Abundance in the ETI Interaction
2.6. Protein Methylation
3. Materials and Methods
3.1. Plants and Infiltration
3.2. Protein Fractionation
3.3. Tryptic Digestion and Mass Spectrometry
3.4. Peptide Data Analysis
3.5. RNA Extraction and Quantitative RT-PCR
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| PAMP | Pathogen-associated molecular pattern |
| PRR | Pattern-recognition receptors |
| PTI | PAMP-triggered immunity |
| ROS | Reactive oxygen species |
| ETS | Effector-triggered susceptibility |
| ETI | Effector-triggered immunity |
| HR | Hypersensitive response |
References
- Zipfel, C. Pattern-recognition receptors in plant innate immunity. Curr. Opin. Immunol. 2008, 20, 10–16. [Google Scholar] [CrossRef] [PubMed]
- Baxter, A.; Mittler, R.; Suzuki, N. ROS as key players in plant stress signalling. J. Exp. Bot. 2013, 65, 1229–1240. [Google Scholar] [CrossRef] [PubMed]
- Pitzschke, A.; Schikora, A.; Hirt, H. MAPK cascade signalling networks in plant defence. Curr. Opin. Plant Biol. 2009, 12, 421–426. [Google Scholar] [CrossRef] [PubMed]
- Jones, J.D.G.; Dangl, J.L. The plant immune system. Nature 2006, 444, 323. [Google Scholar] [CrossRef] [PubMed]
- Coll, N.S.; Epple, P.; Dangl, J.L. Programmed cell death in the plant immune system. Cell Death Differ. 2011, 18, 1247. [Google Scholar] [CrossRef] [PubMed]
- Thomma, B.P.H.J.; Nürnberger, T.; Joosten, M.H.A.J. Of PAMPs and Effectors: The Blurred PTI-ETI Dichotomy. Plant Cell 2011, 23, 4. [Google Scholar] [CrossRef]
- Leibman-Markus, M.; Pizarro, L.; Schuster, S.; Lin, Z.J.D.; Gershony, O.; Bar, M.; Coaker, G.; Avni, A. The intracellular nucleotide-binding leucine-rich repeat receptor (SlNRC4a) enhances immune signalling elicited by extracellular perception. Plant Cell Environ. 2018, 41, 2313–2327. [Google Scholar] [CrossRef]
- Lee, S.; Fu, F.; Xu, S.; Lee, S.Y.; Yun, D.-J.; Mengiste, T. Global Regulation of Plant Immunity by Histone Lysine Methyl Transferases. Plant Cell 2016, 28, 1640. [Google Scholar] [CrossRef]
- Haverkort, A.J.; Struik, P.C.; Visser, R.G.F.; Jacobsen, E. Applied Biotechnology to Combat Late Blight in Potato Caused by Phytophthora Infestans. Potato Res. 2009, 52, 249–264. [Google Scholar] [CrossRef]
- Champouret, N.; Bouwmeester, K.; Rietman, H.; van der Lee, T.; Maliepaard, C.; Heupink, A.; van de Vondervoort, P.J.I.; Jacobsen, E.; Visser, R.G.F.; van der Vossen, E.A.G.; et al. Phytophthora infestans Isolates Lacking Class I ipiO Variants Are Virulent on Rpi-blb1 Potato. Mol. Plant-Microbe Interact. 2009, 22, 1535–1545. [Google Scholar] [CrossRef]
- Lokossou, A.A.; Park, T.-h.; van Arkel, G.; Arens, M.; Ruyter-Spira, C.; Morales, J.; Whisson, S.C.; Birch, P.R.J.; Visser, R.G.F.; Jacobsen, E.; et al. Exploiting Knowledge of R/Avr Genes to Rapidly Clone a New LZ-NBS-LRR Family of Late Blight Resistance Genes from Potato Linkage Group IV. Mol. Plant-Microbe Interact. 2009, 22, 630–641. [Google Scholar] [CrossRef]
- Vleeshouwers, V.G.A.A.; Rietman, H.; Krenek, P.; Champouret, N.; Young, C.; Oh, S.-K.; Wang, M.; Bouwmeester, K.; Vosman, B.; Visser, R.G.F.; et al. Effector Genomics Accelerates Discovery and Functional Profiling of Potato Disease Resistance and Phytophthora Infestans Avirulence Genes. PLoS ONE 2008, 3, e2875. [Google Scholar] [CrossRef]
- Turnbull, D.; Yang, L.; Naqvi, S.; Breen, S.; Welsh, L.; Stephens, J.; Morris, J.; Boevink, P.C.; Hedley, P.E.; Zhan, J.; et al. RXLR Effector AVR2 Up-Regulates a Brassinosteroid-Responsive bHLH Transcription Factor to Suppress Immunity. Plant Physiol. 2017, 174, 356. [Google Scholar] [CrossRef]
- Saunders, D.G.O.; Breen, S.; Win, J.; Schornack, S.; Hein, I.; Bozkurt, T.O.; Champouret, N.; Vleeshouwers, V.G.A.A.; Birch, P.R.J.; Gilroy, E.M.; et al. Host Protein BSL1 Associates with Phytophthora infestans RXLR Effector AVR2 and the Solanum demissum Immune Receptor R2 to Mediate Disease Resistance. Plant Cell 2012, 24, 3420. [Google Scholar] [CrossRef]
- Burra, D.D.; Lenman, M.; Levander, F.; Resjö, S.; Andreasson, E. Comparative Membrane-Associated Proteomics of Three Different Immune Reactions in Potato. Int. J. Mol. Sci. 2018, 19, 538. [Google Scholar] [CrossRef]
- Van Aubel, G.; Buonatesta, R.; Van Cutsem, P. Cos-oga, a new oligosaccharidic elicitor that induces protection against a wide range of plant pathogens. IOBC-WPRS Bull. 2013, 89, 403–407. [Google Scholar]
- Rayapuram, C.; Jensen, M.K.; Maiser, F.; Shanir, J.V.; Hornshøj, H.; Rung, J.H.; Gregersen, P.L.; Schweizer, P.; Collinge, D.B.; Lyngkjær, M.F. Regulation of basal resistance by a powdery mildew-induced cysteine-rich receptor-like protein kinase in barley. Mol. Plant Pathol. 2012, 13, 135–147. [Google Scholar] [CrossRef]
- Xiao, C.; Gao, J.; Zhang, Y.; Wang, Z.; Zhang, D.; Chen, Q.; Ye, X.; Xu, Y.; Yang, G.; Yan, L.; et al. Quantitative Proteomics of Potato Leaves Infected with Phytophthora infestans Provides Insights into Coordinated and Altered Protein Expression during Early and Late Disease Stages. Int. J. Mol. Sci. 2019, 20, 136. [Google Scholar] [CrossRef]
- Afitlhile, M.M.; Fukushige, H.; Nishimura, M.; Hildebrand, D.F. A defect in glyoxysomal fatty acid β-oxidation reduces jasmonic acid accumulation in Arabidopsis. Plant Physiol. Biochem. 2005, 43, 603–609. [Google Scholar] [CrossRef]
- Davidson, R.M.; Reeves, P.A.; Manosalva, P.M.; Leach, J.E. Germins: A diverse protein family important for crop improvement. Plant Sci. 2009, 177, 499–510. [Google Scholar] [CrossRef]
- Lane, B.G. Oxalate, Germins, and Higher-Plant Pathogens. IUBMB Life 2002, 53, 67–75. [Google Scholar] [CrossRef]
- Grosse-Holz, F.; Kelly, S.; Blaskowski, S.; Kaschani, F.; Kaiser, M.; van der Hoorn, R.A.L. The transcriptome, extracellular proteome and active secretome of agroinfiltrated Nicotiana benthamiana uncover a large, diverse protease repertoire. Plant Biotechnol. J. 2018, 16, 1068–1084. [Google Scholar] [CrossRef]
- Roppolo, D.; De Rybel, B.; Tendon, V.D.; Pfister, A.; Alassimone, J.; Vermeer, J.E.M.; Yamazaki, M.; Stierhof, Y.-D.; Beeckman, T.; Geldner, N. A novel protein family mediates Casparian strip formation in the endodermis. Nature 2011, 473, 380. [Google Scholar] [CrossRef]
- Serrazina, S.; Santos, C.; Machado, H.; Pesquita, C.; Vicentini, R.; Pais, M.S.; Sebastiana, M.; Costa, R. Castanea root transcriptome in response to Phytophthora cinnamomi challenge. Tree Genet. Genomes 2015, 11, 6. [Google Scholar] [CrossRef]
- Nes, W.D. Biosynthesis and Requirement for Sterols in the Growth and Reproduction of Oomycetes. In Ecology and Metabolism of Plant Lipids; American Chemical Society: Washington, DC, USA, 1987; Volume 325, pp. 304–328. [Google Scholar]
- Thomas, E.L.; Van der Hoorn, R.A.L. Ten Prominent Host Proteases in Plant-Pathogen Interactions. Int. J. Mol. Sci. 2018, 19, 639. [Google Scholar] [CrossRef]
- Greer, E.L.; Shi, Y. Histone methylation: A dynamic mark in health, disease and inheritance. Nat. Rev. Genet. 2012, 13, 343. [Google Scholar] [CrossRef]
- Ng, A.; Xavier, R.J. Leucine-rich repeat (LRR) proteins: Integrators of pattern recognition and signaling in immunity. Autophagy 2011, 7, 1082–1084. [Google Scholar] [CrossRef]
- Padmanabhan, M.; Cournoyer, P.; Dinesh-Kumar, S.P. The leucine-rich repeat domain in plant innate immunity: A wealth of possibilities. Cell. Microbiol. 2009, 11, 191–198. [Google Scholar] [CrossRef]
- Bengtsson, T.; Weighill, D.; Proux-Wéra, E.; Levander, F.; Resjö, S.; Burra, D.D.; Moushib, L.I.; Hedley, P.E.; Liljeroth, E.; Jacobson, D.; et al. Proteomics and transcriptomics of the BABA-induced resistance response in potato using a novel functional annotation approach. BMC Genom. 2014, 15, 315. [Google Scholar] [CrossRef]
- Lionetti, V.; Cervone, F.; Bellincampi, D. Methyl esterification of pectin plays a role during plant–pathogen interactions and affects plant resistance to diseases. J. Plant Physiol. 2012, 169, 1623–1630. [Google Scholar] [CrossRef]
- Marty, P.; Jouan, B.; Bertheau, Y.; Vian, B.; Goldberg, R. Charge density in stem cell walls of Solanum tuberosum genotypes and susceptibility to blackleg. Phytochemistry 1997, 44, 1435–1441. [Google Scholar] [CrossRef]
- McMillan, G.P.; Hedley, D.; Fyffe, L.; Pérombelon, M.C.M. Potato resistance to soft-rot erwinias is related to cell wall pectin esterification. Physiol. Mol. Plant Pathol. 1993, 42, 279–289. [Google Scholar] [CrossRef]
- Li, J.; Brader, G.; Palva, E.T. Kunitz Trypsin Inhibitor: An Antagonist of Cell Death Triggered by Phytopathogens and Fumonisin B1 in Arabidopsis. Mol. Plant 2008, 1, 482–495. [Google Scholar] [CrossRef]
- Lam, E.; Kato, N.; Lawton, M. Programmed cell death, mitochondria and the plant hypersensitive response. Nature 2001, 411, 848. [Google Scholar] [CrossRef]
- Miller, G.; Shulaev, V.; Mittler, R. Reactive oxygen signaling and abiotic stress. Physiol. Plant. 2008, 133, 481–489. [Google Scholar] [CrossRef]
- Sharma, P.; Jha, A.B.; Dubey, R.S.; Pessarakli, M. Reactive Oxygen Species, Oxidative Damage, and Antioxidative Defense Mechanism in Plants under Stressful Conditions. J. Bot. 2012, 2012, 26. [Google Scholar] [CrossRef]
- Du, Y.-Y.; Wang, P.-C.; Chen, J.; Song, C.-P. Comprehensive Functional Analysis of the Catalase Gene Family in Arabidopsis thaliana. J. Integr. Plant Biol. 2008, 50, 1318–1326. [Google Scholar] [CrossRef]
- Sun, T.; Liu, F.; Wang, W.; Wang, L.; Wang, Z.; Li, J.; Que, Y.; Xu, L.; Su, Y. The Role of Sugarcane Catalase Gene ScCAT2 in the Defense Response to Pathogen Challenge and Adversity Stress. Int. J. Mol. Sci. 2018, 19, 2686. [Google Scholar] [CrossRef]
- Staiger, D.; Korneli, C.; Lummer, M.; Navarro, L. Emerging role for RNA-based regulation in plant immunity. New Phytol. 2013, 197, 394–404. [Google Scholar] [CrossRef]
- Li, D.; Zhang, H.; Liu, H.; Wang, X.; Song, F. OsBIRH1, a DEAD-box RNA helicase with functions in modulating defence responses against pathogen infection and oxidative stress. J. Exp. Bot. 2008, 59, 2133–2146. [Google Scholar] [CrossRef]
- Restrepo, S.; Myers, K.L.; del Pozo, O.; Martin, G.B.; Hart, A.L.; Buell, C.R.; Fry, W.E.; Smart, C.D. Gene Profiling of a Compatible Interaction Between Phytophthora infestans and Solanum tuberosum Suggests a Role for Carbonic Anhydrase. Mol. Plant-Microbe Interact. 2005, 18, 913–922. [Google Scholar] [CrossRef]
- Lukasik, E.; Takken, F.L.W. STANDing strong, resistance proteins instigators of plant defence. Curr. Opin. Plant Biol. 2009, 12, 427–436. [Google Scholar] [CrossRef]
- Real, M.D.; Company, P.; García-Agustín, P.; Bennett, A.B.; González-Bosch, C. Characterization of tomato endo-β-1,4-glucanase Cel1 protein in fruit during ripening and after fungal infection. Planta 2004, 220, 80–86. [Google Scholar] [CrossRef]
- Flors, V.; Leyva, M.d.l.O.; Vicedo, B.; Finiti, I.; Real, M.D.; García-Agustín, P.; Bennett, A.B.; González-Bosch, C. Absence of the endo-β-1,4-glucanases Cel1 and Cel2 reduces susceptibility to Botrytis cinerea in tomato. Plant J. 2007, 52, 1027–1040. [Google Scholar] [CrossRef]
- ZEIER, J. New insights into the regulation of plant immunity by amino acid metabolic pathways. Plant Cell Environ. 2013, 36, 2085–2103. [Google Scholar] [CrossRef]
- Serrano, I.; Buscaill, P.; Audran, C.; Pouzet, C.; Jauneau, A.; Rivas, S. A non canonical subtilase attenuates the transcriptional activation of defence responses in Arabidopsis thaliana. eLife 2016, 5, e19755. [Google Scholar] [CrossRef]
- Mahadevan, C.; Krishnan, A.; Saraswathy, G.G.; Surendran, A.; Jaleel, A.; Sakuntala, M. Transcriptome- Assisted Label-Free Quantitative Proteomics Analysis Reveals Novel Insights into Piper nigrum—Phytophthora capsici Phytopathosystem. Front. Plant Sci. 2016, 7. [Google Scholar] [CrossRef]
- Alban, C.; Tardif, M.; Mininno, M.; Brugière, S.; Gilgen, A.; Ma, S.; Mazzoleni, M.; Gigarel, O.; Martin-Laffon, J.; Ferro, M.; et al. Uncovering the Protein Lysine and Arginine Methylation Network in Arabidopsis Chloroplasts. PLoS ONE 2014, 9, e95512. [Google Scholar] [CrossRef]
- Issakidis-Bourguet, E.; Shen, Y.; Zhou, D.-X. Perspectives on the interactions between metabolism, redox, and epigenetics in plants. J. Exp. Bot. 2016, 67, 5291–5300. [Google Scholar] [CrossRef]
- Locasale, J.W. Serine, glycine and one-carbon units: Cancer metabolism in full circle. Nat. Rev. Cancer 2013, 13, 572. [Google Scholar] [CrossRef]
- Zhang, H.; Deng, X.; Miki, D.; Cutler, S.; La, H.; Hou, Y.-J.; Oh, J.; Zhu, J.-K. Sulfamethazine Suppresses Epigenetic Silencing in Arabidopsis by Impairing Folate Synthesis. Plant Cell 2012, 24, 1230. [Google Scholar] [CrossRef] [PubMed]
- Rojas, C.; Senthil-Kumar, M.; Tzin, V.; Mysore, K. Regulation of primary plant metabolism during plant-pathogen interactions and its contribution to plant defense. Front. Plant Sci. 2014, 5. [Google Scholar] [CrossRef] [PubMed]
- Abreha, K.B.; Alexandersson, E.; Vossen, J.H.; Anderson, P.; Andreasson, E. Inoculation of Transgenic Resistant Potato by Phytophthora infestans Affects Host Plant Choice of a Generalist Moth. PLoS ONE 2015, 10, e0129815. [Google Scholar] [CrossRef] [PubMed]
- Lenman, M.; Ali, A.; Mühlenbock, P.; Carlson-Nilsson, U.; Liljeroth, E.; Champouret, N.; Vleeshouwers, V.G.A.A.; Andreasson, E. Effector-driven marker development and cloning of resistance genes against Phytophthora infestans in potato breeding clone SW93-1015. Theor. Appl. Genet. 2016, 129, 105–115. [Google Scholar] [CrossRef] [PubMed]
- Van Der Vossen, E.; Sikkema, A.; Hekkert, B.t.L.; Gros, J.; Stevens, P.; Muskens, M.; Wouters, D.; Pereira, A.; Stiekema, W.; Allefs, S. An ancient R gene from the wild potato species Solanum bulbocastanum confers broad-spectrum resistance to Phytophthora infestans in cultivated potato and tomato. Plant J. 2003, 36, 867–882. [Google Scholar] [CrossRef] [PubMed]
- Du, J.; Rietman, H.; Vleeshouwers, V.G.A.A. Agroinfiltration and PVX agroinfection in potato and Nicotiana benthamiana. J. Vis. Exp. JoVE 2014, e50971. [Google Scholar] [CrossRef] [PubMed]
- Kessner, D.; Agus, D.; Chambers, M.; Mallick, P.; Burke, R. ProteoWizard: Open source software for rapid proteomics tools development. Bioinformatics 2008, 24, 2534–2536. [Google Scholar] [CrossRef]
- The Potato Genome Sequencing, C.; Xu, X.; Pan, S.; Cheng, S.; Zhang, B.; Mu, D.; Ni, P.; Zhang, G.; Yang, S.; Li, R.; et al. Genome sequence and analysis of the tuber crop potato. Nature 2011, 475, 189. [Google Scholar] [CrossRef]
- Elias, J.E.; Gygi, S.P. Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nat. Methods 2007, 4, 207. [Google Scholar] [CrossRef]
- Häkkinen, J.; Vincic, G.; Månsson, O.; Wårell, K.; Levander, F. The Proteios Software Environment: An Extensible Multiuser Platform for Management and Analysis of Proteomics Data. J. Proteome Res. 2009, 8, 3037–3043. [Google Scholar] [CrossRef]
- Käll, L.; Storey, J.D.; MacCoss, M.J.; Noble, W.S. Assigning Significance to Peptides Identified by Tandem Mass Spectrometry Using Decoy Databases. J. Proteome Res. 2008, 7, 29–34. [Google Scholar] [CrossRef] [PubMed]
- Sandin, M.; Krogh, M.; Hansson, K.; Levander, F. Generic workflow for quality assessment of quantitative label-free LC-MS analysis. Proteomics 2011, 11, 1114–1124. [Google Scholar] [CrossRef] [PubMed]
- Teleman, J.; Chawade, A.; Sandin, M.; Levander, F.; Malmström, J. Dinosaur: A Refined Open-Source Peptide MS Feature Detector. J. Proteome Res. 2016, 15, 2143–2151. [Google Scholar] [CrossRef] [PubMed]
- Sandin, M.; Ali, A.; Hansson, K.; Månsson, O.; Andreasson, E.; Resjö, S.; Levander, F. An Adaptive Alignment Algorithm for Quality-controlled Label-free LC-MS. Mol. Cell. Proteom. 2013, 12, 1407. [Google Scholar] [CrossRef] [PubMed]
- Smyth, G.K. Limma: Linear Models for Microarray Data. Bioinform. Comput. Biol. Solut. Using R Bioconductor 2005, 397–420. [Google Scholar]
- Chawade, A.; Alexandersson, E.; Levander, F. Normalyzer: A Tool for Rapid Evaluation of Normalization Methods for Omics Data Sets. J. Proteome Res. 2014, 13, 3114–3120. [Google Scholar] [CrossRef] [PubMed]
- Taverner, T.; Karpievitch, Y.V.; Polpitiya, A.D.; Brown, J.N.; Dabney, A.R.; Anderson, G.A.; Smith, R.D. DanteR: An extensible R-based tool for quantitative analysis of -omics data. Bioinformatics 2012, 28, 2404–2406. [Google Scholar] [CrossRef] [PubMed]
- Savitski, M.M.; Lemeer, S.; Boesche, M.; Lang, M.; Mathieson, T.; Bantscheff, M.; Kuster, B. Confident Phosphorylation Site Localization Using the Mascot Delta Score. Mol. Amp. Cell. Proteom. 2011, 10, M110.003830. [Google Scholar] [CrossRef] [PubMed]
- Pfaffl, M.W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001, 29, e45. [Google Scholar] [CrossRef] [PubMed]
- Rozen, S.; Skaletsky, H. Primer3 on the WWW for General Users and for Biologist Programmers. Bioinform. Methods Protoc. 2000, 365–386. [Google Scholar]
- Chawade, A.; Alexandersson, E.; Bengtsson, T.; Andreasson, E.; Levander, F. Targeted Proteomics Approach for Precision Plant Breeding. J. Proteome Res. 2016, 15, 638–646. [Google Scholar] [CrossRef] [PubMed]
- Ghislain, M.; Byarugaba, A.A.; Magembe, E.; Njoroge, A.; Rivera, C.; Román, M.L.; Tovar, J.C.; Gamboa, S.; Forbes, G.A.; Kreuze, J.F.; et al. Stacking three late blight resistance genes from wild species directly into African highland potato varieties confers complete field resistance to local blight races. Plant Biotechnol. J. 2019, 17, 1119–1129. [Google Scholar] [CrossRef] [PubMed]
| Protein ID | Protein Name | Degree of Regulation (log2) | ||
|---|---|---|---|---|
| PTI | ETI (Blb1-IpiO) | ETI (AVR2-R2) | ||
| PGSC0003DMP400050772 | Conserved gene of unknown function | 4.57 | 5.07 | 4.63 |
| P00296 | Plastocyanin | 4.47 | 5.10 | 4.88 |
| Q8RXR5 | Polyadenylate-binding protein 2 | 3.20 | 3.79 | 4.20 |
| PGSC0003DMP400046973 | UPF0497 membrane protein | 3.17 | 3.13 | 3.41 |
| Q2XTE0 | Chlorophyll a-b binding protein | 3.07 | 4.51 | 4.81 |
| PGSC0003DMP400047776 | Protein translocase secy subunit | 2.97 | 2.80 | 3.01 |
| PGSC0003DMP400022300 | Chlorophyll binding protein CP24 10B | 2.76 | 3.41 | 3.05 |
| Q2XTC7 | DnaJ-like protein-like | 2.75 | 2.94 | 3.13 |
| P27489 | Chlorophyll a-b binding protein 13 | 2.57 | 4.21 | 4.37 |
| PGSC0003DMP400011243 | Sodium/ exchanger | 2.56 | 2.85 | 2.65 |
| Q2MI72 | Photosystem II reaction center protein | 2.45 | 3.30 | 3.18 |
| Q0PWS7 | Chlorophyll a-b binding protein | 2.22 | 3.51 | 3.67 |
| P10708 | Chlorophyll a-b binding protein 7 | 2.17 | 3.28 | 3.32 |
| C6F3B7 | Protease inhibitor-related protein | 2.08 | 2.46 | 1.83 |
| P32764 | Rubisco small chain 3 | 2.04 | 2.16 | 2.33 |
| P54260 | Aminomethyltransferase | 1.93 | 1.82 | 1.96 |
| Q94KR9 | Translation initiation factor IF-1 | 1.77 | 1.77 | 1.86 |
| Q9FFG6 | AT5g05480/MOP10_2 | 1.75 | 1.10 | 1.24 |
| Q2MI49 | Photosystem I iron-sulfur center | 1.75 | 2.26 | 1.94 |
| D2K7Z2 | Photosystem I reaction center subunit | 1.75 | 2.26 | 2.11 |
| Q7YJ37 | Cytochrome b559 subunit alpha | 1.71 | 2.67 | 2.38 |
| Q2MI71 | Cytochrome b6 | 1.67 | 1.93 | 1.85 |
| PGSC0003DMP400017273 | Brg-1 associated factor | 1.66 | 2.11 | 2.25 |
| E8ZG61 | Pectinesterase | 1.62 | 1.22 | 1.49 |
| PGSC0003DMP400031837 | Germin | 1.57 | 1.43 | 1.36 |
| PGSC0003DMP400033355 | Mitochondrial import inner membrane translocase subunit Tim9 | 1.52 | 1.48 | 1.35 |
| Q0PWS5 | Chlorophyll a-b binding protein | 1.48 | 1.80 | 1.52 |
| PGSC0003DMP400030419 | Heteronuclear ribonucleoprotein A1 | 1.44 | 2.11 | 2.19 |
| P12360 | Chlorophyll a-b binding protein 6A | 1.43 | 1.76 | 1.58 |
| PGSC0003DMP400034002 | Rubisco small chain 1 | 1.40 | 1.73 | 1.73 |
| Q9SL05 | Protein proton gradient regulation 5 | 1.36 | 1.69 | 1.31 |
| P93014 | 30S ribosomal protein S5 | 1.33 | 1.62 | 1.38 |
| Q2VEI0 | Photosystem II CP43 protein | 1.29 | 1.74 | 1.82 |
| Q2MI75 | Photosystem II CP47 protein | 1.24 | 1.80 | 1.93 |
| PGSC0003DMP400053532 | 116 kD U5 small nuclear | 1.15 | 1.24 | 1.48 |
| PGSC0003DMP400038531 | Chloroplast photosystem I, protein V | 1.04 | 1.95 | 1.88 |
| PGSC0003DMP400014027 | Sterol carrier protein 2 | 0.99 | 1.19 | 1.54 |
| Q2UVD9 | PSII cytochrome b559 8kDa subunit | 0.89 | 1.51 | 1.58 |
| Q2VEH0 | ATP synthase subunit beta | 0.51 | 0.88 | 1.10 |
| Q6EIX7 | Potyviral capsid interacting protein 2b | 2.14 | 1.57 | NA |
| B3F8I0 | Glyceraldehyde-3-phosphate dehydrogenase | 1.56 | 1.48 | NA |
| PGSC0003DMP400043522 | Peroxidase 12 | 1.37 | 1.02 | NA |
| PGSC0003DMP400048828 | Splicing factor | 1.29 | 1.25 | NA |
| PGSC0003DMP400039723 | (S)-2-hydroxy-acid oxidase | 1.14 | 0.96 | NA |
| P93363 | Tyrosyl-tRNA synthetase | 1.73 | NA | 1.44 |
| PGSC0003DMP400025801 | Nucellin | 1.48 | NA | 1.48 |
| PGSC0003DMP400044750 | CXE carboxylesterase | 1.07 | NA | 0.87 |
| Q0WVD8 | Adenylate translocator | 0.63 | NA | 0.52 |
| E1AXT8 | Glycolate oxidase | 1.71 | NA | NA |
| PGSC0003DMP400000661 | Beta-galactosidase | 1.42 | NA | NA |
| PGSC0003DMP400010835 | MAR-binding protein | 1.35 | NA | NA |
| PGSC0003DMP400063324 | DUF26 domain-containing protein 1 | 1.33 | NA | NA |
| Q8LK04 | Glyceraldehyde-3-phosphate dehydrogenase | 1.02 | NA | NA |
| PGSC0003DMP400008192 | Glyoxysomal fatty acid beta-oxidation multifunctional protein | 0.75 | NA | NA |
| Protein ID | Protein Name | Degree of Regulation (log2) | ||
|---|---|---|---|---|
| PTI | ETI (Blb1-IpiO) | ETI (AVR2-R2) | ||
| Q38JJ2 | Disulfide-isomerase-like protein | −0.59 | −1.02 | −0.71 |
| Q5M9V4 | ATP synthase subunit alpha | −0.60 | −0.87 | −0.87 |
| Q0WN54 | Molecular chaperone Hsp40/DnaJ | −0.64 | −0.82 | −0.79 |
| Q9ZR78 | ATP synthase subunit beta | −0.66 | −0.91 | −0.87 |
| PGSC0003DMP400042639 | Conserved gene of unknown function | −0.72 | −0.96 | −0.86 |
| PGSC0003DMP400050963 | Chlorophyll a oxygenase | −0.76 | −0.77 | −0.78 |
| PGSC0003DMP400053725 | Ribonucleoprotein. chloroplast | −0.78 | −1.05 | −0.77 |
| PGSC0003DMP400053197 | ATP synthase subunit beta | −0.82 | −0.83 | −0.97 |
| PGSC0003DMP400005664 | Elongation factor Ts | −0.84 | −1.61 | −1.24 |
| PGSC0003DMP400010972 | Hydroxypyruvate reductase | −0.84 | −1.03 | −0.74 |
| PGSC0003DMP400054045 | Alpha-l-fucosidase | −0.87 | −0.90 | −0.80 |
| PGSC0003DMP400002234 | 30S ribosomal protein S1 | −0.89 | −0.94 | −0.74 |
| PGSC0003DMP400008070 | Ubiquitin-associated | −0.93 | −0.84 | −0.76 |
| PGSC0003DMP400001305 | Protein disulfide isomerase family | −0.98 | −1.29 | −0.86 |
| Q2XPV6 | Phosphoglycerate kinase | −1.04 | −0.94 | −0.75 |
| A8MQR4 | 60S acidic ribosomal protein P0 | −1.05 | −1.10 | −0.74 |
| Q9LEB0 | Pectinesterase | −1.05 | −1.04 | −1.08 |
| Q40460 | Ribulose bisphosphate carboxylase | −1.05 | −1.39 | −1.41 |
| PGSC0003DMP400031911 | Pre-mRNA splicing factor | −1.06 | −1.08 | −0.89 |
| C0SUW8 | Eukaryotic translation initiation factor | −1.07 | −0.77 | −0.92 |
| PGSC0003DMP400027836 | Alpha-glucosidase | −1.08 | −1.28 | −1.33 |
| PGSC0003DMP400014012 | Annexin | −1.19 | −1.25 | −1.21 |
| PGSC0003DMP400012806 | Xylem serine proteinase 1 | −1.21 | −1.02 | −1.08 |
| PGSC0003DMP400054512 | Cell division inhibitor | −1.29 | −1.00 | −0.95 |
| PGSC0003DMP400019687 | Translation initiation factor IF-3 | −1.37 | −1.42 | −0.94 |
| PGSC0003DMP400038185 | Transketolase | −1.52 | −1.10 | −0.77 |
| Q9FZ05 | Xyloglucan endotransglucosylase | −1.71 | −0.86 | −1.18 |
| Q84V30 | Phosphatidylserine decarboxylase proenzyme 1 | −2.00 | −1.58 | −1.80 |
| O81394 | Phosphoglycerate kinase | −0.62 | −0.59 | NA |
| Q6J995 | Chloroplast glutamine synthetase | −0.96 | −0.61 | NA |
| PGSC0003DMP400001471 | Multicopper oxidase | −1.00 | −1.03 | NA |
| F4JNJ2 | NAD(P)-binding protein | −1.05 | −0.72 | NA |
| B0FPD8 | Fructose-bisphosphate aldolase | −1.16 | −0.89 | NA |
| P93565 | Fructose-bisphosphate aldolase | −1.17 | −0.98 | NA |
| Q5GM68 | Phosphoenolpyruvate carboxylase 2 | −1.33 | −0.94 | NA |
| F4J912 | Ribosomal protein L5 | −0.93 | NA | −0.73 |
| Q41499 | Spliceosomal protein | −1.03 | NA | −0.53 |
| PGSC0003DMP400048033 | Heme-binding protein | −1.14 | NA | −0.90 |
| PGSC0003DMP400016825 | Kunitz-type protease inhibitor | −1.21 | NA | −1.20 |
| PGSC0003DMP400002269 | Leucine-rich repeat protein | −0.62 | NA | NA |
| Q2MIB4 | ATP synthase subunit b | −0.64 | NA | NA |
| Q9SKI2 | Vacuolar protein | −0.69 | NA | NA |
| PGSC0003DMP400052035 | KH domain-containing protein | −0.72 | NA | NA |
| PGSC0003DMP400032499 | Multicopper oxidase | −0.80 | NA | NA |
| PGSC0003DMP400029544 | Serine-threonine protein kinase | −0.82 | NA | NA |
| PGSC0003DMP400037531 | Small nuclear ribonucleoprotein | −0.87 | NA | NA |
| Q38M62 | Uncharacterized protein | −0.87 | NA | NA |
| PGSC0003DMP400036013 | Nucleic acid binding protein | −1.01 | NA | NA |
| PGSC0003DMP400020414 | Glyceraldehyde-3-phosphate de | −1.13 | NA | NA |
| PGSC0003DMP400011041 | Leucine-rich repeat family protein | −1.23 | NA | NA |
| C0Z2D8 | AT1G20440 protein | −1.38 | NA | NA |
| PGSC0003DMP400004729 | NMDA protein | −1.68 | NA | NA |
| PGSC0003DMP400006728 | Lysine-specific histone demethylase | −2.00 | NA | NA |
| Protein ID | Protein Name | Degree of Regulation (log2) | |
|---|---|---|---|
| ETI (Blb1-IpiO) | ETI (AVR2-R2) | ||
| PGSC0003DMP400024366 | P-rich protein EIG-I30 | 4.45 | 5.3 |
| PGSC0003DMP400015464 | Chlorophyll a/b binding protein | 6.24 | 5.03 |
| PGSC0003DMP400007091 | Acetylglutamate kinase | 2.8 | 3.76 |
| Q40430 | PSI-H | 3.07 | 3.35 |
| Q7M1K8 | Chlorophyll a-b binding protein | 3.01 | 3.18 |
| PGSC0003DMP400029632 | ATP synthase delta chain | 2.49 | 2.69 |
| PGSC0003DMP400002845 | Catalase isozyme 2 | 1.95 | 2.21 |
| P06183 | Photosystem II 10 kDa polypeptide | 2.16 | 2.2 |
| Q9SR73 | 40S ribosomal protein S28-1 | 2.14 | 2.16 |
| P14278 | Chlorophyll a-b binding protein 4 | 2.06 | 2.14 |
| Q9S7N7 | PS I reaction center subunit V | 2.19 | 2.12 |
| Q3S492 | Proteinase inhibitor I | 1.97 | 1.8 |
| Q2MIA5 | Photosystem II D2 protein | 1.75 | 1.78 |
| PGSC0003DMP400022826 | MRNA binding protein | 1.62 | 1.54 |
| PGSC0003DMP400013603 | Translation initiation factor IF-1 | 1.25 | 1.51 |
| Q70PN9 | Putative PSI-D subunit | 1.49 | 1.42 |
| PGSC0003DMP400034978 | PS I reaction center subunit IV isoform | 1.36 | 1.34 |
| B8XLF1 | Chlorophyll a-b binding protein | 1.82 | 1.32 |
| PGSC0003DMP400030353 | Cytosolic acetoacetyl-coenzyme A thiolase | 1.26 | 1.28 |
| F1KC21 | Photosystem II protein D1 | 1.16 | 1.23 |
| PGSC0003DMP400012170 | ATP synthase subunit O | 1.16 | 1.21 |
| PGSC0003DMP400030249 | Chloroplast protease | 0.99 | 1.2 |
| PGSC0003DMP400048099 | Glycolate oxidase | 1.03 | 1.19 |
| PGSC0003DMP400031997 | Photosystem II 11 kDa protein | 1.04 | 1.19 |
| P25079 | Ribulose bisphosphate carboxylase large chain | 0.95 | 1.13 |
| PGSC0003DMP400030421 | Heterogeneous nuclear ribonucleoprotein A1 | 1.37 | 1.11 |
| PGSC0003DMP400025599 | Conserved gene of unknown function | 0.95 | 1.05 |
| Q2MIA8 | DNA-directed RNA polymerase beta | 0.99 | 0.98 |
| Q69GY7 | Cytochrome b6-f iron-sulfur subunit | 0.97 | 0.92 |
| PGSC0003DMP400052418 | Gamma-glutamyl transferase | 1.15 | 0.9 |
| PGSC0003DMP400017746 | Oxygen-evolving enhancer protein 1 | 0.97 | 0.89 |
| Q9S841 | Oxygen-evolving enhancer protein 1-2 | 0.96 | 0.88 |
| PGSC0003DMP400029964 | Heterogeneous nuclear ribonucleoprotein 27C | 0.61 | 0.79 |
| P50433 | Serine hydroxymethyltransferase | 0.62 | 0.74 |
| P93566 | Oxygen-evolving enhancer protein 2 | 0.85 | 0.73 |
| PGSC0003DMP400021624 | PRPL11 | 0.87 | 0.72 |
| Q00321 | Chlorophyll a-b binding protein | 0.83 | 0.72 |
| PGSC0003DMP400046718 | NAD dependent epimerase | 0.75 | 0.58 |
| Q2MI64 | 50S ribosomal protein L14 | 0.82 | 0.58 |
| Q38HV4 | Fructose-bisphosphate aldolase | 1.91 | NA |
| PGSC0003DMP400038572 | Fructose-bisphosphate aldolase | 1.69 | NA |
| PGSC0003DMP400003396 | Non-specific lipid-transfer protein 1 | 1.57 | NA |
| C9EFD1 | Chloroplast ribosomal protein | 1.24 | NA |
| A0MWB6 | Transcriptional coactivator multiprotein bridging factor | 1.09 | NA |
| PGSC0003DMP400041249 | EMB2394 | 0.95 | NA |
| PGSC0003DMP400025698 | High mobility group protein | 0.87 | NA |
| P26320 | Oxygen-evolving enhancer protein 1 | 0.78 | NA |
| PGSC0003DMP400007506 | Photosystem Q | 0.7 | NA |
| C5MR70 | Chloroplast manganese stabilizing protein-II | 0.45 | NA |
| B0ZTE3 | Starch synthase | NA | 4.41 |
| PGSC0003DMP400037406 | 21kD protein | NA | 1.90 |
| PGSC0003DMP400015440 | Ferritin | NA | 1.90 |
| Q6RFS8 | Catalase | NA | 1.42 |
| PGSC0003DMP400048120 | Photosystem I subunit XI | NA | 1.38 |
| Q5NE20 | Carbonic anhydrase | NA | 1.36 |
| P26575 | Rubisco small chain 2A | NA | 1.31 |
| Q308A9 | Ferritin | NA | 1.28 |
| PGSC0003DMP400015694 | Heat shock protein 70 | NA | 1.20 |
| Q8RXK6 | DEAD-box RNA helicase 8 | NA | 1.11 |
| PGSC0003DMP400006368 | Ferredoxin--NADP reductase | NA | 1.10 |
| Q2PYW5 | Catalase | NA | 0.99 |
| PGSC0003DMP400025425 | Gene of unknown function | NA | 0.75 |
| Protein ID | Protein Name | Degree of Regulation (log2) | |
|---|---|---|---|
| ETI (Blb1-IpiO) | ETI (AVR2-R2) | ||
| E1AXT5 | Apoplastic invertase | −0.40 | −0.49 |
| P49316 | Catalase isozyme 2 | −0.66 | −0.63 |
| PGSC0003DMP400032195 | SWIb domain-containing protein | −0.46 | −0.63 |
| PGSC0003DMP400000868 | Actin-11 | −0.70 | −0.65 |
| O49074 | Ribulose bisphosphate carboxylase | −0.64 | −0.66 |
| Q5MA02 | Cytochrome c oxidase subunit 2 | −0.91 | −0.66 |
| PGSC0003DMP400000788 | Inner membrane protein PPF-1 | −0.62 | −0.73 |
| PGSC0003DMP400010545 | O-glycosyl hydrolase | −0.56 | −0.74 |
| PGSC0003DMP400046981 | Kunitz trypsin inhibitor | −0.94 | −0.75 |
| PGSC0003DMP400025093 | ATP-dependent Clp protease | −0.66 | −0.75 |
| PGSC0003DMP400039372 | AGO4-2 | −0.63 | −0.82 |
| Q42871 | Endoglucanase | −1.64 | −0.82 |
| PGSC0003DMP400030598 | Ribonucleoprotein | −0.96 | −0.86 |
| A0A024J2E4 | Putative transcription activator TraR | −0.90 | −1.22 |
| PGSC0003DMP400040149 | Short chain alcohol dehydrogenase | −1.23 | −0.92 |
| PGSC0003DMP400019158 | Aspartate aminotransferase | −0.79 | −0.93 |
| PGSC0003DMP400017124 | Ferredoxin-dependent glutamate synthase 1 | −1.24 | −0.95 |
| PGSC0003DMP400018521 | Subtilase | −0.97 | −1.04 |
| PGSC0003DMP400042601 | Gene of unknown function | −0.66 | −1.04 |
| D0EJY9 | Molecular chaperone Hsp90-3 | −1.29 | −1.08 |
| P32811 | Alpha-glucan phosphorylase | −1.47 | −1.10 |
| Q1EBW2 | Aspartate aminotransferase | −0.91 | −1.13 |
| PGSC0003DMP400018523 | Subtilase | −0.84 | −1.13 |
| PGSC0003DMP400015799 | Basic 7S globulin 2 small subunit | −0.98 | −1.15 |
| A7LKN1 | TAO1 | −1.12 | −1.16 |
| F4HRC1 | THO complex subunit 5A | −0.87 | −1.18 |
| PGSC0003DMP400068875 | Threonyl-tRNA synthetase | −1.21 | −1.19 |
| PGSC0003DMP400039983 | Protein SIS1 | −1.81 | −1.20 |
| PGSC0003DMP400014905 | Polygalacturonase inhibiting protein | −0.88 | −1.24 |
| PGSC0003DMP400011487 | GTP-binding nuclear protein Ran1 | −1.15 | −1.29 |
| PGSC0003DMP400007007 | P69B protein | −1.80 | −1.42 |
| PGSC0003DMP400033260 | Xylem serine proteinase 1 | −2.19 | −1.59 |
| PGSC0003DMP400040582 | Biotin carboxylase carrier protein | −1.18 | −1.61 |
| PGSC0003DMP400032609 | Amidase family protein | −0.50 | NA |
| PGSC0003DMP400044937 | Serine carboxypeptidase | −0.74 | NA |
| PGSC0003DMP400034518 | Chitinase | −0.77 | NA |
| PGSC0003DMP400013944 | Eukaryotic translation initiation factor 4F | −0.88 | NA |
| PGSC0003DMP400046710 | Lysyl-tRNA synthetase | −0.90 | NA |
| PGSC0003DMP400002882 | Glycine-rich protein 2 | −0.90 | NA |
| PGSC0003DMP400001015 | Class III peroxidase | −0.93 | NA |
| PGSC0003DMP400012991 | Apyrase 3 | −0.93 | NA |
| PGSC0003DMP400043401 | NADPH:protochlorophyllide oxidoreductase | −1.00 | NA |
| F4IFG1 | Dynamin related protein | −1.02 | NA |
| PGSC0003DMP400056894 | P69B protein | −1.32 | NA |
| PGSC0003DMP400012143 | Gene of unknown function | −1.39 | NA |
| PGSC0003DMP400009992 | Beta tubulin | −1.43 | NA |
| PGSC0003DMP400036604 | WPP domain-associated protein | −1.45 | NA |
| PGSC0003DMP400033261 | Xylem serine proteinase 1 | −1.55 | NA |
| PGSC0003DMP400032897 | Aspartic proteinase nepenthesin-1 | −2.48 | NA |
| B9JM45 | 6-Phosphogluconate dehydrogenase | NA | −1.31 |
| PGSC0003DMP400020641 | CBL-interacting protein kinase 13 | NA | 0.71 |
| Q8W174 | Peroxidase | NA | 0.70 |
| PGSC0003DMP400016778 | Periplasmic beta-glucosidase | NA | 0.62 |
| Q2MI54 | 30S ribosomal protein S7 | NA | 0.62 |
| PGSC0003DMP400045032 | Pectinesterase | NA | 0.60 |
| E2I6L5 | Polyubiquitin | NA | 0.59 |
| PGSC0003DMP400023756 | GDSL-lipase protein | NA | 0.58 |
| PGSC0003DMP400009676 | Eukaryotic translation initiation factor 2 family protein | NA | 0.58 |
| PGSC0003DMP400045566 | 50S ribosomal protein L19-2 | NA | 0.58 |
| PGSC0003DMP400064549 | Subtilisin-like protease | NA | 0.55 |
| Q3LS00 | Polygalacturonase inhibitor | NA | 0.54 |
| C0Z2Q9 | AT3G13920 protein | NA | 0.53 |
| PGSC0003DMP400014290 | AMP dependent CoA ligase | NA | 0.53 |
| PGSC0003DMP400009317 | Superoxide dismutase | NA | 0.53 |
| F4JWP8 | Homeobox protein knotted-1-like 3 | NA | 0.53 |
| Q9LXG7 | Aldose 1-epimerase family protein | NA | 0.52 |
| PGSC0003DMP400006170 | 60S ribosomal protein L7A | NA | 0.51 |
| Q43286 | Histone H2A | NA | 0.50 |
| PGSC0003DMP400042811 | 60S ribosomal protein L18 | NA | 0.41 |
| PGSC0003DMP400002092 | Histone H1F | NA | 0.37 |
| PGSC0003DMP400011213 | Beta-glucosidase 01 | NA | 0.32 |
| PGSC0003DMP400042879 | Histone H2A.1 | NA | 0.26 |
| PGSC0003DMP400034568 | Histone H1 | NA | 0.07 |
| External Id | Protein Name | Regulation in Immunity | Sequence | Modification |
|---|---|---|---|---|
| P50433 | Serine hydroxymethyltransferase | Up in ETI | YSEGYPGAR | Dimethyl(R) |
| P25079 | Ribulose bisphosphate carboxylase large chain | Up in ETI | DTDILAAFR | Dimethyl(R) |
| DITLGFVDLLR | Dimethyl(R) | |||
| Q2VEH0 | ATP synthase subunit beta | Up in PTI | MRINPTTSGSGVSTLEK | Methyl(R), Oxidation(M) |
| FLSQPFFVAEVFTGSPGKYVGLAETIR | Dimethyl(R) | |||
| FLSQPFFVAEVFTGSPGKYVGLAETIR | Methyl(K), Methyl(R) | |||
| Q9LEB0 | Pectinesterase | Down in PTI | SNTIITGSR | Methyl(R) |
| PGSC0003DMP400046973 | UPF0497 membrane protein | Up in PTI and ETI | YVNGFVDTIETTGIDTFEELR | Dimethyl(R) |
| PGSC0003DMP400002077 | Histone H3.2 | NA | FQSSAVAALQEAAEAYLVGVFEDTNLCAIHAK | Methyl(K) |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Resjö, S.; Zahid, M.A.; Burra, D.D.; Lenman, M.; Levander, F.; Andreasson, E. Proteomics of PTI and Two ETI Immune Reactions in Potato Leaves. Int. J. Mol. Sci. 2019, 20, 4726. https://doi.org/10.3390/ijms20194726
Resjö S, Zahid MA, Burra DD, Lenman M, Levander F, Andreasson E. Proteomics of PTI and Two ETI Immune Reactions in Potato Leaves. International Journal of Molecular Sciences. 2019; 20(19):4726. https://doi.org/10.3390/ijms20194726
Chicago/Turabian StyleResjö, Svante, Muhammad Awais Zahid, Dharani Dhar Burra, Marit Lenman, Fredrik Levander, and Erik Andreasson. 2019. "Proteomics of PTI and Two ETI Immune Reactions in Potato Leaves" International Journal of Molecular Sciences 20, no. 19: 4726. https://doi.org/10.3390/ijms20194726
APA StyleResjö, S., Zahid, M. A., Burra, D. D., Lenman, M., Levander, F., & Andreasson, E. (2019). Proteomics of PTI and Two ETI Immune Reactions in Potato Leaves. International Journal of Molecular Sciences, 20(19), 4726. https://doi.org/10.3390/ijms20194726