PTO-QuickStep: A Fast and Efficient Method for Cloning Random Mutagenesis Libraries
Abstract
1. Introduction
2. Results and Discussion
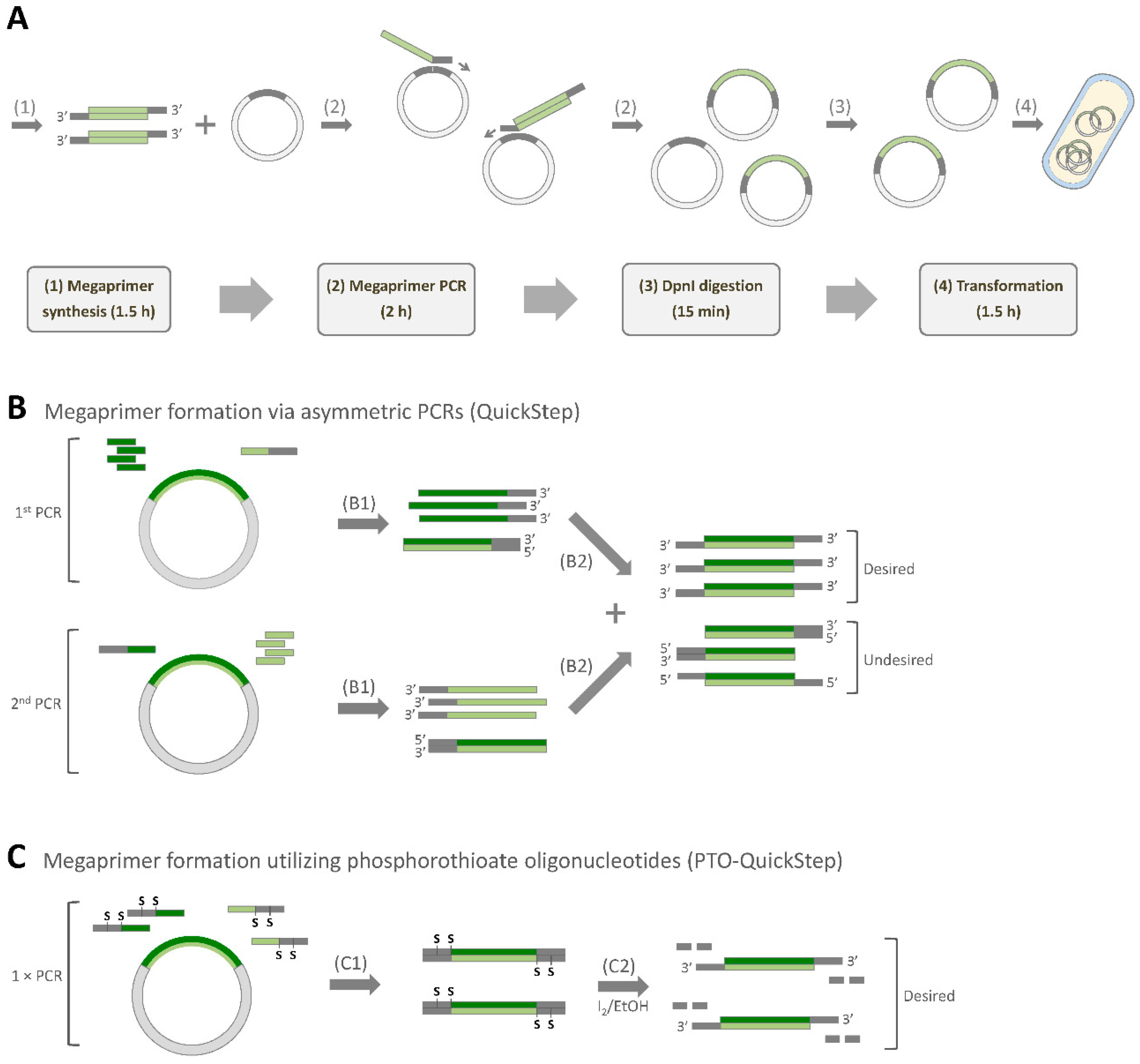
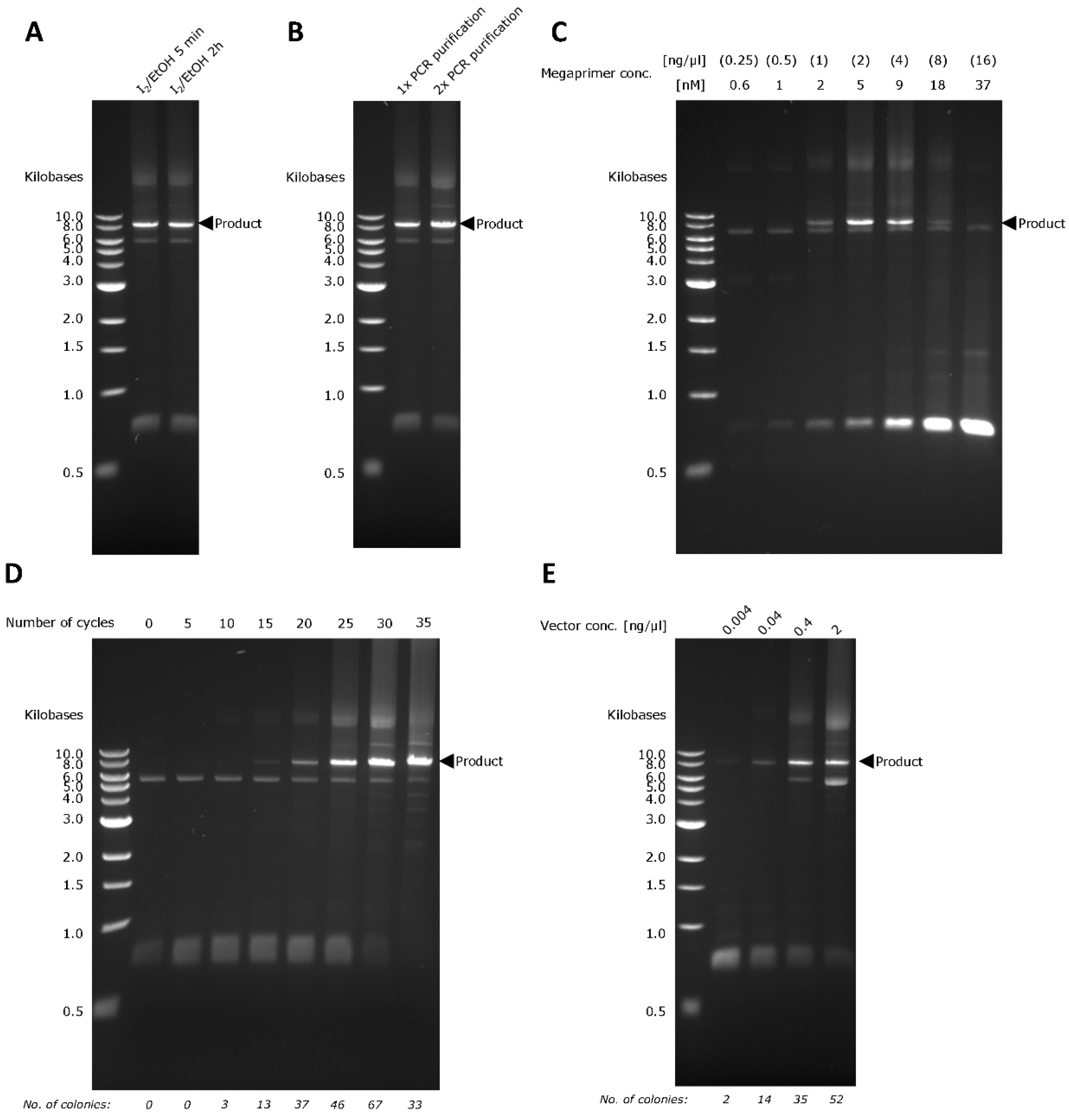
2.1. PTO-QuickStep and its Optimization
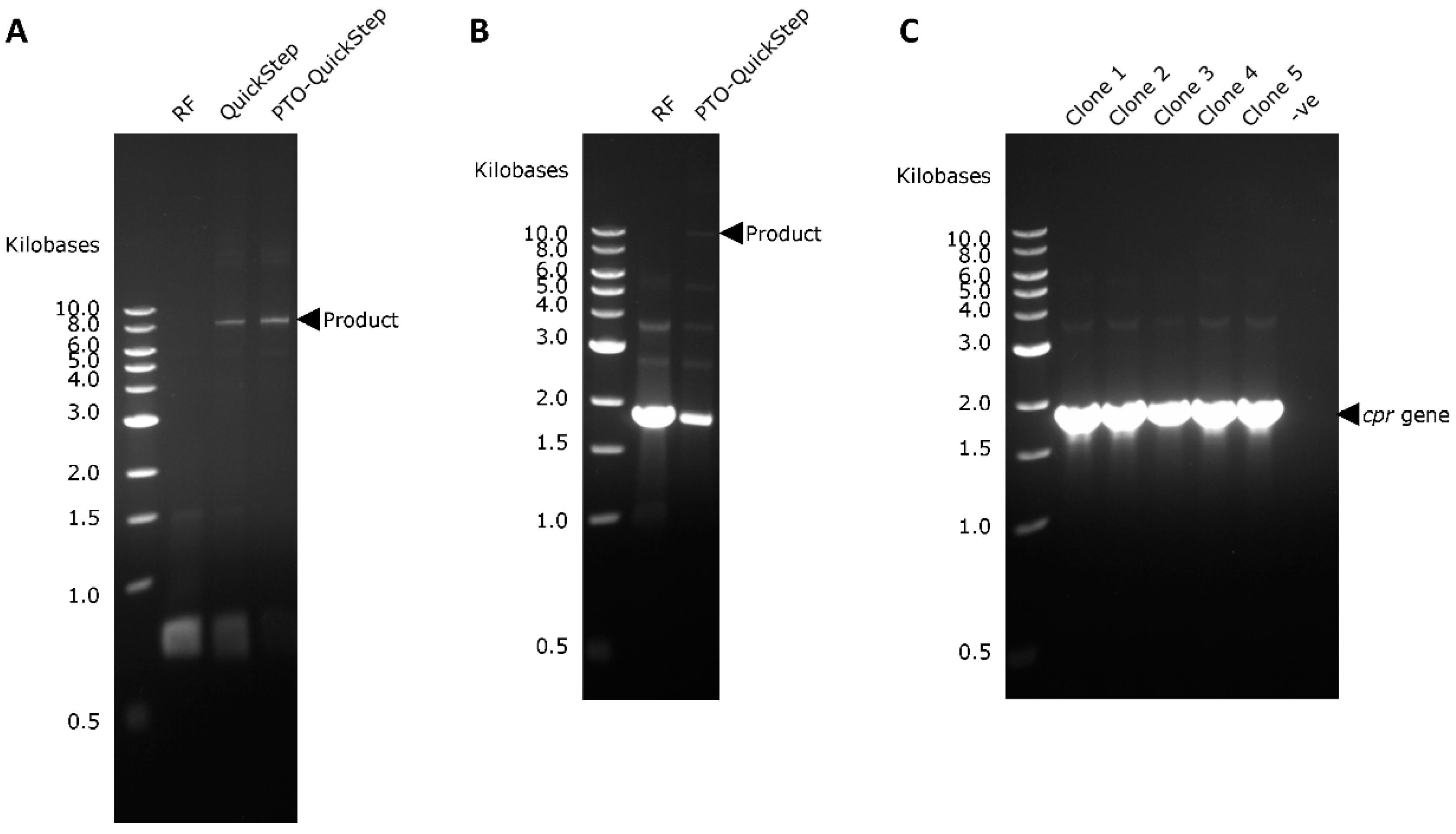
2.2. PTO-QuickStep is Superior to QuickStep-Cloning and RF Cloning
2.3. Right-First-Time Cloning of P450 BM-3 Reductase Gene (cpr)
2.4. Applying PTO-QuickStep for Directed Protein Evolution
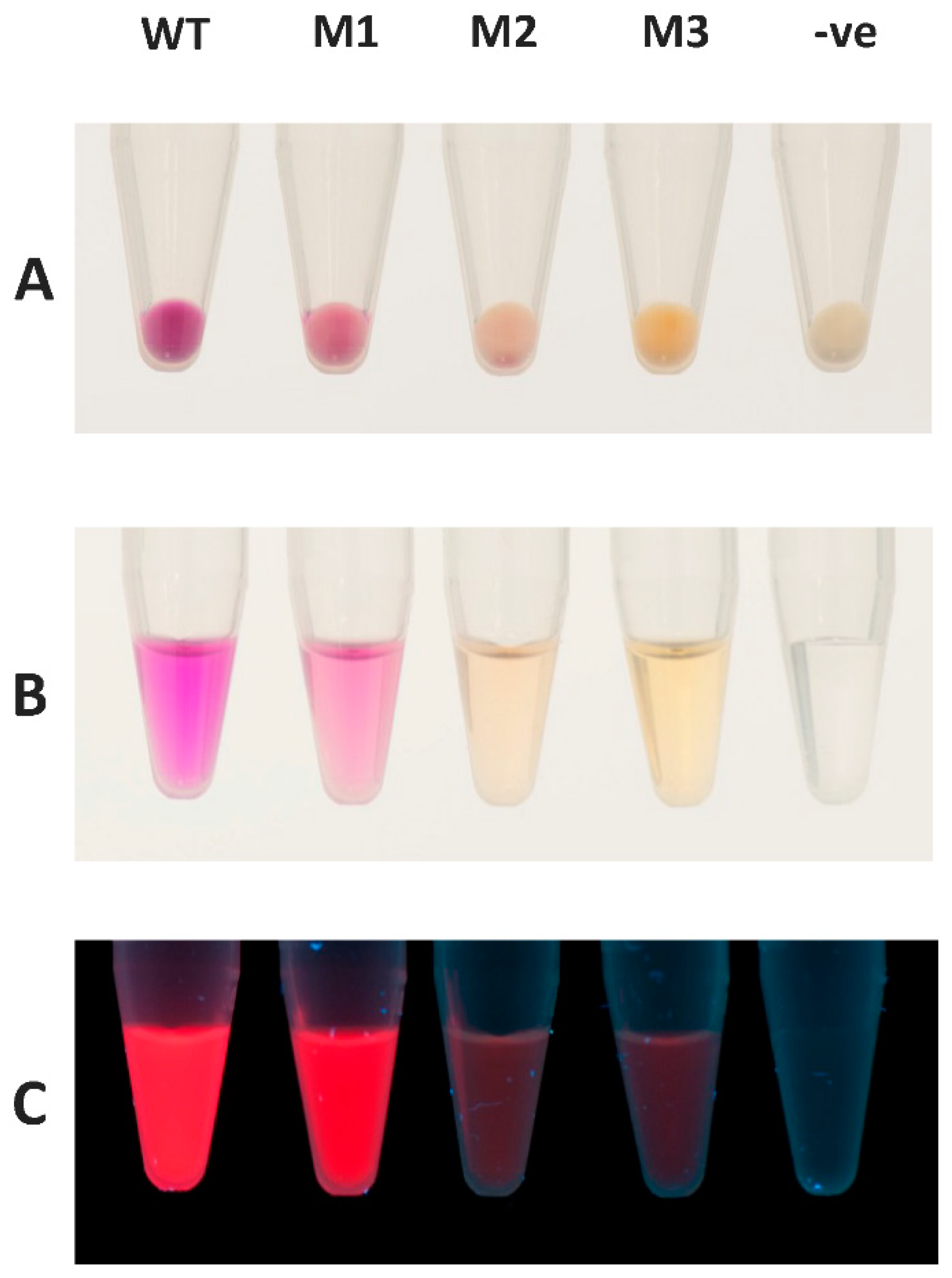
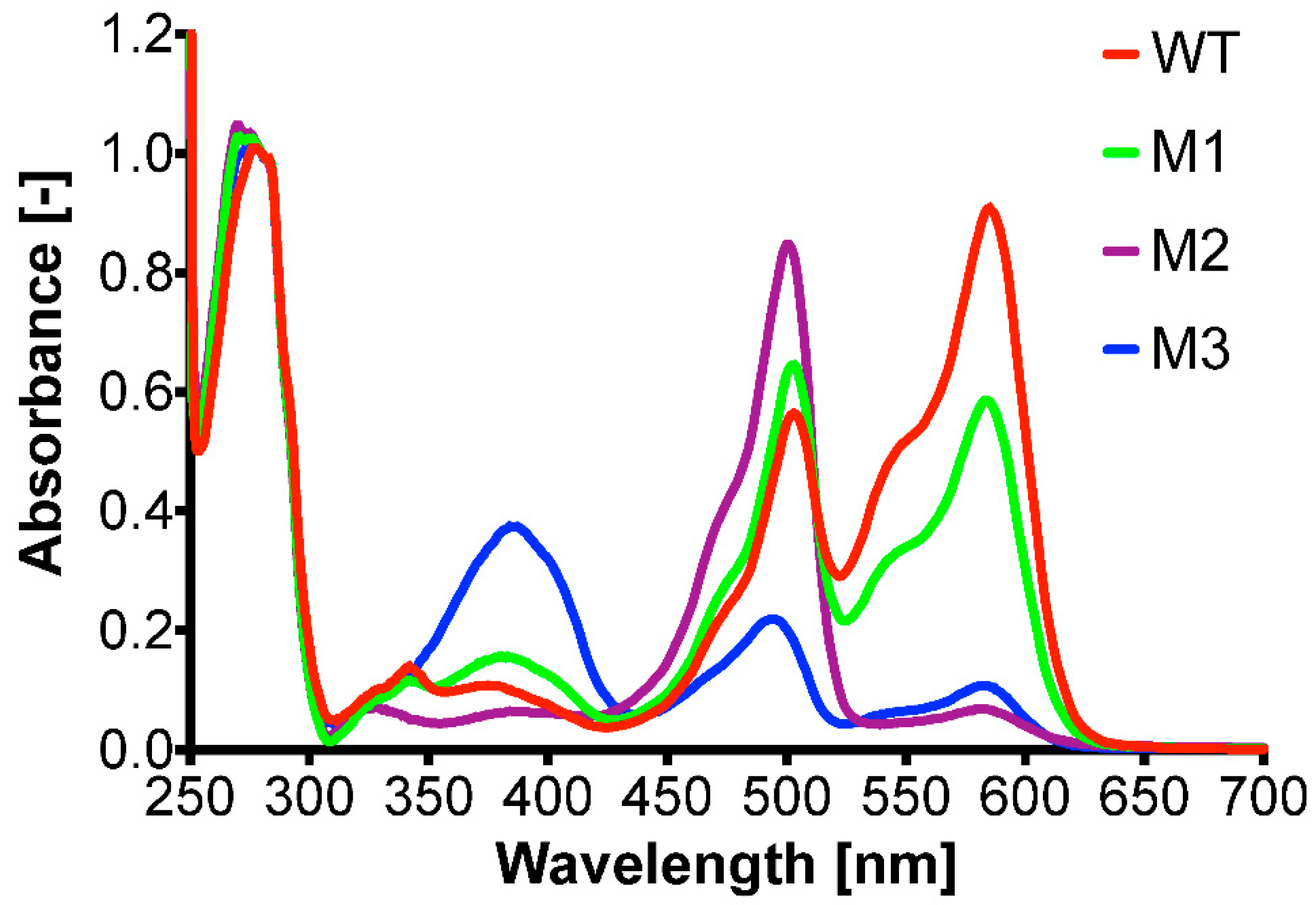
2.5. mRFP1 Variants Isolated from PTO-QuickStep Library
2.6. PTO-QuickStep: Potential Limitations and Mitigation
3. Materials and Methods
3.1. Materials
3.2. Primers
3.3. QuickStep-Cloning
3.4. Restriction-Free (RF) Cloning
3.5. PTO-QuickStep
3.6. DNA Gel Electrophoresis
3.7. Chemical Transformation and Clone Analysis
3.8. Error-Prone PCR
3.9. MEGAWHOP
3.10. Transformation of mRFP1 Library
3.11. mRFP1 Expression and Purification
4. Conclusions
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| bp | base pair |
| cfu | colony-forming unit |
| DNA | deoxyribonucleic acid |
| dATP | deoxyadenosine triphosphate |
| dCTP | deoxycytidine triphosphate |
| dGTP | deoxyguanosine triphosphate |
| dNTP | deoxynucleoside triphosphate |
| dTTP | deoxythymidine triphosphate |
| epPCR | error-prone polymerase chain reaction |
| IPTG | isopropyl β-D-1-thiogalactopyranoside |
| kb | kilobase |
| LB | Luria–Bertani |
| MEGAWHOP | megaprimer PCR of whole plasmid |
| PCR | polymerase chain reaction |
| PTO | phosphorothioate |
| RF | restriction-free |
| RFP | red fluorescent protein |
| ssDNA | single-stranded deoxyribonucleic acid |
| TBE | tris-borate-ethylenediaminetetraacetic acid |
References
- Tee, K.L.; Wong, T.S. Polishing the craft of genetic diversity creation in directed evolution. Biotechnol Adv. 2013, 31, 1707–1721. [Google Scholar] [CrossRef] [PubMed]
- Tee, K.L.; Wong, T.S. Basic to basics: Creating genetic diversity. In Directed Enzyme Evolution: Advances and Applications; Alcalde, M., Ed.; Springer, Cham: Basel, Switzerland, 2017; pp. 201–227. [Google Scholar]
- Lehman, I.R. DNA ligase: Structure, mechanism, and function. Science 1974, 186, 790–797. [Google Scholar] [CrossRef] [PubMed]
- Smith, H.O.; Wilcox, K.W. A restriction enzyme from hemophilus influenzae. I. Purification and general properties. J. Mol. Biol. 1970, 51, 379–391. [Google Scholar] [CrossRef]
- Hughes, R.A.; Ellington, A.D. Synthetic DNA synthesis and assembly: Putting the synthetic in synthetic biology. Cold Spring Harb Perspect Biol. 2017, 9. [Google Scholar] [CrossRef] [PubMed]
- Jajesniak, P.; Wong, T.S. Quickstep-cloning: A sequence-independent, ligation-free method for rapid construction of recombinant plasmids. J. Biol. Eng. 2015, 9, 15. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Jajesniak, P.; Wong, T.S. Rapid construction of recombinant plasmids by quickstep-cloning. Methods Mol. Biol. 2017, 1472, 205–214. [Google Scholar] [PubMed]
- Putney, S.D.; Benkovic, S.J.; Schimmel, P.R. A DNA fragment with an alpha-phosphorothioate nucleotide at one end is asymmetrically blocked from digestion by exonuclease iii and can be replicated in vivo. Proc Natl. Acad. Sci. USA 1981, 78, 7350–7354. [Google Scholar] [CrossRef] [PubMed]
- Wan, W.B.; Seth, P.P. The medicinal chemistry of therapeutic oligonucleotides. J. Med. Chem. 2016, 59, 9645–9667. [Google Scholar] [CrossRef]
- Gish, G.; Eckstein, F. DNA and rna sequence determination based on phosphorothioate chemistry. Science 1988, 240, 1520–1522. [Google Scholar] [CrossRef]
- Wong, T.S.; Tee, K.L.; Hauer, B.; Schwaneberg, U. Sequence saturation mutagenesis (sesam): A novel method for directed evolution. Nucleic Acids Res. 2004, 32, e26. [Google Scholar] [CrossRef]
- Wong, T.S.; Roccatano, D.; Loakes, D.; Tee, K.L.; Schenk, A.; Hauer, B.; Schwaneberg, U. Transversion-enriched sequence saturation mutagenesis (sesam-tv+): A random mutagenesis method with consecutive nucleotide exchanges that complements the bias of error-prone pcr. Biotechnol. J. 2008, 3, 74–82. [Google Scholar] [CrossRef] [PubMed]
- Wong, T.S.; Tee, K.L.; Hauer, B.; Schwaneberg, U. Sequence saturation mutagenesis with tunable mutation frequencies. Anal. Biochem. 2005, 341, 187–189. [Google Scholar] [CrossRef] [PubMed]
- Dennig, A.; Marienhagen, J.; Ruff, A.J.; Schwaneberg, U. Omnichange: Simultaneous site saturation of up to five codons. Methods Mol. Biol. 2014, 1179, 139–149. [Google Scholar] [PubMed]
- Blanusa, M.; Schenk, A.; Sadeghi, H.; Marienhagen, J.; Schwaneberg, U. Phosphorothioate-based ligase-independent gene cloning (plicing): An enzyme-free and sequence-independent cloning method. Anal. Biochem. 2010, 406, 141–146. [Google Scholar] [CrossRef] [PubMed]
- Shaner, N.C.; Campbell, R.E.; Steinbach, P.A.; Giepmans, B.N.; Palmer, A.E.; Tsien, R.Y. Improved monomeric red, orange and yellow fluorescent proteins derived from discosoma sp. Red fluorescent protein. Nat. Biotechnol. 2004, 22, 1567–1572. [Google Scholar] [CrossRef] [PubMed]
- van den Ent, F.; Lowe, J. Rf cloning: A restriction-free method for inserting target genes into plasmids. J. Biochem. Biophys Methods 2006, 67, 67–74. [Google Scholar] [CrossRef] [PubMed]
- Miyazaki, K.; Takenouchi, M. Creating random mutagenesis libraries using megaprimer pcr of whole plasmid. Biotechniques 2002, 33, 1033–1034, 1036–1038. [Google Scholar] [CrossRef] [PubMed]
- Jach, G.; Pesch, M.; Richter, K.; Frings, S.; Uhrig, J.F. An improved mrfp1 adds red to bimolecular fluorescence complementation. Nat. Methods 2006, 3, 597–600. [Google Scholar] [CrossRef]
- Campbell, R.E.; Tour, O.; Palmer, A.E.; Steinbach, P.A.; Baird, G.S.; Zacharias, D.A.; Tsien, R.Y. A monomeric red fluorescent protein. Proc. Natl. Acad. Sci. USA 2002, 99, 7877–7882. [Google Scholar] [CrossRef]
- Wong, T.S.; Zhurina, D.; Schwaneberg, U. The diversity challenge in directed protein evolution. Comb. Chem. High Throughput Screen 2006, 9, 271–288. [Google Scholar] [CrossRef]
- Li, Y.; Tsien, R.W. Phtomato, a red, genetically encoded indicator that enables multiplex interrogation of synaptic activity. Nat. Neurosci. 2012, 15, 1047–1053. [Google Scholar] [CrossRef] [PubMed]
| Name | Length [bp] | Sequence (5′→3′) |
|---|---|---|
| RFP-Fwd | 19 | ATGGCGAGTAGCGAAGACG |
| RFP-Rev | 21 | TTAAGCACCGGTGGAGTGACG |
| IntA-RFP-Fwd | 47 | cgaaaacctgtacttccagggtggatccATGGCGAGTAGCGAAGACG |
| IntB-RFP-Rev | 47 | ctaggatctgactgcggctcctccatTTAAGCACCGGTGGAGTGACG |
| IntA-RFP-Fwd-PTO | 47 | cgaaaacctgtact*tccagggtggatcc*ATGGCGAGTAGCGAAGACG |
| IntB-RFP-Rev-PTO | 47 | ctaggatctgact*gcggctcctccat*TTAAGCACCGGTGGAGTGACG |
| IntA-CPR-Fwd | 56 | ggtaaaagcaaaatcgaaaaaaattccgcttGGCGGTATTCCTTCACCTAGCACTG |
| IntB-CPR-Rev | 43 | gtcgacggagctcgaattcttaCCCAGCCCACACGTCTTTTGC |
| IntA-CPR-Fwd-PTO | 56 | ggtaaaagcaaaatc*gaaaaaaattccgctt*GGCGGTATTCCTTCACCTAGCACTG |
| IntB-CPR-Rev-PTO | 43 | gtcgacggagc*tcgaattctta*CCCAGCCCACACGTCTTTTGC |
| Target Gene/ Recipient Plasmid | Strain/ Transformation Method | QS* | RF* | PTO-QS (25 cycles)* | PTO-QS (30 cycles)* | MEGAWHOP | Transformation Efficiency [cfu/µg]** |
|---|---|---|---|---|---|---|---|
| rfp/ pET24a-HLTev-p53 | E. coli DH5α/ Chemical | 9 (9) | 0 (0) | 19 (16) | 43 (41) | N/A | 2.0 × 104 |
| cpr/ pETM11-BMP-WT | E. coli DH5α/ Chemical | N/D | 0 | 49 | N/D | N/A | 4.0 × 104 |
| rfp library/ pET24a-HLTev-p53 | E. coli C41 (DE3)/ Electroporation | N/D | N/A | 1.0 × 104 | N/D | 1.5 × 104 | 2.0 × 106 |
| RFP Variants | Nucleotide Substitution | Amino Acid Substitution |
|---|---|---|
| M1 | ACC → TCC | T195S |
| M2 | TTC → CTC | F91L |
| M3 | CAG → CTG | Q66L |
| ACC → GCC | T202A |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jajesniak, P.; Tee, K.L.; Wong, T.S. PTO-QuickStep: A Fast and Efficient Method for Cloning Random Mutagenesis Libraries. Int. J. Mol. Sci. 2019, 20, 3908. https://doi.org/10.3390/ijms20163908
Jajesniak P, Tee KL, Wong TS. PTO-QuickStep: A Fast and Efficient Method for Cloning Random Mutagenesis Libraries. International Journal of Molecular Sciences. 2019; 20(16):3908. https://doi.org/10.3390/ijms20163908
Chicago/Turabian StyleJajesniak, Pawel, Kang Lan Tee, and Tuck Seng Wong. 2019. "PTO-QuickStep: A Fast and Efficient Method for Cloning Random Mutagenesis Libraries" International Journal of Molecular Sciences 20, no. 16: 3908. https://doi.org/10.3390/ijms20163908
APA StyleJajesniak, P., Tee, K. L., & Wong, T. S. (2019). PTO-QuickStep: A Fast and Efficient Method for Cloning Random Mutagenesis Libraries. International Journal of Molecular Sciences, 20(16), 3908. https://doi.org/10.3390/ijms20163908







