Chromatin Remodeling and Epigenetic Regulation in Plant DNA Damage Repair
Abstract
1. Introduction
2. DNA Damage Signaling in the Context of Chromatin
3. DNA Damage Repair in the Context of Chromatin
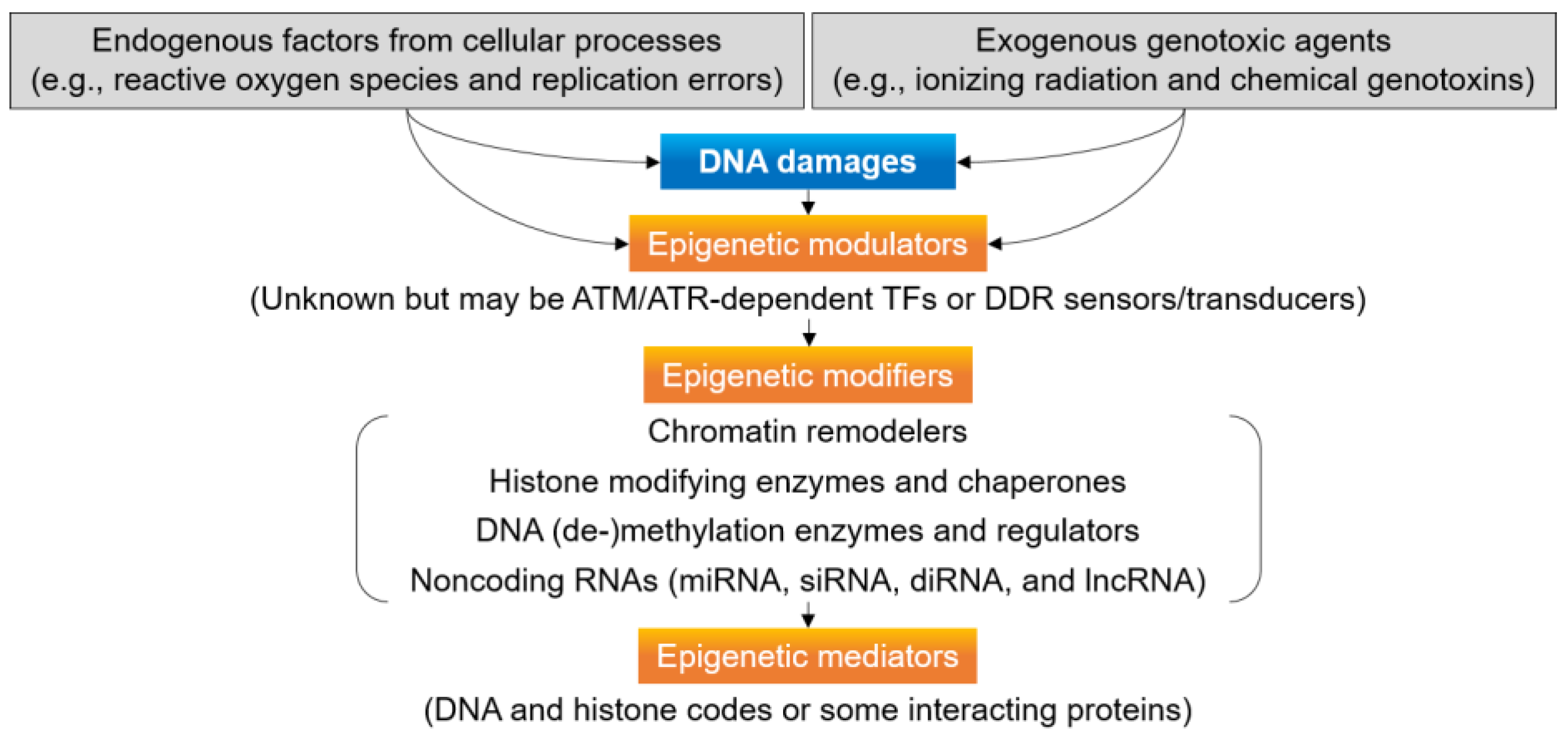
4. Epigenetic Regulation for DNA Damage Signaling and Repair
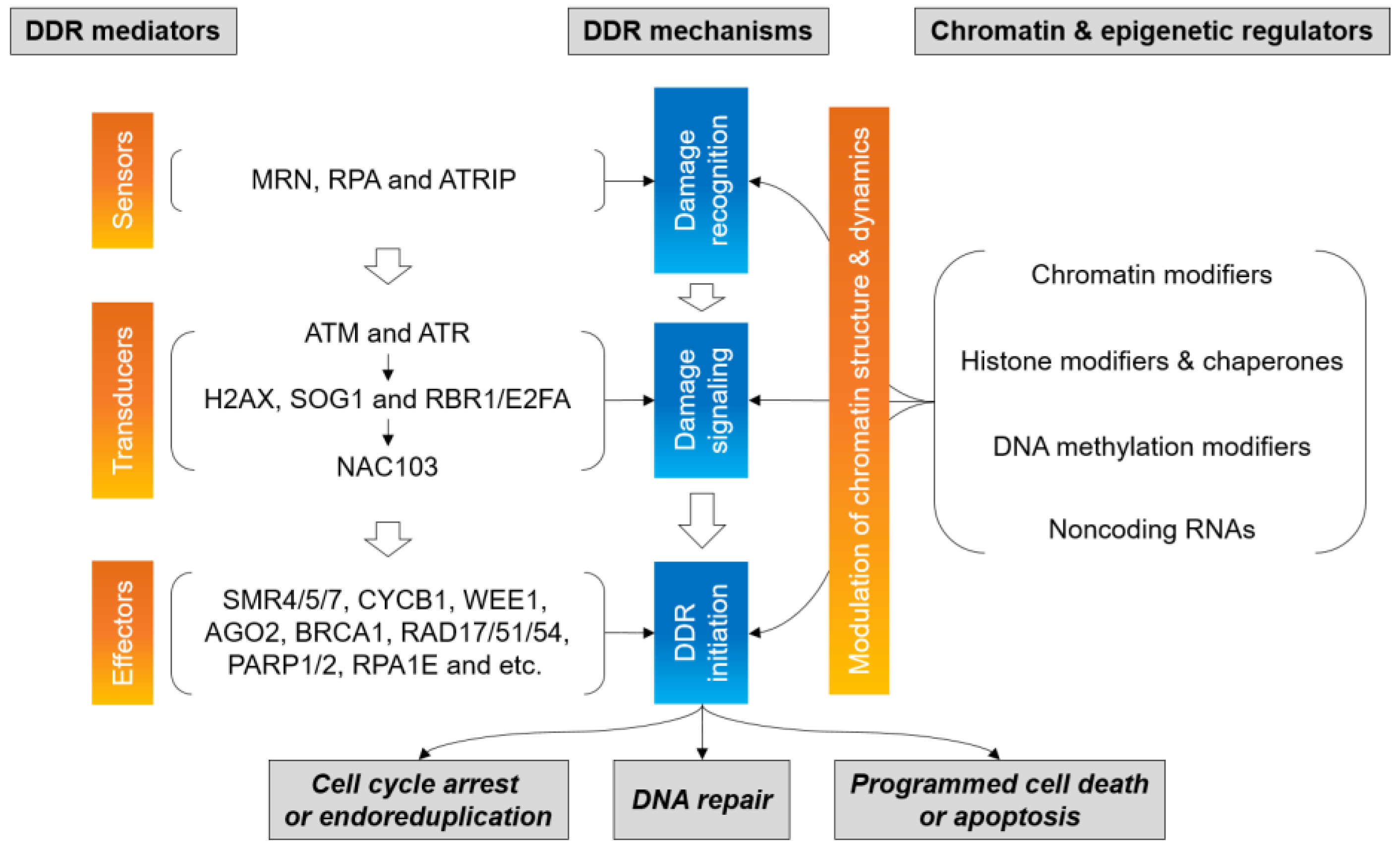
5. Chromatin and Epigenetic Modifiers for DDR
5.1. Chromatin Remodelers
5.2. Histone Modifiers
5.3. DNA (De-)Methylation Enzymes
5.4. Noncoding RNAs
6. Concluding Remarks and Perspectives
Funding
Conflicts of Interest
Abbreviations
| ACRs | ATP-dependent chromatin remodelers |
| AGO | Argonaute |
| ATM/ATR | Ataxia telangiectasia mutated /ATM and rad3-related |
| BER or NER | Base or nucleotide excision repair |
| CHD | Chromodomain helicase DNA |
| DDR | DNA damage response |
| diRNAs or DDRNAs | DSB-induced RNAs or DNA damage response RNAs |
| DME/DML2/DML3 | Demeter/Demeter-like2/Demeter-like3 |
| DSBs | DNA double-strand breaks |
| HDACs | Histone deacetylases |
| HR | Homologous recombination |
| INO80 | Inositol-requiring mutant80 |
| ISWI | Imitation switch |
| MET1/CMT2/3/DRM2 | Methyltransferase1/Chromomethylase2/3/Domains rearranged methyltransferase2 |
| miRNAs | MicroRNAs |
| MMR | Mismatch repair |
| NHEJ | Non-homologous end-joining |
| PTMs | Post-translational modifications |
| RAD54 | Radiation-sensitive54 |
| RBR1 | Retinoblastoma related1 |
| RdDM | RNA-directed DNA methylation |
| ROS1 | Repressor of silencing1 |
| siRNAs | Small interfering RNAs |
| sncRNAs or lncRNA | Small or long noncoding RNAs |
| SOG1 | Suppressor of gamma response1 |
| SWI/SNF | Switch/Sucrose nonfermentable |
| TGS | Transcriptional gene silencing |
References
- Dion, V.; Gasser, S.M. Chromatin movement in the maintenance of genome stability. Cell 2013, 152, 1355–1364. [Google Scholar] [CrossRef]
- Nair, N.; Shoaib, M.; Sørensen, C.S. Chromatin dynamics in genome stability: Roles in suppressing endogenous DNA damage and facilitating DNA repair. Int. J. Mol. Sci. 2017, 18, 1486. [Google Scholar] [CrossRef] [PubMed]
- Bakkenist, C.J.; Kastan, M.B. Chromatin perturbations during the DNA damage response in higher eukaryotes. DNA Repair 2015, 36, 8–12. [Google Scholar] [CrossRef]
- Sinha, M.; Peterson, C.L. Chromatin dynamics during repair of chromosomal DNA double-strand breaks. Epigenomics 2009, 1, 371–385. [Google Scholar] [CrossRef]
- Shi, L.; Oberdoerffer, P. Chromatin dynamics in DNA double-strand break repair. Biochim. Biophys. Acta 2012, 1819, 811–819. [Google Scholar] [CrossRef] [PubMed]
- Caridi, P.C.; Delabaere, L.; Zapotoczny, G.; Chiolo, I. And yet, it moves: nuclear and chromatin dynamics of a heterochromatic double-strand break. Phil. Trans. R. Soc. B 2017, 372, 20160291. [Google Scholar] [CrossRef] [PubMed]
- Hauer, M.H.; Seeber, A.; Singh, V.; Thierry, R.; Sack, R.; Amitai, A.; Kryzhanovska, M.; Eglinger, J.; Holcman, D.; Owen-Hughes, T.; et al. Histone degradation in response to DNA damage enhances chromatin dynamics and recombination rates. Nat. Struct. Mol. Biol. 2017, 24, 99–107. [Google Scholar] [CrossRef]
- Hauer, M.H.; Gasser, S.M. Chromatin and nucleosome dynamics in DNA damage and repair. Gene. Dev. 2017, 31, 2204–2221. [Google Scholar] [CrossRef]
- Papamichos-Chronakis, M.; Peterson, C.L. Chromatin and the genome integrity network. Nat. Rev. Genet. 2013, 14, 62–75. [Google Scholar] [CrossRef]
- Ikura, T.; Tashiro, S.; Kakino, A.; Shima, H.; Jacob, N.; Amunugama, R.; Yoder, K.; Izumi, S.; Kuraoka, I.; Tanaka, K.; et al. DNA damage-dependent acetylation and ubiquitination of H2AX enhances chromatin dynamics. Mol. Cell. Biol. 2007, 27, 7028–7040. [Google Scholar] [CrossRef]
- Morrison, A.J. Genome maintenance functions of the INO80 chromatin remodeller. Phil. Trans. R. Soc. B 2017, 372, 20160289. [Google Scholar] [CrossRef]
- Zhang, H.; Lang, Z.; Zhu, J.K. Dynamics and function of DNA methylation in plants. Nat. Rev. Mol. Cell Biol. 2018, 19, 489–506. [Google Scholar] [PubMed]
- Van Wolfswinkel, J.C.; Ketting, R.F. The role of small non-coding RNAs in genome stability and chromatin organization. J. Cell Sci. 2010, 123, 1825–1839. [Google Scholar] [CrossRef]
- Marchese, F.P.; Raimondi, I.; Huarte, M. The multidimensional mechanisms of long noncoding RNA function. Genome Biol. 2017, 18, 206. [Google Scholar] [CrossRef]
- Desvoyes, B.; Fernández-Marcos, M.; Sequeira-Mendes, J.; Otero, S.; Vergara, Z.; Gutierrez, C. Looking at plant cell cycle from the chromatin window. Front. Plant Sci. 2014, 5, 369. [Google Scholar]
- Vergara, Z.; Gutierrez, C. Emerging roles of chromatin in the maintenance of genome organization and function in plants. Genome Biol. 2017, 18, 96. [Google Scholar] [CrossRef] [PubMed]
- To, T.K.; Kim, J.M. Epigenetic regulation of gene responsiveness in Arabidopsis. Front. Plant Sci. 2014, 4, 548. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Santos, A.P.; Ferreira, L.J.; Oliveira, M.M. Concerted flexibility of chromatin structure, methylome, and histone modifications along with plant stress responses. Biology 2017, 6, 3. [Google Scholar] [CrossRef]
- Yoshiyama, K.O.; Sakaguchi, K.; Kimura, S. DNA damage response in plants: Conserved and variable response compared to animals. Biology 2013, 2, 1338–1356. [Google Scholar] [CrossRef]
- Roy, S. Maintenance of genome stability in plants: Repairing DNA double strand breaks and chromatin structure stability. Front. Plant Sci. 2014, 5, e487. [Google Scholar] [CrossRef] [PubMed]
- Donà, M.; Mittelsten Scheid, O. DNA damage repair in the context of plant chromatin. Plant Physiol. 2015, 168, 1206–1218. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.-H.; Ryu, T.H.; Lee, S.S.; Lee, S.; Chung, B.Y. Ionizing radiation manifesting DNA damage response in plants: An overview of DNA damage signaling and repair mechanisms in plants. Plant Sci. 2019, 278, 44–53. [Google Scholar] [CrossRef]
- Maréchal, A.; Zou, L. DNA damage sensing by the ATM and ATR kinases. Cold Spring Harb. Perspect. Biol. 2013, 5, a012716. [Google Scholar] [CrossRef] [PubMed]
- Agarwal, P.; Miller, K.M. The nucleosome: orchestrating DNA damage signaling and repair within chromatin. Biochem. Cell Biol. 2016, 94, 381–395. [Google Scholar] [CrossRef]
- Dupré, A.; Boyer-Chatenet, L.; Gautier, J. Two-step activation of ATM by DNA and the Mre11-Rad50-Nbs1 complex. Nat. Struct. Mol. Biol. 2006, 13, 451–457. [Google Scholar] [CrossRef]
- Cortez, D.; Guntuku, S.; Qin, J.; Elledge, S.J. ATR and ATRIP: Partners in checkpoint signaling. Science 2001, 294, 1713–1716. [Google Scholar] [CrossRef]
- Zou, L.; Elledge, S.J. Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes. Science 2003, 300, 1542–1548. [Google Scholar] [CrossRef]
- Yoshiyama, K.O.; Kobayashi, J.; Ogita, N.; Ueda, M.; Kimura, S.; Maki, H.; Umeda, M. ATM-mediated phosphorylation of SOG1 is essential for the DNA damage response in Arabidopsis. EMBO Rep. 2013, 14, 817–822. [Google Scholar] [CrossRef] [PubMed]
- Horvath, B.M.; Kourova, H.; Nagy, S.; Nemeth, E.; Magyar, Z.; Papdi, C.; Ahmad, Z.; Sanchez-Perez, G.F.; Perilli, S.; Blilou, I.; et al. Arabidopsis RETINOBLASTOMA RELATED directly regulates DNA damage responses through functions beyond cell cycle control. EMBO J. 2017, 36, 1261–1278. [Google Scholar] [CrossRef] [PubMed]
- Yoshiyama, K.; Conklin, P.A.; Huefner, N.D.; Britta, A.B. Suppressor of gamma response 1 (SOG1) encodes a putative transcription factor governing multiple responses to DNA damage. Proc. Natl. Acad. Sci. USA 2009, 106, 12843–12848. [Google Scholar] [CrossRef] [PubMed]
- Ryu, T.H.; Go, Y.S.; Choi, S.H.; Kim, J.-I.; Chung, B.Y.; Kim, J.-H. SOG1-dependent NAC103 modulates the DNA damage response as a transcriptional regulator in Arabidopsis. Plant J. 2019, 98, 83–96. [Google Scholar] [CrossRef] [PubMed]
- Ogita, N.; Okushima, Y.; Tokizawa, M.; Yamamoto, Y.Y.; Tanaka, M.; Seki, M.; Makita, Y.; Matsui, M.; Okamoto-Yoshiyama, K.; Sakamoto, T.; et al. Identifying the target genes of SUPPRESSOR OF GAMMA RESPONSE 1, a master transcription factor controlling DNA damage response in Arabidopsis. Plant J. 2018, 94, 439–453. [Google Scholar] [PubMed]
- Widlak, P.; Pietrowska, M.; Lanuszewska, J. The role of chromatin proteins in DNA damage recognition and repair. Histochem. Cell Biol. 2006, 125, 119–126. [Google Scholar] [CrossRef] [PubMed]
- Rossetto, D.; Truman, A.W.; Kron, S.J.; Côté, J. Epigenetic modifications in double-strand break DNA damage signaling and repair. Clin. Cancer Res. 2010, 16, 4543–4552. [Google Scholar] [CrossRef] [PubMed]
- Smeenk, G.; Wiegant, W.W.; Vrolijk, H.; Solari, A.P.; Pastink, A.; van Attikum, H. The NuRD chromatin-remodeling complex regulates signaling and repair of DNA damage. J. Cell Biol. 2010, 190, 741–749. [Google Scholar] [CrossRef]
- Smeenk, G.; Wiegant, W.W.; Marteijn, J.A.; Luijsterburg, M.S.; Sroczynski, N.; Costelloe, T.; Romeijn, R.J.; Pastink, A.; Mailand, N.; Vermeulen, W.; et al. Poly(ADP-ribosyl)ation links the chromatin remodeler SMARCA5/SNF2H to RNF168-dependent DNA damage signaling. J. Cell Sci. 2013, 126, 889–903. [Google Scholar] [CrossRef] [PubMed]
- Han, S.-K.; Wu, M.-F.; Cui, S.; Wagner, D. Roles and activities of chromatin remodeling ATPases in plants. Plant J. 2015, 83, 62–77. [Google Scholar]
- Price, B.D.; D’Andrea, A.D. Chromatin remodeling at DNA double-strand breaks. Cell 2013, 152, 1344–1354. [Google Scholar] [CrossRef] [PubMed]
- Choi, S.H.; Ryu, T.H.; Kim, J.-I.; Lee, S.; Kim, J.-H. Mutation in DDM1 inhibits the homology directed repair of double strand breaks. PLoS ONE 2019, 14, e0211878. [Google Scholar] [CrossRef]
- House, N.C.M.; Koch, M.R.; Freudenreich, C.H. Chromatin modifications and DNA repair: Beyond double-strand breaks. Front. Genet. 2014, 5, 296. [Google Scholar] [PubMed]
- Feng, W.; Hale, C.J.; Over, R.S.; Cokus, S.J.; Jacobsen, S.E.; Michaels, S.D. Large-scale heterochromatin remodeling linked to overreplication-associated DNA damage. Proc. Natl. Acad. Sci. USA 2017, 114, 406–411. [Google Scholar] [CrossRef] [PubMed]
- Manova, V.; Gruszka, D. DNA damage and repair in plants—From models to crops. Front. Plant Sci. 2015, 6, 885. [Google Scholar]
- Lans, H.; Marteijn, J.A.; Vermeulen, W. ATP-dependent chromatin remodeling in the DNA-damage response. Epigenet. Chromatin 2012, 5, 4. [Google Scholar] [CrossRef] [PubMed]
- Osakabe, K.; Abe, K.; Yoshioka, T.; Osakabe, Y.; Todoriki, S.; Ichikawa, H.; Hohn, B.; Toki, S. Isolation and characterization of the RAD54 gene from Arabidopsis thaliana. Plant J. 2006, 48, 827–842. [Google Scholar] [CrossRef]
- Xu, P.; Yuan, D.; Liu, M.; Li, C.; Liu, Y.; Zhang, S.; Yao, N.; Yang, C. AtMMS21, an SMC5/6 complex subunit, is involved in stem cell niche maintenance and DNA damage responses in Arabidopsis roots. Plant Physiol. 2013, 161, 1755–1768. [Google Scholar] [CrossRef] [PubMed]
- Yuan, D.; Lai, J.; Xu, P.; Zhang, S.; Zhang, J.; Li, C.; Wang, Y.; Du, J.; Liu, Y.; Yang, C. AtMMS21 regulates DNA damage response and homologous recombination repair in Arabidopsis. DNA Repair 2014, 21, 140–147. [Google Scholar] [CrossRef]
- Oliveira, D.V.; Kato, A.; Nakamura, K.; Ikura, T.; Okada, M.; Kobayashi, J.; Yanagihara, H.; Saito, Y.; Tauchi, H.; Komatsu, K. Histone chaperone FACT regulates homologous recombination by chromatin remodeling through interaction with RNF20. J. Cell Sci. 2014, 127, 763–772. [Google Scholar] [CrossRef]
- Li, X.; Tyler, J.K. Nucleosome disassembly during human non-homologous end joining followed by concerted HIRA- and CAF-1-dependent reassembly. Elife 2016, 5, e15129. [Google Scholar] [CrossRef]
- Shim, E.Y.; Ma, J.L.; Oum, J.H.; Yanez, Y.; Lee, S.E. The yeast chromatin remodeler RSC complex facilitates end joining repair of DNA double-strand breaks. Mol. Cell Biol. 2005, 25, 3934–3944. [Google Scholar] [CrossRef]
- Czaja, W.; Mao, P.; Smerdon, M.J. The emerging roles of ATP-dependent chromatin remodeling Enzymes in nucleotide excision repair. Int. J. Mol. Sci. 2012, 13, 11954–11973. [Google Scholar] [CrossRef]
- Gong, F.; Fahy, D.; Smerdon, M.J. Rad4-Rad23 interaction with SWI/SNF links ATP-dependent chromatin remodeling with nucleotide excision repair. Nat. Struct. Mol. Biol. 2006, 13, 902–907. [Google Scholar] [CrossRef]
- Sarkar, S.; Kiely, R.; McHugh, P.J. The Ino80 chromatin-remodeling complex restores chromatin structure during UV DNA damage repair. J. Cell Biol. 2010, 191, 1061–1068. [Google Scholar] [CrossRef]
- Jiang, Y.; Wang, X.; Bao, S.; Guo, R.; Johnson, D.G.; Shen, X.; Li, L. INO80 chromatin remodeling complex promotes the removal of UV lesions by the nucleotide excision repair pathway. Proc. Natl. Acad. Sci. USA 2010, 107, 17274–17279. [Google Scholar] [CrossRef]
- Odell, I.D.; Wallace, S.S.; Pederson, D.S. Rules of engagement for base excision repair in chromatin. J. Cell. Physiol. 2013, 228, 258–266. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez, Y.; Hinz, J.M.; Smerdon, M.J. Accessing DNA damage in chromatin: Preparing the chromatin landscape for base excision repair. DNA Repair 2015, 32, 113–119. [Google Scholar] [CrossRef] [PubMed]
- Balliano, A.J.; Hayes, J.J. Base excision repair in chromatin: Insights from reconstituted systems. DNA Repair 2015, 36, 77–85. [Google Scholar] [CrossRef]
- Menoni, H.; Gasparutto, D.; Hamiche, A.; Cadet, J.; Dimitrov, S.; Bouvet, P.; Angelov, D. ATP-dependent chromatin remodeling is required for base excision repair in conventional but not in variant H2A.Bbd nucleosomes. Mol. Cell. Biol. 2007, 27, 5949–5956. [Google Scholar] [CrossRef] [PubMed]
- Menoni, H.; Di Mascio, P.; Cadet, J.; Dimitrov, S.; Angelov, D. Chromatin associated mechanisms in base excision repair-nucleosome remodeling and DNA transcription, two key players. Free Radic. Biol. Med. 2017, 107, 159–169. [Google Scholar] [CrossRef]
- Li, G.-M. New insights and challenges in mismatch repair: getting over the chromatin hurdle. DNA Repair 2014, 19, 48–54. [Google Scholar] [CrossRef]
- Schöpf, B.; Bregenhorn, S.; Quivy, J.P.; Kadyrov, F.A.; Almouzni, G.; Jiricny, J. Interplay between mismatch repair and chromatin assembly. Proc. Natl. Acad. Sci. USA 2012, 109, 1895–1900. [Google Scholar] [CrossRef] [PubMed]
- Shaked, H.; Avivi-Ragolsky, N.; Levy, A.A. Involvement of the Arabidopsis SWI2/SNF2 chromatin remodeling gene family in DNA damage response and recombination. Genetics 2006, 173, 985–994. [Google Scholar] [CrossRef] [PubMed]
- Diaz, M.; Pecinka, A. Scaffolding for repair: Understanding molecular functions of the SMC5/6 complex. Genes 2018, 9, 36. [Google Scholar] [CrossRef] [PubMed]
- Díaz, M.; Pečinková, P.; Nowicka, A.; Baroux, C.; Sakamoto, T.; Gandha, P.Y.; Jeřábková, H.; Matsunaga, S.; Grossniklaus, U.; Pecinka, A. The SMC5/6 complex subunit NSE4A is involved in DNA damage repair and seed development. Plant Cell 2019, 31, 1579–1597. [Google Scholar] [CrossRef] [PubMed]
- Dabin, J.; Fortuny, A.; Polo, S.E. Epigenome maintenance in response to DNA damage. Mol. Cell. 2016, 62, 712–727. [Google Scholar] [CrossRef]
- Kovalchuk, O.; Baulch, J.E. Epigenetic changes and nontargeted radiation effects—Is there a link? Environ. Mol. Mutagen. 2008, 49, 16–25. [Google Scholar] [CrossRef] [PubMed]
- Yin, H.; Zhang, X.; Liu, J.; Wang, Y.; He, J.; Yang, T.; Hong, X.; Yang, Q.; Gong, Z. Epigenetic regulation, somatic homologous recombination, and abscisic acid signaling are influenced by DNA polymerase ε mutation in Arabidopsis. Plant Cell 2009, 21, 386–402. [Google Scholar] [CrossRef]
- Fleming, A.M.; Ding, Y.; Burrows, C.J. Oxidative DNA damage is epigenetic by regulating gene transcription via base excision repair. Proc. Natl. Acad. Sci. USA 2017, 114, 2604–2609. [Google Scholar] [CrossRef] [PubMed]
- Chinnusamy, V.; Zhu, J.K. Epigenetic regulation of stress responses in plants. Curr. Opin. Plant Biol. 2009, 12, 133–139. [Google Scholar] [CrossRef] [PubMed]
- Boyko, A.; Kovalchuk, I. Genome instability and epigenetic modification—heritable responses to environmental stress. Curr. Opin. Plant Biol. 2011, 14, 1–7. [Google Scholar] [CrossRef]
- Feinberg, A.P.; Koldobskiy, M.A.; Göndör, A. Epigenetic modulators, modifiers and mediators in cancer aetiology and progression. Nat. Rev. Genet. 2016, 17, 284–299. [Google Scholar] [PubMed]
- Mailand, N.; Bekker-Jensen, S.; Faustrup, H.; Melander, F.; Bartek, J.; Lukas, C.; Lukas, J. RNF8 ubiquitylates histones at DNA double-strand breaks and promotes assembly of repair proteins. Cell 2007, 131, 887–900. [Google Scholar] [CrossRef] [PubMed]
- Mattiroli, F.; Vissers, J.H.A.; van Dijk, W.J.; Ikpa, P.; Citterio, E.; Vermeulen, W.; Marteijn, J.A.; Sixma, T.K. RNF168 ubiquitinates K13-15 on H2A/H2AX to drive DNA damage signaling. Cell 2012, 150, 1182–1195. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Mao, G.; Tong, D.; Huang, J.; Gu, L.; Yang, W.; Li, G.-M. The histone mark H3K36me3 regulates human DNA mismatch repair through its interaction with MutSα. Cell 2013, 153, 590–600. [Google Scholar] [CrossRef]
- Huang, Y.; Gu, L.; Li, G.-M. H3K36me3-mediated mismatch repair preferentially protects actively transcribed genes from mutation. J. Biol. Chem. 2018, 293, 7811–7823. [Google Scholar] [CrossRef] [PubMed]
- Francia, S. Non-coding RNA: Sequence-specific guide for chromatin modification and DNA damage signaling. Front. Genet. 2015, 6, 320. [Google Scholar] [CrossRef]
- Pikaard, C.S.; Mittelsten Scheid, O. Epigenetic regulation in plants. Cold Spring Harb. Perspect. Biol. 2014, 6, a019315. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.E.; Lee, M.H.; Cho, E.J.; Kim, J.H.; Chung, B.Y.; Kim, J.-H. Characterization of non-CG genomic hypomethylation associated with gamma-ray-induced suppression of CMT3 transcription in Arabidopsis thaliana. Radiat. Res. 2013, 180, 638–648. [Google Scholar] [CrossRef]
- Mondal, S.; Go, Y.S.; Lee, S.S.; Chung, B.Y.; Kim, J.-H. Characterization of histone modifications associated with DNA damage repair genes upon exposure to gamma rays in Arabidopsis seedling. J. Radiat. Res. 2016, 57, 646–654. [Google Scholar] [CrossRef] [PubMed]
- Joly-Tonetti, N.; Lamartine, J. The role of microRNAs in the cellular response to ionizing radiations. In Current Topics in Ionizing Radiation Research; InTechOpen: Rijeka, Croatia, 2012; ISBN 978-953-51-0196-3. [Google Scholar]
- Kim, J.-H.; Go, Y.S.; Kim, J.K.; Chung, B.Y. Characterization of microRNAs and their target genes associated with transcriptomic changes in gamma-irradiated Arabidopsis. Genet. Mol. Res. 2016, 15. [Google Scholar] [CrossRef] [PubMed]
- Ehrenhofer-Murray, A.E. Chromatin dynamics at DNA replication, transcription and repair. Eur. J. Biochem. 2004, 271, 2335–2349. [Google Scholar] [CrossRef]
- Rothbart, S.B.; Strahl, B.D. Interpreting the language of histone and DNA modifications. Biochim. Biophys. Acta 2014, 1839, 627–643. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.; Yip, R.K.H.; Zhou, Z. Chromatin remodeling, DNA damage repair and aging. Curr. Genom. 2012, 13, 533–547. [Google Scholar] [CrossRef] [PubMed]
- Van Attikum, H.; Fritsch, O.; Hohn, B.; Gasser, S.M. Recruitment of the INO80 complex by H2A phosphorylation links ATP-dependent chromatin remodeling with DNA double-strand break repair. Cell 2004, 119, 777–788. [Google Scholar] [CrossRef] [PubMed]
- Morrison, A.J.; Kim, J.A.; Person, M.D.; Highland, J.; Xiao, J.; Wehr, T.S.; Hensley, S.; Bao, Y.; Shen, J.; Collins, S.R.; et al. Mec1/Tel1 phosphorylation of the INO80 chromatin remodeling complex influences DNA damage checkpoint responses. Cell 2007, 130, 499–511. [Google Scholar] [CrossRef] [PubMed]
- Saravanan, M.; Wuerges, J.; Bose, D.; McCormack, E.A.; Cook, N.J.; Zhang, X.; Wigley, D.B. Interactions between the nucleosome histone core and Arp8 in the INO80 chromatin remodeling complex. Proc. Natl. Acad. Sci. USA 2012, 109, 20883–20888. [Google Scholar] [CrossRef] [PubMed]
- Fritsch, O.; Benvenuto, G.; Bowler, C.; Molinier, J.; Hohn, B. The INO80 protein controls homologous recombination in Arabidopsis thaliana. Mol. Cell. 2004, 16, 479–485. [Google Scholar] [CrossRef]
- Zhang, C.; Cao, L.; Rong, L.; An, Z.; Zhou, W.; Ma, J.; Shen, W.-H.; Zhu, Y.; Dong, A. The chromatin-remodeling factor AtINO80 plays crucial roles in genome stability maintenance and in plant development. Plant J. 2015, 82, 655–668. [Google Scholar] [CrossRef]
- Kandasamy, M.K.; McKinney, E.C.; Deal, R.B.; Smith, A.P.; Meagher, R.B. Arabidopsis actin-related protein ARP5 in multicellular development and DNA repair. Dev. Biol. 2009, 335, 22–32. [Google Scholar] [CrossRef][Green Version]
- van Attikum, H.; Fritsch, O.; Gasser, S.M. Distinct roles for SWR1 and INO80 chromatin remodeling complexes at chromosomal double-strand breaks. EMBO J. 2007, 26, 4113–4125. [Google Scholar] [CrossRef]
- Noh, Y.-S.; Amasino, R.M. PIE1, an ISWI family gene, is required for FLC activation and floral repression in Arabidopsis. Plant Cell 2003, 15, 1671–1682. [Google Scholar] [CrossRef]
- March-Díaz, R.; García-Domínguez, M.; Florencio, F.J.; Reyes, J.C. SEF, a new protein required for flowering repression in Arabidopsis, interacts with PIE1 and ARP6. Plant Physiol. 2007, 143, 893–901. [Google Scholar] [CrossRef] [PubMed]
- Rosa, M.; Von Harder, M.; Cigliano, R.A.; Schlögelhofer, P.; Mittelsten Scheida, O. The Arabidopsis SWR1 chromatin-remodeling complex is important for DNA repair, somatic recombination, and meiosis. Plant Cell 2013, 25, 1990–2001. [Google Scholar] [CrossRef] [PubMed]
- Berriri, S.; Gangappa, S.N.; Kumar, S.V. SWR1 chromatin-remodeling complex subunits and H2A.Z have non-overlapping functions in immunity and gene regulation in Arabidopsis. Mol. Plant. 2016, 9, 1051–1065. [Google Scholar] [CrossRef]
- Hu, Y.; Lai, Y.; Zhu, D. Transcription regulation by CHD proteins to control plant development. Front Plant Sci. 2014, 5, 223. [Google Scholar] [CrossRef] [PubMed]
- Zou, B.; Sun, Q.; Zhang, W.; Ding, Y.; Yang, D.-L.; Shi, Z.; Hua, J. The Arabidopsis chromatin-remodeling factor CHR5 regulates plant immune responses and nucleosome occupancy. Plant Cell Physiol. 2017, 58, 2202–2216. [Google Scholar] [CrossRef]
- Li, G.; Liu, S.; Wang, J.; He, J.; Huang, H.; Zhang, Y.; Xu, L. ISWI proteins participate in the genome-wide nucleosome distribution in Arabidopsis. Plant J. 2014, 78, 706–714. [Google Scholar] [CrossRef]
- Zemach, A.; Kim, M.Y.; Hsieh, P.H.; Coleman-Derr, D.; Eshed-Williams, L.; Thao, K.; Harmer, S.L.; Zilberman, D. The Arabidopsis nucleosome remodeler DDM1 allows DNA methyltransferases to access H1-containing heterochromatin. Cell 2013, 153, 193–205. [Google Scholar] [CrossRef] [PubMed]
- Kozak, J.; West, C.E.; White, C.; da Costa-Nunes, J.A.; Angelis, K.J. Rapid repair of DNA double strand breaks in Arabidopsis thaliana is dependent on proteins involved in chromosome structure maintenance. DNA Repair 2009, 8, 413–419. [Google Scholar] [CrossRef] [PubMed]
- Groth, M.; Stroud, H.; Feng, S.; Greenberg, M.V.; Vashisht, A.A.; Wohlschlegel, J.A.; Jacobsen, S.E.; Ausin, I. SNF2 chromatin remodeler-family proteins FRG1 and -2 are required for RNA-directed DNA methylation. Proc. Natl. Acad. Sci. USA 2014, 111, 17666–17671. [Google Scholar] [CrossRef]
- Redon, C.; Pilch, D.; Rogakou, E.; Sedelnikova, O.; Newrock, K.; Bonner, W. Histone H2A variants H2AX and H2AZ. Curr. Opin. Genet. Dev. 2002, 12, 162–169. [Google Scholar] [CrossRef]
- Friesner, J.D.; Liu, B.; Culligan, K.; Britt, A.B. Ionizing radiation-dependent γ-H2AX focus formation requires ataxia telangiectasia mutated and ataxia telangiectasia mutated and Rad3-related. Mol. Biol. Cell 2005, 16, 2566–2576. [Google Scholar] [CrossRef] [PubMed]
- Vidanes, G.M.; Bonilla, C.Y.; Toczyski, D.P. Complicated tails: histone modifications and the DNA damage response. Cell 2005, 121, 973–976. [Google Scholar] [CrossRef] [PubMed]
- Chen, N.; Zhou, W.-B.; Wang, Y.-X.; Dong, A.-W.; Yu, Y. Polycomb-group histone methyltransferase CLF is required for proper somatic recombination in Arabidopsis. J. Integr. Plant Biol. 2014, 56, 550–558. [Google Scholar] [CrossRef] [PubMed]
- Jacob, Y.; Bergamin, E.; Donoghue, M.T.; Mongeon, V.; LeBlanc, C.; Voigt, P.; Underwood, C.J.; Brunzelle, J.S.; Michaels, S.D.; Reinberg, D.; et al. Selective methylation of histone H3 variant H3.1 regulates heterochromatin replication. Science 2014, 343, 1249–1253. [Google Scholar] [CrossRef] [PubMed]
- Campi, M.; D’Andrea, L.; Emiliani, J.; Casati, P. Participation of chromatin-remodeling proteins in the repair of ultraviolet-B-damaged DNA. Plant Physiol. 2012, 158, 981–995. [Google Scholar] [CrossRef] [PubMed]
- Fina, J.P.; Casati, P. HAG3, a histone acetyltransferase, affects UV-B responses by negatively regulating the expression of DNA repair enzymes and sunscreen content in Arabidopsis thaliana. Plant Cell Physiol. 2015, 56, 1388–1400. [Google Scholar] [CrossRef] [PubMed]
- Fina, J.P.; Masotti, F.; Rius, S.P.; Crevacuore, F.; Casati, P. HAC1 and HAF1 histone acetyltransferases have different roles in UV-B responses in Arabidopsis. Front. Plant Sci. 2017, 8, 1179. [Google Scholar] [CrossRef] [PubMed]
- Luo, M.; Cheng, K.; Xu, Y.; Yang, S.; Wu, K. Plant responses to abiotic stress regulated by histone deacetylases. Front. Plant Sci. 2017, 8, 2147. [Google Scholar] [CrossRef]
- Gao, J.; Zhu, Y.; Zhou, W.; Molinier, J.; Dong, A.; Shen, W.-H. NAP1 family histone chaperones are required for somatic homologous recombination in Arabidopsis. Plant Cell 2012, 24, 1437–1447. [Google Scholar] [CrossRef] [PubMed]
- González-Arzola, K.; Díaz-Quintana, A.; Rivero-Rodríguez, F.; Velázquez-Campoy, A.; De la Rosa, M.A.; Díaz-Moreno, I. Histone chaperone activity of Arabidopsis thaliana NRP1 is blocked by cytochrome c. Nucleic Acids Res. 2017, 45, 2150–2165. [Google Scholar] [CrossRef]
- Frost, J.M.; Kim, M.Y.; Park, G.T.; Hsieh, P.-H.; Nakamura, M.; Lin, S.J.H.; Yoo, H.; Choi, J.; Ikeda, Y.; Kinoshita, T.; et al. FACT complex is required for DNA demethylation at heterochromatin during reproduction in Arabidopsis. Proc. Natl. Acad. Sci. USA 2018, 115, E4720–E4729. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Weng, M.; Yang, Y.; Zhang, C.; Li, Z.; Shen, W.H.; Dong, A. Arabidopsis homologues of the histone chaperone ASF1 are crucial for chromatin replication and cell proliferation in plant development. Plant J. 2011, 66, 443–455. [Google Scholar] [CrossRef] [PubMed]
- Schönrock, N.; Exner, V.; Probst, A.; Gruissem, W.; Hennig, L. Functional genomic analysis of CAF-1 mutants in Arabidopsis thaliana. J. Biol. Chem. 2006, 281, 9560–9568. [Google Scholar] [CrossRef] [PubMed]
- Varas, J.; Santos, J.L.; Pradillo, M. The absence of the Arabidopsis chaperone complex CAF-1 produces mitotic chromosome abnormalities and changes in the expression profiles of genes involved in DNA repair. Front. Plant Sci. 2017, 8, 525. [Google Scholar] [CrossRef] [PubMed]
- Takeda, S.; Tadele, Z.; Hofmann, I.; Probst, A.V.; Angelis, K.J.; Kaya, H.; Araki, T.; Mengiste, T.; Mittelsten Scheid, O.; Shibahara, K.; et al. BRU1, a novel link between responses to DNA damage and epigenetic gene silencing in Arabidopsis. Gene. Dev. 2004, 18, 782–793. [Google Scholar] [CrossRef] [PubMed]
- Adam, S.; Polo, S.E.; Almouzni, G. Transcription recovery after DNA damage requires chromatin priming by the H3.3 histone chaperone HIRA. Cell 2013, 155, 94–106. [Google Scholar] [CrossRef] [PubMed]
- Nie, X.; Wang, H.; Li, J.; Holec, S.; Berger, F. The HIRA complex that deposits the histone H3.3 is conserved in Arabidopsis and facilitates transcriptional dynamics. Biol. Open 2014, 3, 794–802. [Google Scholar] [CrossRef]
- Gehring, M.; Henikoff, S. DNA methylation and demethylation in Arabidopsis. Arabidopsis Book 2008, 6, e0102. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.-K. Active DNA demethylation mediated by DNA glycosylases. Annu. Rev. Genet. 2009, 43, 143–166. [Google Scholar] [CrossRef]
- Widman, N.; Jacobsen, S.E.; Pellegrini, M. Determining the conservation of DNA methylation in Arabidopsis. Epigenetics 2009, 4, 119–124. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Mirouze, M.; Lieberman-Lazarovich, M.; Aversano, R.; Bucher, E.; Nicolet, J.; Reinders, J.; Paszkowski, J. Loss of DNA methylation affects the recombination landscape in Arabidopsis. Proc. Natl. Acad. Sci. USA 2012, 109, 5880–5885. [Google Scholar] [CrossRef] [PubMed]
- Georgieva, M.; Rashydov, N.M.; Hajduch, M. DNA damage, repair monitoring and epigenetic DNA methylation changes in seedlings of Chernobyl soybeans. DNA Repair 2017, 50, 14–21. [Google Scholar] [CrossRef] [PubMed]
- Gong, Z.; Morales-Ruiz, T.; Ariza, R.R.; Roldán-Arjona, T.; David, L.; Zhu, J.K. ROS1, a repressor of transcriptional gene silencing in Arabidopsis, encodes a DNA glycosylase/lyase. Cell 2002, 111, 803–814. [Google Scholar] [CrossRef]
- Elmayan, T.; Proux, F.; Vaucheret, H. Arabidopsis RPA2: a genetic link among transcriptional gene silencing, DNA repair, and DNA replication. Curr. Biol. 2005, 15, 1919–1925. [Google Scholar] [CrossRef] [PubMed]
- Groth, M.; Moissiard, G.; Wirtz, M.; Wang, H.; Garcia-Salinas, C.; Ramos-Parra, P.A.; Bischof, S.; Feng, S.; Cokus, S.J.; John, A.; et al. MTHFD1 controls DNA methylation in Arabidopsis. Nat. Commun. 2016, 7, 11640. [Google Scholar] [CrossRef] [PubMed]
- Wollmann, H.; Stroud, H.; Yelagandula, R.; Tarutani, Y.; Jiang, D.; Jing, L.; Jamge, B.; Takeuchi, H.; Holec, S.; Nie, X.; et al. The histone H3 variant H3.3 regulates gene body DNA methylation in Arabidopsis thaliana. Genome Biol. 2017, 18, 94. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Macías, M.I.; Córdoba-Cañero, D.; Ariza, R.R.; Roldán-Arjona, T. The DNA repair protein XRCC1 functions in the plant DNA demethylation pathway by stimulating cytosine methylation (5-meC) excision, gap tailoring, and DNA ligation. J. Biol. Chem. 2013, 288, 5496–5505. [Google Scholar] [CrossRef] [PubMed]
- Schalk, C.; Drevensek, S.; Kramdi, A.; Kassam, M.; Ahmed, I.; Cognat, V.; Graindorge, S.; Bergdoll, M.; Baumberger, N.; Heintz, D.; et al. DNA DAMAGE BINDING PROTEIN2 shapes the DNA methylation landscape. Plant Cell 2016, 28, 2043–2059. [Google Scholar] [CrossRef] [PubMed]
- Córdoba-Cañero, D.; Cognat, V.; Ariza, R.R.; Roldán Arjona, T.; Molinier, J. Dual control of ROS1-mediated active DNA demethylation by DNA damage-binding protein 2 (DDB2). Plant J. 2017, 92, 1170–1181. [Google Scholar] [CrossRef] [PubMed]
- Khraiwesh, B.; Zhu, J.-K.; Zhu, J. Role of miRNAs and siRNAs in biotic and abiotic stress responses of plants. Biochim. Biophys. Acta 2012, 1819, 137–148. [Google Scholar] [CrossRef]
- Schalk, C.; Cognat, V.; Graindorge, S.; Vincent, T.; Voinnet, O.; Molinier, J. Small RNA-mediated repair of UV-induced DNA lesions by the DNA DAMAGE-BINDING PROTEIN 2 and ARGONAUTE 1. Proc. Natl. Acad. Sci. USA 2017, 114, E2965–E2974. [Google Scholar] [CrossRef]
- Wei, W.; Ba, Z.; Gao, M.; Wu, Y.; Ma, Y.; Amiard, S.; White, C.I.; Rendtlew Danielsen, J.M.; Yang, Y.-G.; Qi, Y. A role for small RNAs in DNA double-strand break repair. Cell 2012, 149, 101–112. [Google Scholar] [CrossRef] [PubMed]
- d’Adda di Fagagna, F. A direct role for small non-coding RNAs in DNA damage response. Trends Cell Biol. 2014, 24, 171–178. [Google Scholar] [CrossRef] [PubMed]
- Cifuentes-Rojas, C.; Nelson, A.D.; Boltz, K.A.; Kannan, K.; She, X.; Shippen, D.E. An alternative telomerase RNA in Arabidopsis modulates enzyme activity in response to DNA damage. Genes Dev. 2012, 26, 2512–2523. [Google Scholar] [CrossRef] [PubMed]
- Hunt, C.R.; Ramnarain, D.; Horikoshi, N.; Iyengar, P.; Pandita, R.K.; Shay, J.W.; Pandita, T.K. Histone modifications and DNA double-strand break repair after exposure to ionizing radiations. Radiat. Res. 2013, 179, 383–392. [Google Scholar] [CrossRef] [PubMed]
- Williamson, E.A.; Wray, J.W.; Bansal, P.; Hromas, R. Overview for the histone codes for DNA repair. Prog. Mol. Biol. Transl. Sci. 2012, 110, 207–227. [Google Scholar] [PubMed]
- Yun, M.; Wu, J.; Workman, J.L.; Li, B. Readers of histone modifications. Cell Res. 2011, 21, 564–578. [Google Scholar] [CrossRef] [PubMed]
- Musselman, C.A.; Lalonde, M.E.; Côté, J.; Kutateladze, T.G. Perceiving the epigenetic landscape through histone readers. Nat. Struct. Mol. Biol. 2012, 19, 1218–1227. [Google Scholar]
- Bowman, G.D.; Poirier, M.G. Post-translational modifications of histones that influence nucleosome dynamics. Chem. Rev. 2015, 115, 2274–2295. [Google Scholar] [CrossRef]
- Li, W.; Liu, H.; Cheng, Z.J.; Su, Y.H.; Han, H.N.; Zhang, Y.; Zhang, X.S. DNA methylation and histone modifications regulate de novo shoot regeneration in Arabidopsis by modulating WUSCHEL expression and auxin signaling. PLoS Genet. 2011, 7, e1002243. [Google Scholar] [CrossRef]
- Bilichak, A.; Ilnystkyy, Y.; Hollunder, J.; Kovalchuk, I. The progeny of Arabidopsis thaliana plants exposed to salt exhibit changes in DNA methylation, histone modifications and gene expression. PLoS ONE 2012, 7, e30515. [Google Scholar] [CrossRef] [PubMed]
- Drury, G.E.; Dowle, A.A.; Ashford, D.A.; Waterworth, W.M.; Thomas, J.; West, C.E. Dynamics of plant histone modifications in response to DNA damage. Biochem. J. 2012, 445, 393–401. [Google Scholar] [CrossRef] [PubMed]
- Zhao, S.; Zhang, B.; Yang, M.; Zhu, J.; Li, H. Systematic profiling of histone readers in Arabidopsis thaliana. Cell Rep. 2018, 22, 1090–1102. [Google Scholar] [CrossRef] [PubMed]
- Sanders, S.L.; Portoso, M.; Mata, J.; Bähler, J.; Allshire, R.C.; Kouzarides, T. Methylation of histone H4 lysine 20 controls recruitment of Crb2 to sites of DNA damage. Cell 2004, 119, 603–614. [Google Scholar] [CrossRef] [PubMed]
- Faucher, D.; Wellinger, R.J. Methylated H3K4, a transcription-associated histone modification, is involved in the DNA damage response pathway. PLoS Genet. 2010, 6, e1001082. [Google Scholar] [CrossRef]
- Sharma, V.M.; Tomar, R.S.; Dempsey, A.E.; Reese, J.C. Histone deacetylases RPD3 and HOS2 regulate the transcriptional activation of DNA damage-inducible genes. Mol. Cell. Biol. 2007, 27, 3199–3210. [Google Scholar] [CrossRef] [PubMed]
- Sharma, G.G.; So, S.; Gupta, A.; Kumar, R.; Cayrou, C.; Avvakumov, N.; Bhadra, U.; Pandita, R.K.; Porteus, M.H.; Chen, D.J.; et al. MOF and histone H4 acetylation at lysine 16 are critical for DNA damage response and double-strand break repair. Mol. Cell. Biol. 2010, 30, 3582–3595. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Wang, Y. Cross-talk between the H3K36me3 and H4K16ac histone epigenetic marks in DNA double-strand break repair. J. Biol. Chem. 2017, 292, 11951–11959. [Google Scholar] [CrossRef]
- Trejo-Arellano, M.S.; Mahrez, W.; Nakamura, M.; Moreno-Romero, J.; Nanni, P.; Köhler, C.; Hennig, L. H3K23me1 is an evolutionarily conserved histone modification associated with CG DNA methylation in Arabidopsis. Plant J. 2017, 90, 293–303. [Google Scholar] [CrossRef]
- Cuozzo, C.; Porcellini, A.; Angrisano, T.; Morano, A.; Lee, B.; Di Pardo, A.; Messina, S.; Iuliano, R.; Fusco, A.; Santillo, M.R.; et al. DNA damage, homology-directed repair, and DNA methylation. PLoS Genet. 2007, 3, e110. [Google Scholar] [CrossRef]
- Russo, G.; Landi, R.; Pezone, A.; Morano, A.; Zuchegna, C.; Romano, A.; Muller, M.T.; Gottesman, M.E.; Porcellini, A.; Avvedimento, E.V. DNA damage and repair modify DNA methylation and chromatin domain of the targeted locus: Mechanism of allele methylation polymorphism. Sci. Rep. 2016, 6, 33222. [Google Scholar] [CrossRef] [PubMed]
- Mortusewicz, O.; Schermelleh, L.; Walter, J.; Cardoso, M.C.; Leonhardt, H. Recruitment of DNA methyltransferase I to DNA repair sites. Proc. Natl. Acad. Sci. USA 2005, 102, 8905–8909. [Google Scholar] [CrossRef] [PubMed]
- Wojtczyk-Miaskowska, A.; Presler, M.; Michajlowski, J.; Matuszewski, M.; Schlichtholz, B. Gene expression, DNA methylation and prognostic significance of DNA repair genes in human bladder cancer. Cell. Physiol. Biochem. 2017, 42, 2404–2417. [Google Scholar] [CrossRef] [PubMed]
- Palii, S.S.; Van Emburgh, B.O.; Sankpal, U.T.; Brown, K.D.; Robertson, K.D. DNA methylation inhibitor 5-aza-2’-deoxycytidine induces reversible genome-wide DNA damage that is distinctly influenced by DNA methyltransferases 1 and 3B∇. Mol. Cell. Biol. 2008, 28, 752–771. [Google Scholar] [CrossRef]
- Liu, C.H.; Finke, A.; Díaz, M.; Rozhon, W.; Poppenberger, B.; Baubec, T.; Pecinka, A. Repair of DNA damage induced by the cytidine analog zebularine requires ATR and ATM in Arabidopsis. Plant Cell 2015, 27, 1788–1800. [Google Scholar] [CrossRef]
- Kim, H.J.; Kim, J.H.; Chie, E.K.; Young, P.D.; Kim, I.A.; Kim, I.H. DNMT (DNA methyltransferase) inhibitors radiosensitize human cancer cells by suppressing DNA repair activity. Radiat. Oncol. 2012, 7, 39. [Google Scholar] [CrossRef]
- Armstrong, C.A.; Jones, G.D.; Anderson, R.; Iyer, P.; Narayanan, D.; Sandhu, J.; Singh, R.; Talbot, C.J.; Tufarelli, C. DNMTs are required for delayed genome instability caused by radiation. Epigenetics 2012, 7, 892–902. [Google Scholar] [CrossRef]
- Thalheim, T.; Herberg, M.; Galle, J. Linking DNA damage and age-related promoter DNA hyper-methylation in the intestine. Genes 2018, 9, 17. [Google Scholar] [CrossRef]
- Brocklehurst, S.; Watson, M.; Carr, I.M.; Out, S.; Heidmann, I.; Meyer, P. Induction of epigenetic variation in Arabidopsis by over-expression of DNA METHYLTRANSFERASE1 (MET1). PLoS ONE 2018, 13, e0192170. [Google Scholar] [CrossRef]
- Ho, J.J.; Cattoglio, C.; McSwiggen, D.T.; Tjian, R.; Fong, Y.W. Regulation of DNA demethylation by the XPC DNA repair complex in somatic and pluripotent stem cells. Genes Dev. 2017, 31, 830–844. [Google Scholar] [CrossRef] [PubMed]
- Sharma, V.; Misteli, T. Non-coding RNAs in DNA damage and repair. FEBS Lett. 2013, 587, 1832–1839. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Peng, G. Non-coding RNAs: an emerging player in DNA damage response. Mutat. Res. 2015, 763, 202–211. [Google Scholar] [CrossRef]
- Hawley, B.R.; Lu, W.-T.; Wilczynska, A.; Bushell, M. The emerging role of RNAs in DNA damage repair. Cell Death Differ. 2017, 24, 580–587. [Google Scholar] [CrossRef] [PubMed]
- Thapar, R. Regulation of DNA double-strand break repair by non-coding RNAs. Molecules 2018, 23, 2789. [Google Scholar] [CrossRef] [PubMed]
- Tehrani, S.S.; Karimian, A.; Parsian, H.; Majidinia, M.; Yousefi, B. Multiple functions of long non-coding RNAs in oxidative stress, DNA damage response and cancer progression. J. Cell. Biochem. 2018, 119, 223–236. [Google Scholar] [CrossRef] [PubMed]
- Cho, J. Transposon-Derived Non-coding RNAs and Their Function in Plants. Front. Plant Sci. 2018, 9, 600. [Google Scholar] [CrossRef]
- Yi, X.; Zhang, Z.; Ling, Y.; Xu, W.; Su, Z. PNRD: a plant non-coding RNA database. Nucleic Acids Res. 2015, 43, D982–D989. [Google Scholar] [CrossRef]
- Simone, N.L.; Soule, B.P.; Ly, D.; Saleh, A.D.; Savage, J.E.; Degraff, W.; Cook, J.; Harris, C.C.; Gius, D.; Mitchell, J.B. Ionizing radiation-induced oxidative stress alters miRNA expression. PLoS ONE 2009, 4, e6377. [Google Scholar] [CrossRef] [PubMed]
- Lhakhang, T.W.; Chaudhry, M.A. Interactome of radiation-induced microRNA-predicted target genes. Comp. Funct. Genom. 2012, 2012, 569731. [Google Scholar] [CrossRef] [PubMed]
- Choi, Y.E.; Pan, Y.; Park, E.; Konstantinopoulos, P.; De, S.; D’Andrea, A.; Chowdhury, D. MicroRNAs down-regulate homologous recombination in the G1 phase of cycling cells to maintain genomic stability. Elife 2014, 3, e02445. [Google Scholar] [CrossRef]
- Choi, Y.E.; Meghani, K.; Brault, M.E.; Leclerc, L.; He, Y.J.; Day, T.A.; Elias, K.M.; Drapkin, R.; Weinstock, D.M.; Dao, F.; et al. Platinum and PARP inhibitor resistance due to overexpression of microRNA-622 in BRCA1-mutant ovarian cancer. Cell. Rep. 2016, 14, 429–439. [Google Scholar] [CrossRef]
- Zhou, X.; Wang, G.; Zhang, W. UV-B responsive microRNA genes in Arabidopsis thaliana. Mol. Syst. Biol. 2007, 3, 103. [Google Scholar] [CrossRef] [PubMed]
- Sunkar, R.; Kapoor, A.; Zhu, J.K. Posttranscriptional induction of two Cu/Zn superoxide dismutase genes in Arabidopsis is mediated by downregulation of miR398 and important for oxidative stress tolerance. Plant Cell 2006, 18, 2051–2065. [Google Scholar] [CrossRef] [PubMed]
- Patchsung, M.; Settayanon, S.; Pongpanich, M.; Mutirangura, D.; Jintarith, P.; Mutirangura, A. Alu siRNA to increase Alu element methylation and prevent DNA damage. Epigenomics 2018, 10, 175–185. [Google Scholar] [CrossRef] [PubMed]
- Francia, S.; Michelini, F.; Saxena, A.; Tang, D.; de Hoon, M.; Anelli, V.; Mione, M.; Carninci, P.; d’Adda di Fagagna, F. Site-specific DICER and DROSHA RNA products control the DNA-damage response. Nature 2012, 488, 231–235. [Google Scholar] [CrossRef]
- Oliver, C.; Santos, J.L.; Pradillo, M. On the role of some ARGONAUTE proteins in meiosis and DNA repair in Arabidopsis thaliana. Front. Plant Sci. 2014, 5, 177. [Google Scholar] [CrossRef]
- Bajczyk, M.; Bhat, S.S.; Szewc, L.; Szweykowska-Kulinska, Z.; Jarmolowski, A.; Dolata, J. Novel nuclear functions of Arabidopsis ARGONAUTE1: Beyond RNA interference. Plant Physiol. 2019, 179, 1030–1039. [Google Scholar] [CrossRef] [PubMed]
- Bai, Y.; Dai, X.; Harrison, A.P.; Chen, M. RNA regulatory networks in animals and plants: a long noncoding RNA perspective. Brief. Funct. Genom. 2015, 14, 91–101. [Google Scholar] [CrossRef]
- Liu, J.; Wang, H.; Chua, N.H. Long noncoding RNA transcriptome of plants. Plant Biotechnol. J. 2015, 13, 319–328. [Google Scholar]
- Rinn, J.L.; Chang, H.Y. Genome regulation by long noncoding RNAs. Annu. Rev. Biochem. 2012, 81, 145–166. [Google Scholar] [CrossRef] [PubMed]
- Deniz, E.; Erman, B. Long noncoding RNA (lincRNA), a new paradigm in gene expression control. Funct. Integr. Genom. 2017, 17, 135–143. [Google Scholar] [CrossRef] [PubMed]
- Jiao, Y.; Liu, C.; Cui, F.-M.; Xu, J.-Y.; Tong, J.; Qi, X.-F.; Wang, L.-L.; Zhu, W. Long intergenic non-coding RNA induced by X-ray irradiation regulates DNA damage response signaling in the human bronchial epithelial BEAS-2B cell line. Oncol. Lett. 2015, 9, 169–176. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Michelini, F.; Pitchiaya, S.; Vitelli, V.; Sharma, S.; Gioia, U.; Pessina, F.; Cabrini, M.; Wang, Y.; Capozzo, I.; Iannelli, F.; et al. Damage-induced lncRNAs control the DNA damage response through interaction with DDRNAs at individual double-strand breaks. Nat. Cell Biol. 2017, 19, 1400–1411. [Google Scholar] [CrossRef] [PubMed]
- Dianatpour, A.; Ghafouri-Fard, S. The role of long noncoding RNAs in the repair of DNA double strand breaks. Int. J. Mol. Cell. Med. 2017, 6, 1–12. [Google Scholar] [PubMed]
- Wang, Z.; Schwacke, R.; Kunze, R. DNA damage-induced transcription of transposable elements and long non-coding RNAs in Arabidopsis is rare and ATM-dependent. Mol. Plant 2016, 9, 1142–1155. [Google Scholar] [CrossRef] [PubMed]
| DNA Repair | Chromatin Modifier | Action Mechanisms | Reference |
|---|---|---|---|
| HR | 1–3. RAD54, 1. DDM1 | Induce chromatin remodeling through interaction with RAD51 | [39,43,44] |
| 1. MMS21 | Function as a critical subunit of the SMC5/6 complex | [45,46] | |
| 2. RNF20 | Ubiquitylate H2B through interaction with FACT | [47] | |
| NHEJ | 1–3. INO80 | Involved in nucleosome disassembly around DSBs | [21,43,48] |
| 2. ASF1A, HIRA, CAF-1 | Involved in nucleosome reassembly | [48] | |
| 3. RSC | Facilitate NHEJ through interaction with MRE11 and KU70/80 | [49] | |
| NER | 1–3. ACRs | Reorganize chromatin structure and control DNA accessibility | [43,50] |
| 2–3. SWI, INO80, ARP5 | Promote the removal of UV lesions through interaction with RAD4/23 | [51,52,53] | |
| BER | 1–3. ACRs | Affect accessibility of BER enzymes to nucleosomal DNA | [54,55,56] |
| 3. SWI/SNF | Induce chromatin remodeling to facilitate the removal of oxidative 8-oxoG lesions | [57,58] | |
| MMR | 1–3. Chromatin and histone modifiers | Affect nucleosome assembly and disassembly | [59] |
| 2. PCNA | Involved in chromatin assembly through interaction with MSH6 and CAF-1 | [60] |
| Epigenetic Modifier | Member or Subunit | Functions | Reference |
|---|---|---|---|
| Chromatin remodeler | |||
| INO80/SWR1 | INO80, ARP5, ARP8 | Involved in HR repair of DNA damage, maintenance of genome stability, and somatic HR and meiosis | [87,88,89,93] |
| SWR1, PIE1, ARP6, SWC6 | Involved in substitution of nucleosomal H2A by H2AZ and gene regulation | [91,92,94] | |
| SWI/SNF | BRM, SWI3, CHC1, ARP4, BSH | Function in DDR and HR repair through unknown mechanisms | [21,37,61] |
| CHD | CHR5, PKL, PKR, PKR2 | Unknown in DDR, but involved in nucleosome remodeling and gene regulation | [21,37,95,96] |
| ISWI | ISWI, CHR11, CHR17 | Unknown in DDR, but involved in nucleosome distribution | [21,37,97] |
| Uncategorized | RAD54 | Involved in DDR and HR repair by modulating chromatin structure through interaction with RAD51 | [44,61] |
| DDM1 | Involved in DDR and HR repair as well as in methylation and silencing of transposable elements | [39,98] | |
| MIM/RAD18, RAD21.1 | Involved in KU-independent NHEJ repair by constituting SMC complex | [99] | |
| DRD1, FRG1/FRG2 | Unknown in DDR, but involved in chromatin remodeling as RdDM components | [37,61,100] | |
| Histone modifier | |||
| Kinase | ATM, ATR | Facilitate recruitment of repair machineries at DSB sites by phosphorylating H2AX and start DDR | [101,102,103] |
| Methyltransferase | CLF, ATXR5, ATXR6 | Involved in regulation of somatic and meiotic HR repair or in preventing overreplication-associated heterochromatic DNA damage as a H3K27 methyltransferase | [41,104,105] |
| Acetyltransferase/deacetylase | HAM1, HAG3, HAC1, HAF1 | Involved in UV-B-induced DDR signaling and/or DNA repair | [106,107,108] |
| HDA2, ADA6, ADA19 | Unknown in DDR, but involved in gene regulation in abiotic stress responses as a H3K9 deacetylase | [109] | |
| Chaperone | NAP1, NRP1, NRP2, FACT | Involved in nucleosome remodeling for somatic HR or in targeting of DME as a H2A-H2B chaperone | [47,110,111,112] |
| CAF-1, ASF1, HIRA | Contribute to genome integrity/stability and transcriptional regulation of HR/NHEJ genes as a H3-H4 chaperone | [113,114,115,116,117,118] | |
| DNA (de-)methylation enzyme | |||
| Methyltransferase | MET1, CMT2, CMT3, DRM2 | Unknown in DDR, but correlate with meiotic recombination landscape, global non-CG hypomethylation after γ-irradiation, or increased CG/non-CG methylation in Chernobyl soybean seedlings | [77,98,119,120,121,122,123] |
| Demethylase | ROS1, DME, DML2, DML3 | ROS influences DDR to genotoxic stress as a TGS repressor | [120,124] |
| Regulator | RPA2, MTHFD1, H3.3 | Unknown in DDR, but involved in DNA methylation for TGS or transcriptional regulation | [125,126,127] |
| XRCC1, FACT | Unknown in DDR, but involved in active DNA demethylation by interacting with ROS1 or DME | [112,128] | |
| DDB2 | Involved in DNA methylation by interacting with AGO4-siRNA or active DNA demethylation by regulating ROS1 | [129,130] | |
| ncRNA | |||
| miRNA | miR156/159/160/166/390/393, miR398, miR840, miR850 | Unknown in DDR, but involved in stress responses as well as development and maintenance of genome integrity | [80,131] |
| siRNA | 24-nt siRNA, 21-nt siRNA | Involved in de novo DNA methylation with DDB2-AGO4 or repair of DNA photoproducts with DDB2-AGO1 | [132] |
| diRNA | 21-nt diRNA | Involved in DSB repair or DDR activation via interaction with AGO2 | [133,134] |
| lncRNA | TER2 | Involved in maintenance of genome integrity by inhibiting TERT under genotoxic stress | [135] |
© 2019 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, J.-H. Chromatin Remodeling and Epigenetic Regulation in Plant DNA Damage Repair. Int. J. Mol. Sci. 2019, 20, 4093. https://doi.org/10.3390/ijms20174093
Kim J-H. Chromatin Remodeling and Epigenetic Regulation in Plant DNA Damage Repair. International Journal of Molecular Sciences. 2019; 20(17):4093. https://doi.org/10.3390/ijms20174093
Chicago/Turabian StyleKim, Jin-Hong. 2019. "Chromatin Remodeling and Epigenetic Regulation in Plant DNA Damage Repair" International Journal of Molecular Sciences 20, no. 17: 4093. https://doi.org/10.3390/ijms20174093
APA StyleKim, J.-H. (2019). Chromatin Remodeling and Epigenetic Regulation in Plant DNA Damage Repair. International Journal of Molecular Sciences, 20(17), 4093. https://doi.org/10.3390/ijms20174093




