Directed Evolution of an Improved Rubisco; In Vitro Analyses to Decipher Fact from Fiction
Abstract
1. Introduction
2. Results
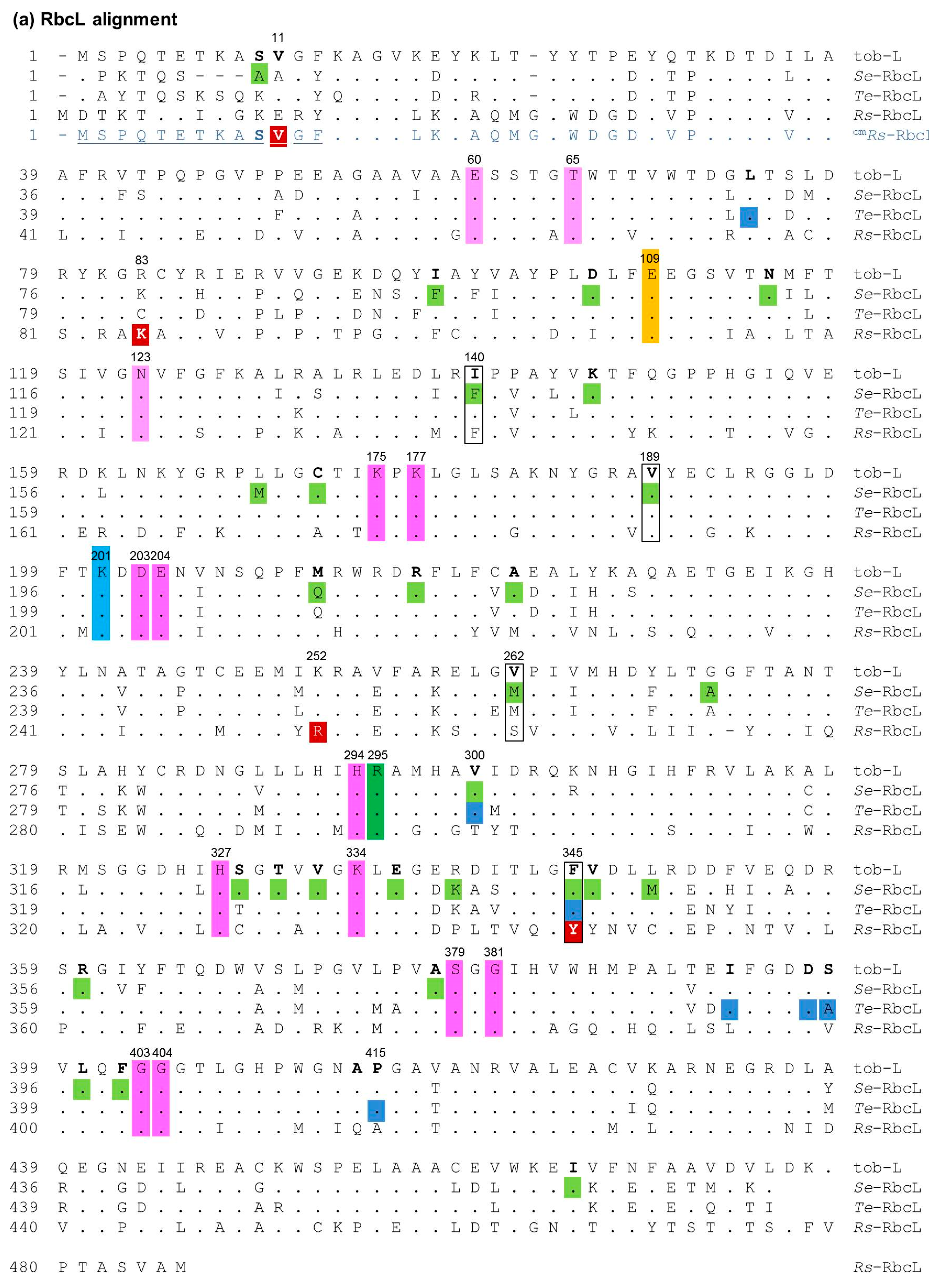
2.1. Ambiguity in How Cyanobacteria Rubisco Mutations Improve RDE Fitness
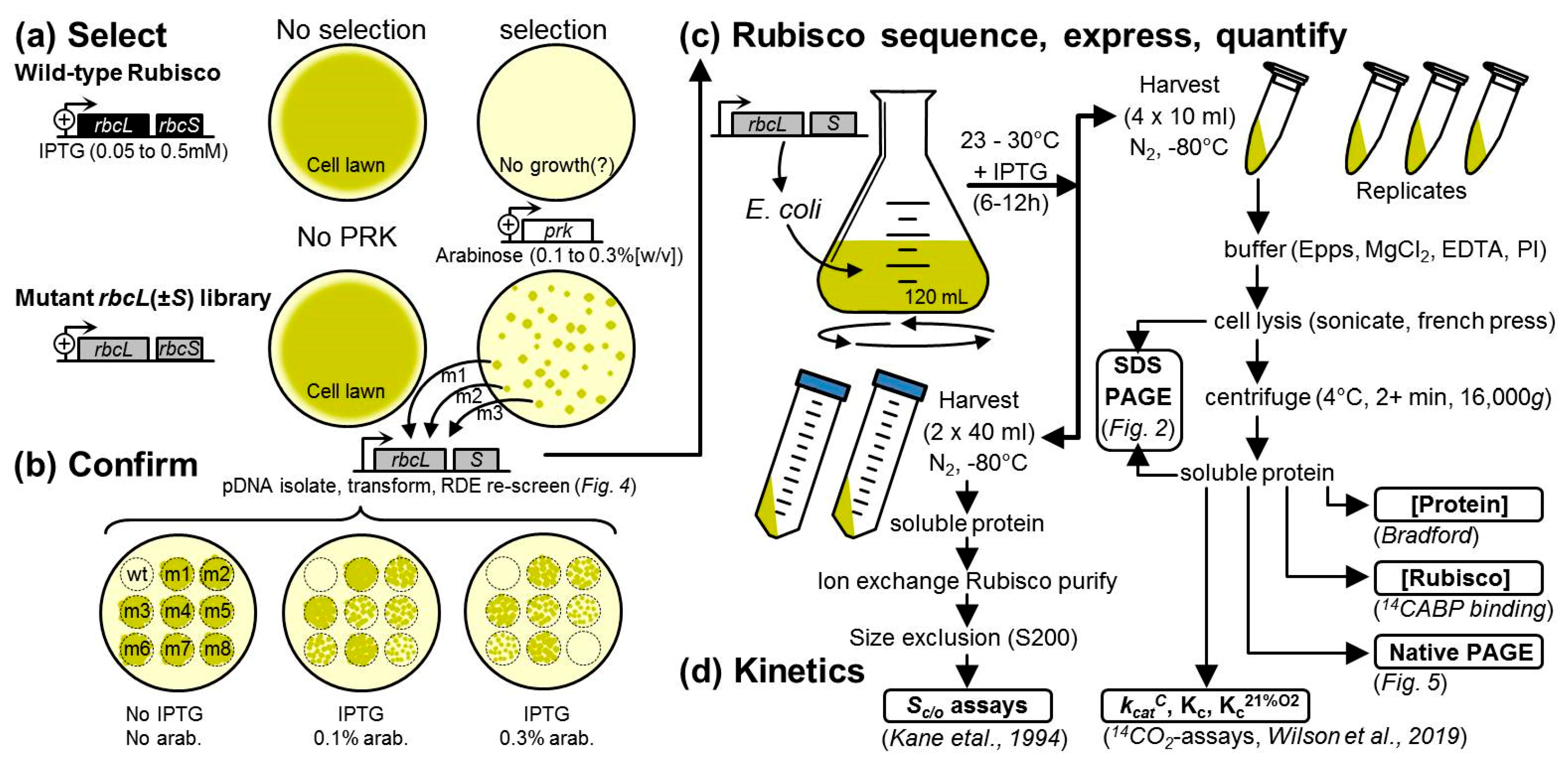
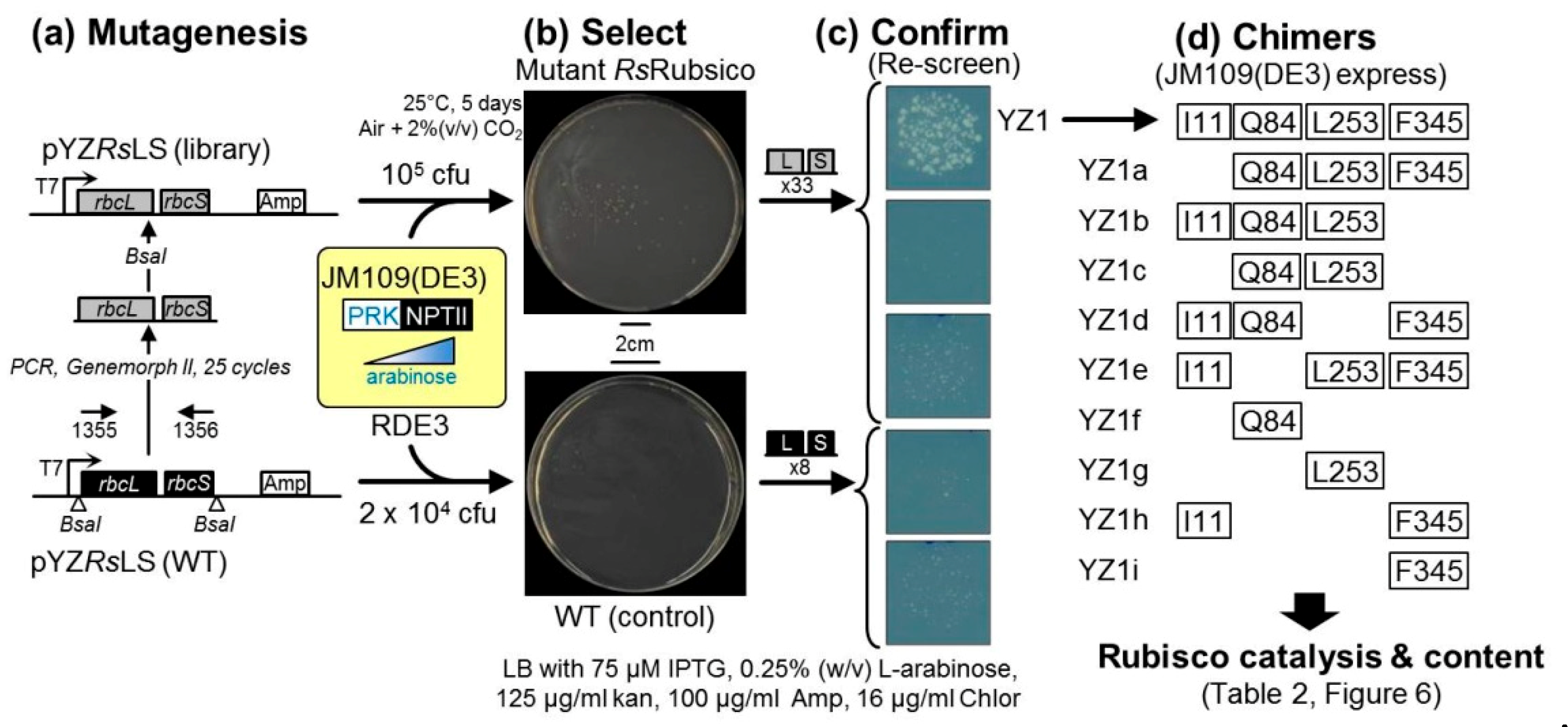
2.2. An Analytical Pathway for the Directed Evolution of Rubisco Using an RDE Screen
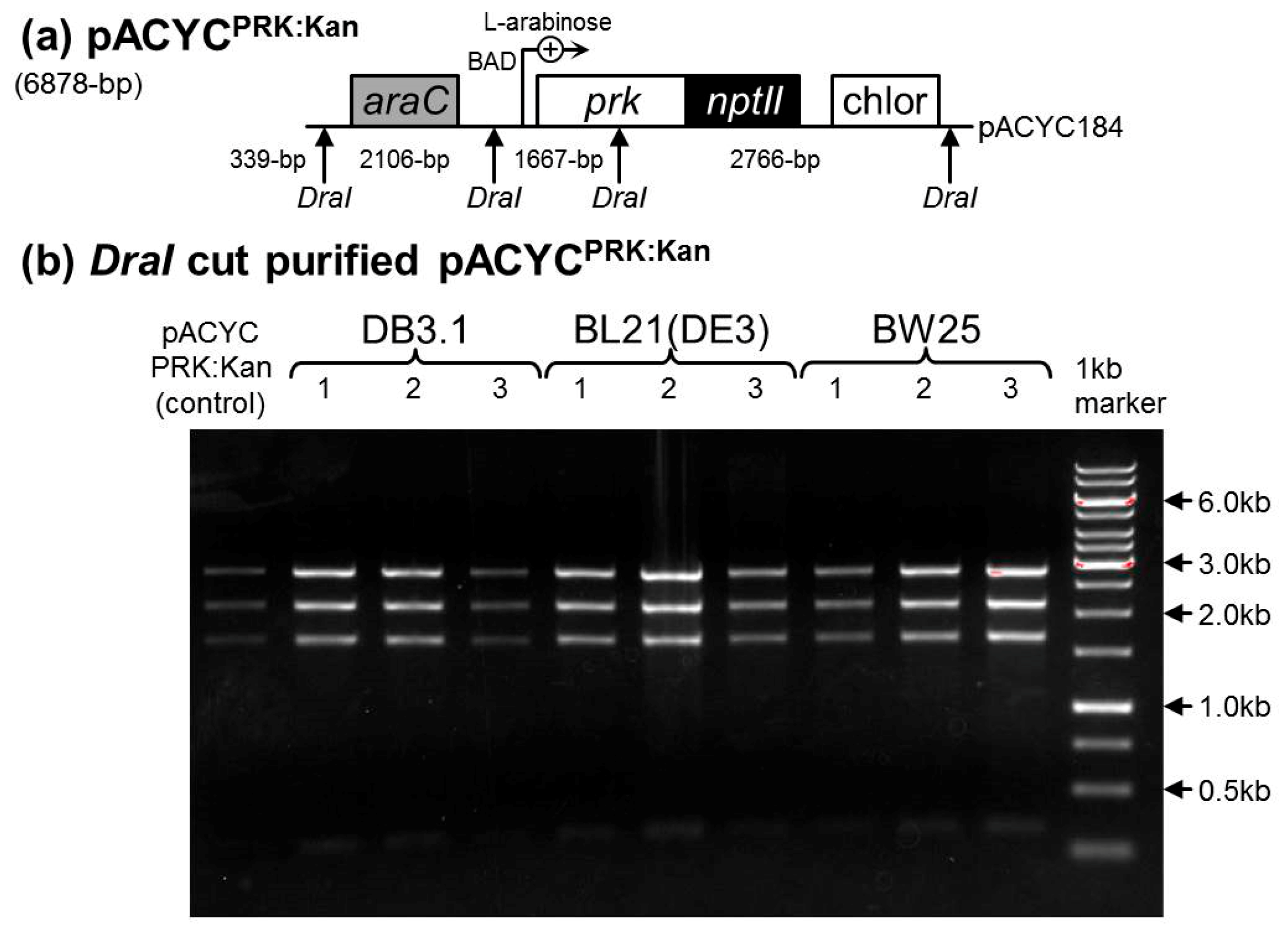
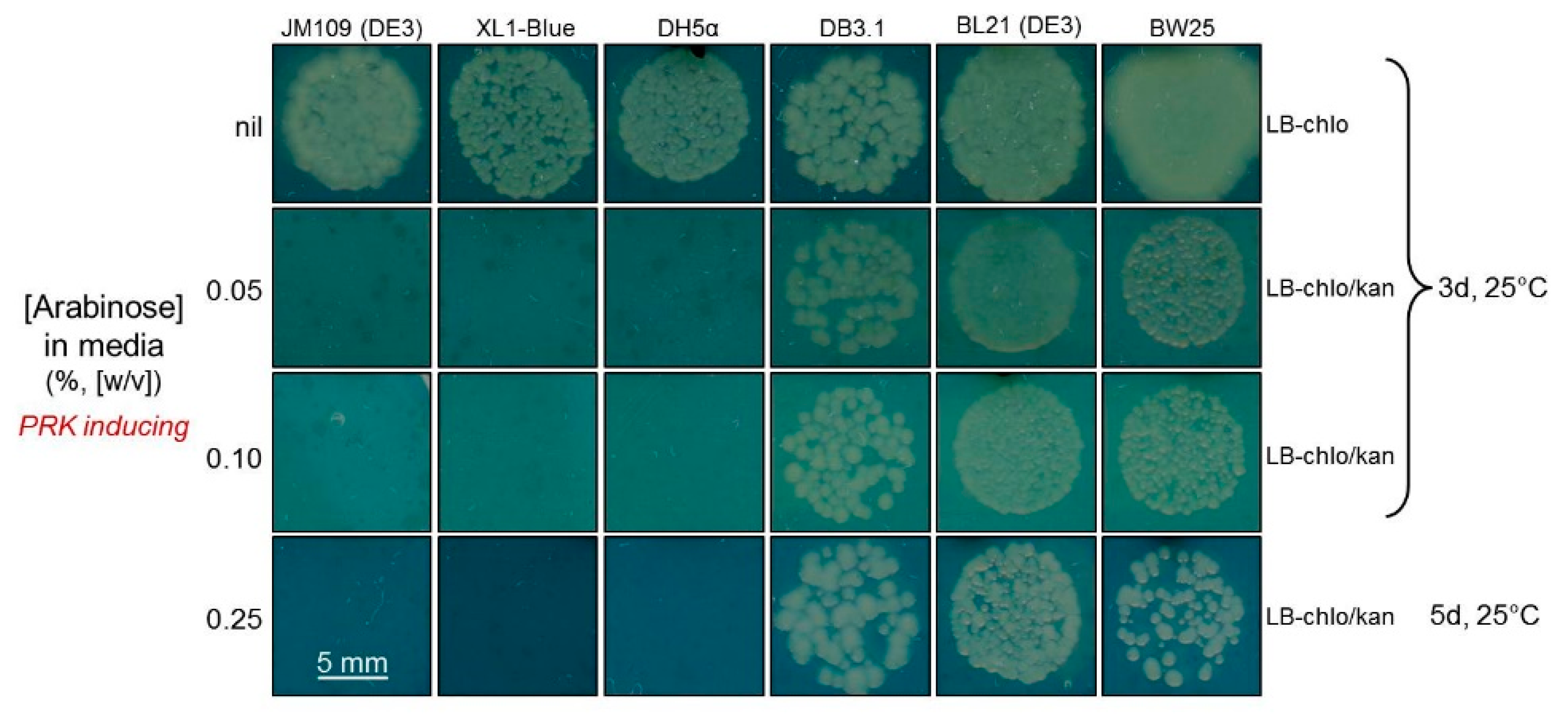
2.3. Determination of a Suitable E. coli Strain for RDE3
2.4. Directed Evolution of RsRubisco in the RDE3 Screen
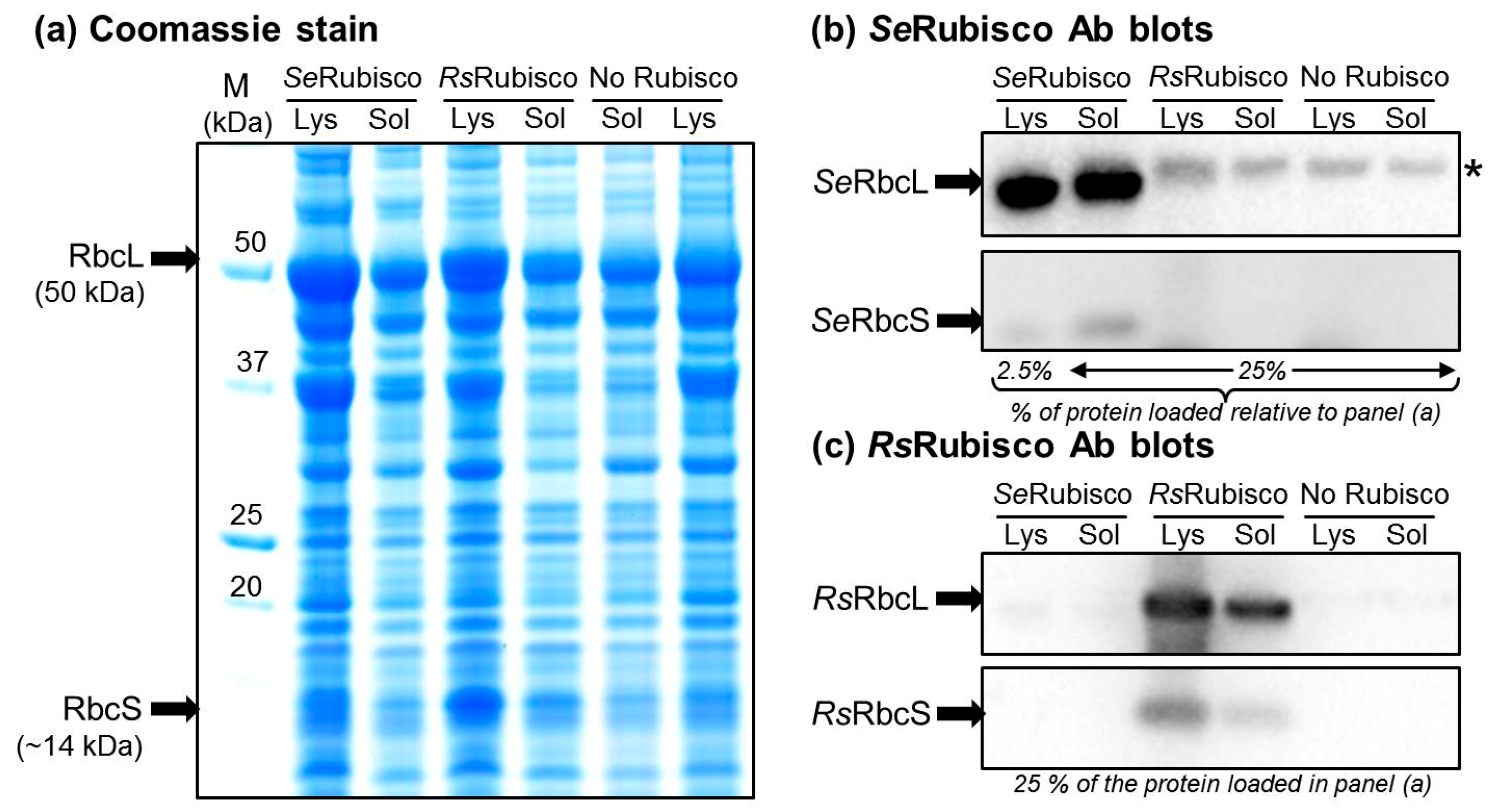
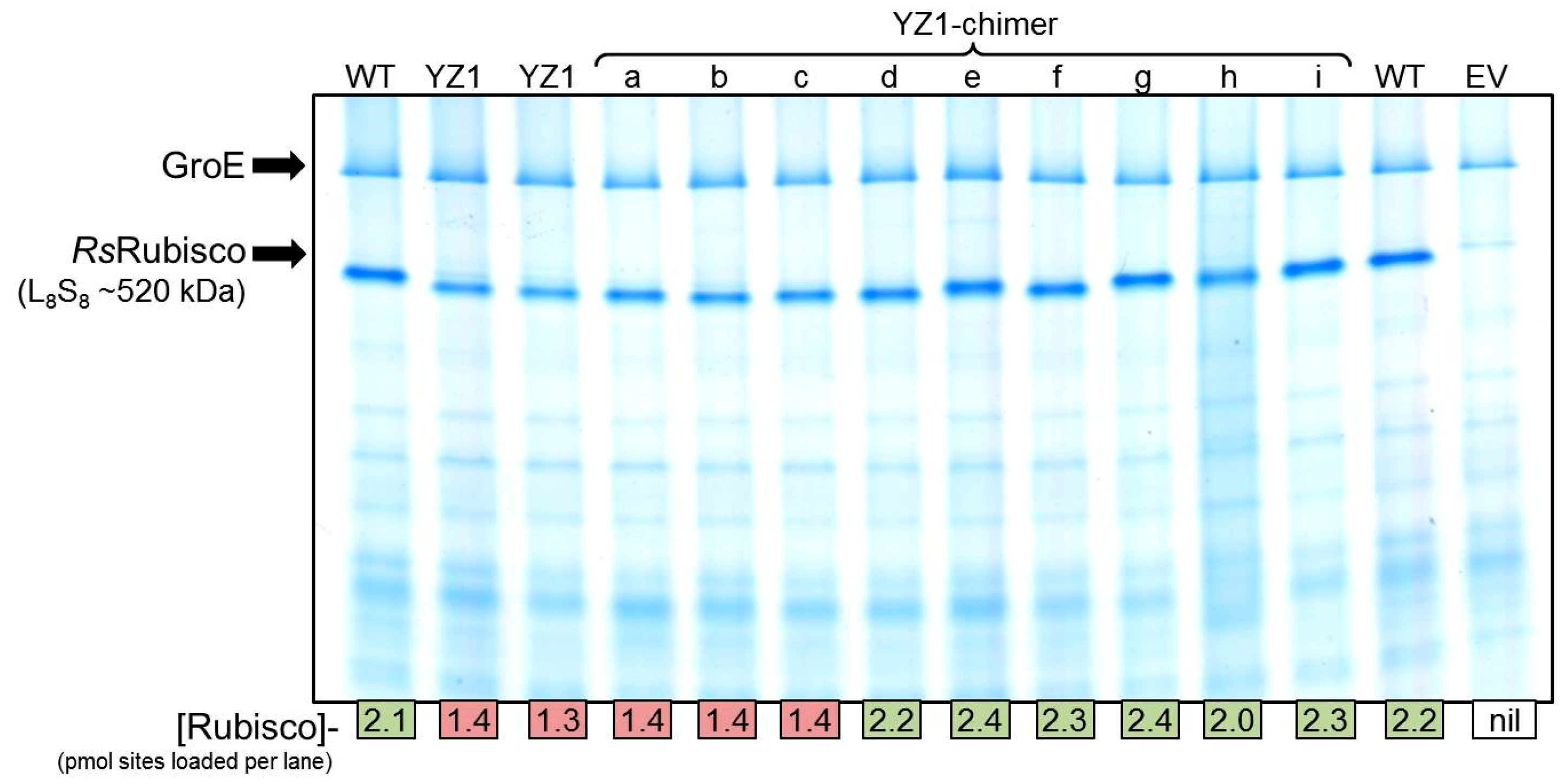
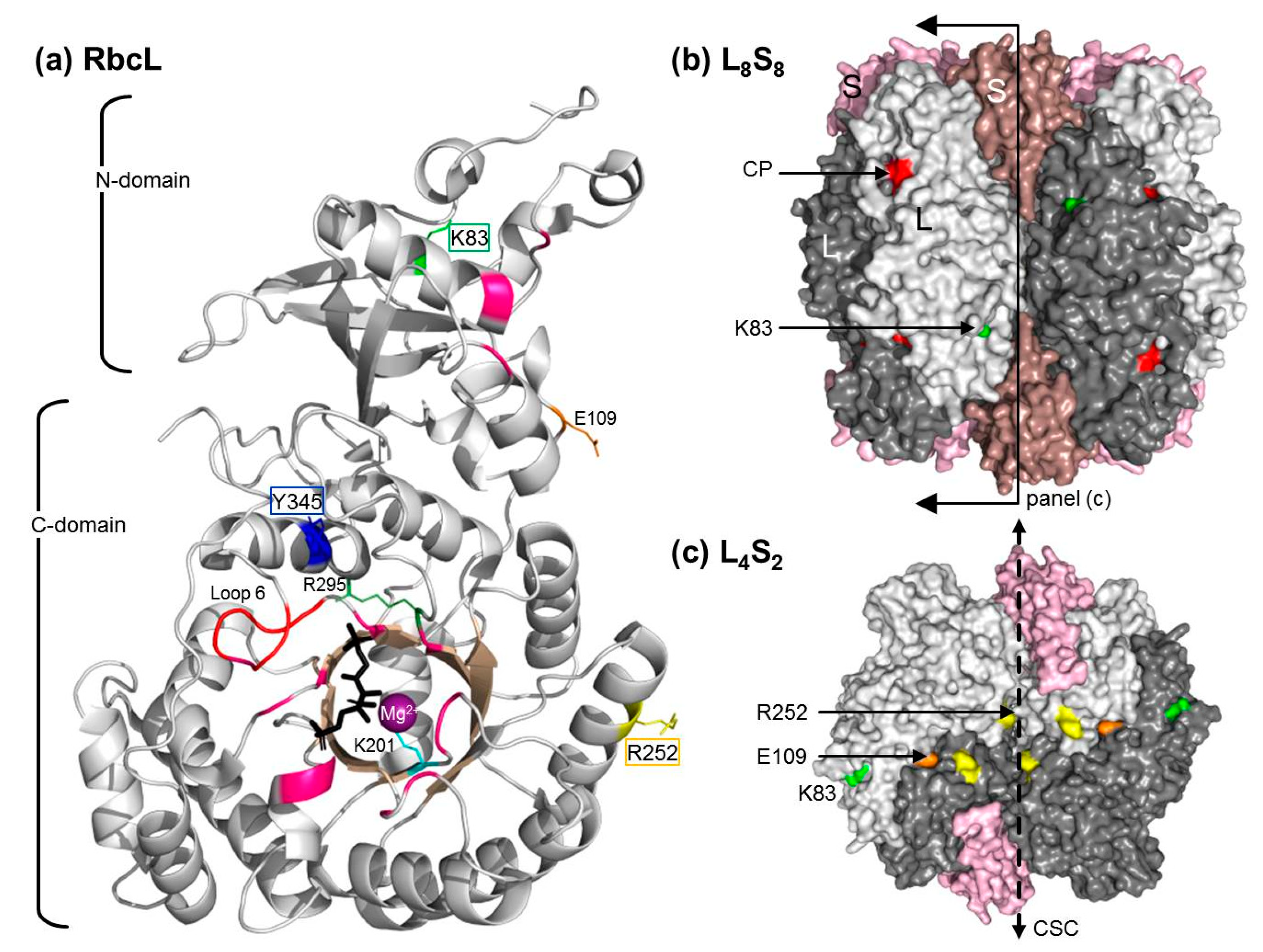
2.5. The RbcL Substitution Y345F Enhances RsRubisco Carboxylation without Impairing Enzyme Biogenesis
3. Discussion
3.1. Mutating RbcL Amino Acid 345 Has Alternate Effects on Form I Rubisco Biogenesis and Catalysis
3.2. How Do the K83Q andR252L Substitutions Function Cooperativly to Reduce RsRubisco Biogenesis?
3.3. Understanding the Basis of RDE Selection
3.4. Pathways for Evolving RsRubisco Further—Why and How?
4. Materials and Methods
4.1. Comparing the PRK Sensitivity of Different E. coli Strains
4.2. Library Construction and Rubisco Selection Using the RDE Screening
4.3. Rubisco Content and Catalysis
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| Raf1 | Rubisco accumulation factor 1 |
| Raf2 | Rubisco accumulation factor 2 |
| BSD2 | Bundle Sheath Defective II Rubisco chaperone |
| RbcX | Rubisco specific chaperone |
| cfu | Colony forming units |
| RuBP | Ribulose-1,5-bisphosphate |
| SC/O | Specificity for CO2 over O2 |
| kcatC | Substrate saturated carboxylation rate |
| kcatO | Substrate saturated oxygenation rate |
| KC | Apparent Km for CO2 |
| KC21%O2 | Apparent Km for CO2 under ambient (20.6% (v/v)) O2 |
| KO | Apparent Km for O2 |
| PRK | Phosphoribulokinase |
Appendix A

| Rubisco Origin | Mutation (s) | Plant RbcL Numbering | KC (μM) | KC21%O2 (μM) | kcatC (s−1) | kcatC/KC (s−1·mM−1) | kcatC/KC21%O2 (s−1· mM−1) | KO (μM) | kcatO (s−1) | kcatO/KO | SC/O | KRuBP (μM) | Rubisco (%CSP) | RDE Strain |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Synchococcus PCC6301 (L8S8) | WT | - | 273 | - | 11.4 | 42 | - | - | - | - | 44.0 | 64 | 1.0 | E. coli K12 Ref [29] |
| M259T | 262 | 237 | - | 12.8 | 54 | - | - | - | - | 43.0 | 67 | 6.0 | ||
| A8S/M259T | 10/262 | 216 | - | 11.4 | 53 | - | - | - | - | 41.0 | 48 | 5.5 | ||
| F342S/M259T | 345/262 | 207 | - | 10.0 | 48 | - | - | - | - | 38.0 | 34 | 3.5 | ||
| Synchococcus PCC6301 (L8S8) | WT | - | 200 | 242 | 11.8 | 59 | 48.7 | 1191 | 1.7 | 1.42 | 41.5 | 44 | 0.2 | MM1 Ref [30] |
| F345I | 345 | 208 | 266 | 10.1 | 49 | 38.0 | 908 | 1.2 | 1.33 | 36.6 | 31 | 3.4 | ||
| I174V | 174 | 233 | 283 | 10.2 | 44 | 36.0 | 1174 | 1.2 | 1.02 | 42.9 | 33 | 1.1 | ||
| F345L | 345 | 245 | - | 9.8 | 40 | - | - | - | - | 40.0 | 93 | 1.9 | ||
| Q212L | 212 | 370 | 444 | 9.9 | 27 | 22.3 | 1252 | 0.9 | 0.72 | 37.4 | 31 | 1.7 | ||
| L348M | 348 | 287 | - | 11.3 | 39 | - | - | - | - | 35.1 | 84 | - | ||
| N115S | 115 | 191 | - | 9.0 | 47 | - | - | - | - | 43.5 | 44 | - | ||
| A272G | 272 | 244 | - | 9.7 | 40 | - | - | - | - | 40.2 | 34 | - | ||
| K339R | 339 | 148 | - | 10.1 | 68 | - | - | - | - | 43.0 | 35 | - | ||
| M262T | 262 | - | - | - | - | - | - | - | - | - | - | 0.9 | ||
| M169L | 169 | - | - | - | - | - | - | - | - | - | - | 0.8 | ||
| L161M | 161 | - | - | - | - | - | - | - | - | - | - | 1.1 | ||
| Synech.PCC7002 (L8S8) | WT | 132 | - | 6.7 | 51 | - | - | - | - | - | 52 | - | BL21(DE3) Ref [58] | |
| E49VS/D82GS | E51VS/D96GS | 200 | - | 14.6 | 74 | - | - | - | - | - | 50 | - | ||
| Synchococcus PCC6301 (L8S8) | WT | - | 248 | - | 12.9 | 52 | - | - | - | 0.9 | 56.1 | - | - | MM1 Ref [27] |
| F140I | 140 | 129 | - | 19.4 | 150 | - | - | - | 2.9 | 51.3 | - | - | ||
| C172G | 172 | 493 | - | 12.8 | 26 | - | - | - | 0.4 | 59.6 | - | - | ||
| V189I | 189 | 160 | - | 14.3 | 89 | - | - | - | 1.8 | 49.2 | - | - | ||
| V221I | 221 | 181 | - | 2.5 | 14 | - | - | - | 0.3 | 57.9 | - | - | ||
| V346L | 346 | 256 | - | 4.3 | 17 | - | - | - | 0.3 | 56.9 | - | - | ||
| V346I | 346 | 274 | - | 7.2 | 26 | - | - | - | 0.5 | 54.4 | - | - | ||
| S398C | 398 | 322 | - | 18.6 | 58 | - | - | - | 1.0 | 56.2 | - | - | ||
| I465V | 465 | 298 | - | 14.3 | 48 | - | - | - | 0.9 | 53.7 | - | - | ||
| F345I | 345 | 274 | - | 11.2 | 41 | - | - | - | 0.8 | 52.1 | - | - | ||
| F140I/F345I | 140/345 | 218 | - | 15.6 | 72 | - | - | - | 1.2 | 59.3 | - | - | ||
| Thermosynech elongatus (L8S8) | WT | - | - | 107 | 6.6 | - | 70 | - | - | - | 53.3 | 36 | 6.6 | RDE2 XL1-B Ref [22] |
| F345I | 345 | - | 101 | 6.9 | - | 47 | - | - | - | 48.0 | - | 14.6 | ||
| P415A | 415 | - | 111 | 6.9 | - | 62 | - | - | - | 52.6 | - | 10.4 | ||
| V300A | 300 | - | 135 | 6.6 | - | 49 | - | - | - | 51.9 | - | 7.1 | ||
| F345I/P415A | 345I/415 | - | 102 | 4.8 | - | 47 | - | - | - | 49.2 | - | 17.5 | ||
| P415A/V98MS | 415/112S | - | 93 | 9.5 | - | 100 | - | - | - | 56.3 | - | 20.4 | ||
| P415A/A48VS | 415/58S | - | 105 | 8.8 | - | 84 | - | - | - | 48.0 | - | 9.7 | ||
| P415A/H37LS | 415/39S | - | 100 | 8.4 | - | 84 | - | - | - | 49.7 | - | 11.1 | ||
| P415A/Y36NS/G112DS | 415/38S/122S/123S | - | 94 | 9.3 | - | 99 | - | - | - | 47.7 | - | 9.7 | ||
| P415A/Y36NS/R51HS | 415/38S/65S | - | 120 | 8.0 | - | 68 | - | - | - | 51.8 | - | 13.6 | ||
| P415A/A398T/A48VS | 415/398/58S | - | 105 | 8.1 | - | 75 | - | - | - | 45.5 | - | 13.3 | ||
| P415A/L74M/D397N | 415/74/397 | - | 92 | 8.2 | - | 90 | - | - | - | 51.8 | - | 12.5 | ||
| P415A/A414T | 415/414 | - | 102 | 8.0 | - | 78 | - | - | - | 55.8 | - | 13.9 | ||
| P415A/I393M | 415/393 | - | 90 | 7.8 | - | 87 | - | - | - | 48.1 | - | 16.6 | ||
| P415A/A398T | 415/398 | - | 110 | 6.4 | - | 59 | - | - | - | 50.5 | - | 16.6 | ||
| Synchococcus PCC6301 (L8S8) | WT | - | - | 251 | 12.6 | - | 50 | - | - | - | 46.0 | 44 | 1.3 | |
| F345I | 345 | - | 285 | 10.3 | - | 36 | - | - | - | 42.0 | - | 14.4 | ||
| F140I | 140 | - | 211 | 11.1 | - | 53 | - | - | - | 46.0 | - | 6.3 | ||
| V189I | 189 | - | 186 | 10.6 | - | 57 | - | - | - | 39.0 | NA | 6.2 | ||
| RhodospirIllum Rubrum (L2) | WT | - | 149 | - | 12.3 | 83 | - | 159 | 1.4 | 9.0 | 9.0 | 63 | 0.2 | MM1 Ref [26] |
| D117V | 129 | 196 | - | 7.3 | 38 | - | 199 | 1.5 | 8.0 | 5.3 | 55 | 0.6 | ||
| D117H | 129 | 192 | - | 7.5 | 39 | - | 153 | 1.1 | 7.0 | 5.3 | 48 | 0.5 | ||
| H44Q | 56 | 301 | - | 9.3 | 31 | - | 185 | 1.2 | 6.0 | 5.3 | 56 | 0.9 | ||
| H44N | 56 | 204 | - | 9.8 | 48 | - | 116 | 1.0 | 9.0 | 5.5 | 59 | 0.7 | ||
| Methanococcoides burtonii (L2–L10) | WT | - | 57 | 1337 | 0.6 | 11 | 0.5 | 11 | 0.1 | 8.0 | 1.3 | 4 | 7.0 | MM1 Ref [40] |
| E138V | 146 | 66 | 543 | 1.0 | 15 | 1.8 | 35 | 0.4 | 11.0 | 1.5 | 2 | 8.0 | ||
| K332E | 336 | 78 | 892 | 1.2 | 15 | 1.4 | 24 | 0.3 | 13.0 | 1.3 | 5 | 6.0 | ||
| T421A | 400 | - | - | 1.3 | - | - | - | - | - | 1.7 | - | 9.0 | ||
| M423V | 402 | - | - | 1.1 | - | - | - | - | - | - | - | 7.0 | ||
| H379R | 360 | - | - | 0.6 | - | - | - | - | - | - | - | 6.0 | ||
| Y471N | 471 | - | - | 0.9 | - | - | - | - | - | - | - | 4.0 | ||
| G327S | 331/insert\332 | - | - | 0.7 | - | - | - | - | - | - | - | 6.0 | ||
| Synechococcus PCC6301 (L8S8) | WT | - | 186 | - | 3.5 | 22 | - | - | - | - | - | 38 | - | R.capsulatus SBI/II-mutant Ref [31] |
| F342V | 345 | 256 | - | 2.4 | 12 | - | - | - | - | - | 21 | - | ||
| D103V | 106 | 459 | - | 2.4 | 7 | - | - | - | - | - | 34 | - | ||
| D103N | 106 | 325 | - | 1.8 | 6 | - | - | - | - | - | 36 | - | ||
| D103E | 106 | 301 | - | 1.3 | 3 | - | - | - | - | - | 33 | - | ||
| Synechococcus PCC6301 (L8S8) | WT | - | 186 | - | 4.9 | 26 | - | 850 | - | - | - | 27 | - | R.capsulatus SBI/II-mutant Ref [43] |
| D103V | 106 | 388 | - | 4.9 | 13 | - | 866 | - | - | - | 25 | - | ||
| A375V | 378 | 143 | - | 0.6 | 4 | - | 1088 | - | - | - | 34 | - | ||
| D103V/A375V | 106/378 | 294 | - | 3.1 | 10 | - | 1153 | - | - | - | 269 | - | ||
| Synechococcus PCC6301 (L8S8) | WT | - | 190 | 247 | 4.3 | 21 | - | 841 | - | - | - | 29 | - | R.capsulatus SBI/II-mutant Ref [28] |
| A375T | 378 | 146 | - | 0.5 | 3 | - | 1076 | - | - | - | 37 | - | ||
| M259T | 262 | 147 | 209 | 4.4 | 27 | - | 595 | - | - | - | 30 | - | ||
| A375V/M259T/M57IS | 378/262/81S | 93 | 126 | 2.2 | 22 | - | 716 | - | - | - | 64 | - | ||
| R214H | 217 | 682 | - | 3.6 | 6 | - | 2152 | - | - | - | 30 | - | ||
| R214H/A375S | 217/378 | 264 | - | 0.6 | 2 | - | 1731 | - | - | - | 25 | - | ||
| T327A | 330 | 273 | - | 2.8 | 11 | - | 1303 | - | - | - | 13 | - | ||
| V186I | 189 | 106 | 145 | 2.8 | 28 | - | 686 | - | - | - | 25 | - | ||
| V186I/T327A | 189/330 | 110 | 133 | 2.1 | 18 | - | 1189 | - | - | - | 25 | - | ||
| S325L | 328 | 111 | - | 4.0 | 36 | - | 931 | - | - | - | 251 | - | ||
| S325L/T327A | 328/330 | 142 | - | 2.6 | 21 | - | 577 | - | - | - | 77 | - | ||
| Ralstonia eutropha (Cupriavidus necator; L8S8) | WT | - | 37 | - | 3.8 | 102 | - | 1149 | - | - | - | 34 | - | R.eutropha H16ΔLS mutant Ref [44] |
| Y347V | 345 | 35 | - | 4.1 | 117 | - | 1139 | - | - | - | - | - | ||
| A380V | 378 | 34 | - | 0.3 | 9 | - | 1435 | - | - | - | 939 | - | ||
| T330A | 328 | 44 | - | 2.8 | 64 | - | 1087 | - | - | - | 29 | - | ||
| Y348C | 346 | 24 | - | 1.6 | 25 | - | 674 | - | - | - | 36 | - | ||
| T330A/A380V | 328/378 | 28 | - | 0.7 | 25 | - | 1069 | - | - | - | 769 | - | ||
| Y348C/A380V | 346/378 | 20 | - | 0.7 | 35 | - | 652 | - | - | - | 581 | - |
References
- Sharwood, R.E. Engineering chloroplasts to improve Rubisco catalysis: Prospects for translating improvements into food and fiber crops. New Phytol. 2017, 213, 494–510. [Google Scholar] [CrossRef]
- Ellis, R.J. The most abundant protein in the world. Trends Biochem. Sci. 1979, 4, 241–244. [Google Scholar] [CrossRef]
- Young, J.N.; Heureux, A.M.C.; Sharwood, R.E.; Rickaby, R.E.M.; Morel, F.M.M.; Whitney, S.M. Large variation in the Rubisco kinetics of diatoms reveals diversity among their carbon-concentrating mechanisms. J. Exp. Bot. 2016, 67, 3445–3456. [Google Scholar] [CrossRef] [PubMed]
- Makino, A. Rubisco and nitrogen relationships in rice: Leaf photosynthesis and plant growth. Soil Sci. Plant Nutr. 2003, 49, 319–327. [Google Scholar] [CrossRef]
- Bauwe, H.; Hagemann, M.; Kern, R.; Timm, S. Photorespiration has a dual origin and manifold links to central metabolism. Curr. Opin. Plant Biol. 2012, 15, 269–275. [Google Scholar] [CrossRef] [PubMed]
- Sage, R.F.; Sage, T.L.; Kocacinar, F. Photorespiration and the evolution of C4 photosynthesis. Annu. Rev. Plant Biol. 2012, 63, 19–47. [Google Scholar] [CrossRef] [PubMed]
- Carmo-Silva, E.; Scales, J.C.; Madgwick, P.J.; Parry, M.A.J. Optimizing Rubisco and its regulation for greater resource use efficiency. Plantcell Environ. 2015, 38, 1817–1832. [Google Scholar] [CrossRef] [PubMed]
- Parry, M.A.J.; Andralojc, P.J.; Scales, J.C.; Salvucci, M.E.; Carmo-Silva, A.E.; Alonso, H.; Whitney, S.M. Rubisco activity and regulation as targets for crop improvement. J. Exp. Bot. 2013, 64, 717–730. [Google Scholar] [CrossRef] [PubMed]
- Andersson, I.; Backlund, A. Structure and function of Rubisco. Plant Physiol. Biochem. 2008, 46, 275–291. [Google Scholar] [CrossRef]
- Andersson, I. Catalysis and regulation in Rubisco. J. Exp. Bot. 2008, 59, 1555–1568. [Google Scholar] [CrossRef]
- Cummins, P.L.; Kannappan, B.; Gready, J.E. Revised mechanism of carboxylation of ribulose-1,5-biphosphate by rubisco from large scale quantum chemical calculations. J. Comput. Chem. 2018, 39, 1656–1665. [Google Scholar] [CrossRef] [PubMed]
- Ishikawa, C.; Hatanaka, T.; Misoo, S.; Miyake, C.; Fukayama, H. Functional incorporation of sorghum small subunit increases the catalytic turnover rate of Rubisco in transgenic rice. Plant Physiol. 2011, 156, 1603–1611. [Google Scholar] [CrossRef] [PubMed]
- Whitney, S.M.; Sharwood, R.E.; Orr, D.; White, S.J.; Alonso, H.; Galmés, J. Isoleucine 309 acts as a C4 catalytic switch that increases ribulose-1,5-bisphosphate carboxylase/oxygenase (rubisco) carboxylation rate in Flaveria. Proc. Natl. Acad. Sci. USA 2011, 108, 14688–14693. [Google Scholar] [CrossRef] [PubMed]
- Spreitzer, R.J.; Peddi, S.R.; Satagopan, S. Phylogenetic engineering at an interface between large and small subunits imparts land-plant kinetic properties to algal Rubisco. Proc. Nat. Acad. Sci. 2005, 102, 17225–17230. [Google Scholar] [CrossRef] [PubMed]
- Farquhar, G.D.; von Caemmerer, S.; Berry, J.A. A biochemical model of photosynthetic CO2 assimilation in leaves of C3 species. Planta 1980, 149, 78–90. [Google Scholar] [CrossRef] [PubMed]
- Andrews, T.J.; Whitney, S.M. Manipulating ribulose bisphosphate carboxylase/oxygenase in the chloroplasts of higher plants. Arch. Biochem. Biophys. 2003, 414, 159–169. [Google Scholar] [CrossRef]
- Long, S.P.; Marshall-Colon, A.; Zhu, X.-G. Meeting the global food demand of the future by engineering crop photosynthesis and yield potential. Cell 2015, 161, 56–66. [Google Scholar] [CrossRef] [PubMed]
- Whitney, S.M.; Baldet, P.; Hudson, G.S.; Andrews, T.J. Form I Rubiscos from non-green algae are expressed abundantly but not assembled in tobacco chloroplasts. Plant J. 2001, 26, 535–547. [Google Scholar] [CrossRef]
- Lin, M.T.; Hanson, M.R. Red algal Rubisco fails to accumulate in transplastomic tobacco expressing Griffithsia monilis RbcL and RbcS genes. Plant Direct. 2018, 2, e00045. [Google Scholar] [CrossRef]
- Long, B.M.; Hee, W.Y.; Sharwood, R.E.; Rae, B.D.; Kaines, S.; Lim, Y.-L.; Nguyen, N.D.; Massey, B.; Bala, S.; von Caemmerer, S.; et al. Carboxysome encapsulation of the CO2-fixing enzyme Rubisco in tobacco chloroplasts. Nat. Commun. 2018, 9, 3570. [Google Scholar] [CrossRef]
- Occhialini, A.; Lin, M.T.; Andralojc, P.J.; Hanson, M.R.; Parry, M.A.J. Transgenic tobacco plants with improved cyanobacterial Rubisco expression but no extra assembly factors grow at near wild-type rates if provided with elevated CO(2). Plant J. 2016, 85, 148–160. [Google Scholar] [CrossRef] [PubMed]
- Wilson, R.H.; Martin-Avila, E.; Conlan, C.; Whitney, S.M. An improved Escherichia coli screen for Rubisco identifies a protein–protein interface that can enhance CO2-fixation kinetics. J. Biol. Chem. 2018, 293, 18–27. [Google Scholar] [CrossRef] [PubMed]
- Sharwood, R.E.; Ghannoum, O.; Kapralov, M.V.; Gunn, L.H.; Whitney, S.M. Temperature responses of Rubisco from Paniceae grasses provide opportunities for improving C3 photosynthesis. Nat. Plants 2016, 2, 16186. [Google Scholar] [CrossRef] [PubMed]
- Mueller-Cajar, O.; Whitney, S.M. Directing the evolution of Rubisco and Rubisco activase: First impressions of a new tool for photosynthesis research. Photosynth. Res. 2008, 98, 667–675. [Google Scholar] [CrossRef] [PubMed]
- Wilson, R.H.; Whitney, S.M. Improving CO2 fixation by enhancing Rubisco performance. In Directed Enzyme Evolution: Advances and Applications; Alcalde, M., Ed.; Springer International Publishing: Berlin, Germany, 2017; pp. 101–126. [Google Scholar]
- Mueller-Cajar, O.; Morell, M.; Whitney, S.M. Directed evolution of Rubisco in Escherichia coli reveals a specificity-determining hydrogen bond in the form II enzyme. Biochemistry 2007, 46, 14067–14074. [Google Scholar] [CrossRef]
- Durao, P.; Aigner, H.; Nagy, P.; Mueller-Cajar, O.; Hartl, F.U.; Hayer-Hartl, M. Opposing effects of folding and assembly chaperones on evolvability of Rubisco. Nat. Chem. Biol. 2015, 11, 148–155. [Google Scholar] [CrossRef]
- Satagopan, S.; Huening, K.A.; Tabita, F.R. Selection of cyanobacterial Synechococcus sp. Strain PCC 6301 Rubisco variants with improved functional properties that confer enhanced CO2-dependent growth of Rhodobacter capsulatus, a photosynthetic bacterium. mBio 2019, 10, e01537-19. [Google Scholar] [CrossRef]
- Greene, D.N.; Whitney, S.M.; Matsumura, I. Artificially evolved Synechococcus PCC6301 Rubisco variants exhibit improvements in folding and catalytic efficiency. Biochem. J. 2007, 404, 517–524. [Google Scholar] [CrossRef]
- Mueller-Cajar, O.; Whitney, S.M. Evolving improved Synechococcus Rubisco functional expression in Escherichia coli. Biochem. J. 2008, 414, 205–214. [Google Scholar] [CrossRef]
- Smith, S.A.; Tabita, F.R. Positive and negative selection of mutant forms of prokaryotic (cyanobacterial) ribulose-1, 5-bisphosphate carboxylase/oxygenase. J. Mol. Biol. 2003, 331, 557–569. [Google Scholar] [CrossRef]
- Emlyn-Jones, D.; Woodger, F.J.; Price, G.D.; Whitney, S.M. RbcX can function as a Rubisco-chaperonin, but is non essential in Synechococcus PCC7942. Plant Cell Physiol. 2006, 47, 1630–1640. [Google Scholar] [CrossRef] [PubMed]
- Andrews, T.J.; Ballment, B. The function of the small subunits of ribulose bisphosphate carboxylase-oxygenase. J. Biol. Chem. 1983, 258, 7514–7518. [Google Scholar] [PubMed]
- Bracher, A.; Whitney, S.M.; Hartl, F.U.; Hayer-Hartl, M. Biogenesis and metabolic maintenance of Rubisco. Annu. Rev. Plant Biol. 2017, 68, 29–60. [Google Scholar] [CrossRef]
- Liu, D.; Ramya, R.C.S.; Mueller-Cajar, O. Surveying the expanding prokaryotic Rubisco multiverse. FEMS Microbiol. Lett. 2017, 364, 16. [Google Scholar] [CrossRef] [PubMed]
- Parikh, M.R.; Greene, D.N.; Woods, K.K.; Matsumura, I. Directed evolution of Rubisco hypermorphs through genetic selection in engineered E. coli. Protein Eng. Des. Sel. 2006, 19, 113–119. [Google Scholar] [CrossRef] [PubMed]
- Aigner, H.; Wilson, R.H.; Bracher, A.; Calisse, L.; Bhat, J.Y.; Hartl, F.U.; Hayer-Hartl, M. Plant Rubisco assembly in E. coli with five chloroplast chaperones including BSD2. Science 2017, 358, 1272–1278. [Google Scholar] [CrossRef]
- Whitney, S.M.; Sharwood, R.E. Plastid transformation for Rubisco engineering and protocols for assessing expression. Methods Mol. Biol. 2014, 1132, 245–262. [Google Scholar]
- Blayney, M.; Whitney, S.; Beck, J. NanoESI mass spectrometry of Rubisco and Rubisco activase structures and their Interactions with nucleotides and sugar phosphates. J. Am. Soc. Mass Spectrom. 2011, 22, 1588–1601. [Google Scholar] [CrossRef]
- Wilson, R.H.; Alonso, H.; Whitney, S.M. Evolving Methanococcoides burtonii archaeal Rubisco for improved photosynthesis and plant growth. Sci. Rep. 2016, 6, 22284. [Google Scholar] [CrossRef]
- Kane, H.J.; Viil, J.; Entsch, B.; Paul, K.; Morell, M.K.; Andrews, T.J. An improved method for measuring the CO2/O2 specificity of ribulosebisphosphate carboxylase-oxygenase. Aust. J. Plant Physiol. 1994, 21, 449–461. [Google Scholar] [CrossRef]
- Sharwood, R.E.; von Caemmerer, S.; Maliga, P.; Whitney, S.M. The catalytic properties of hybrid Rubisco comprising tobacco small and sunflower large Subunits mirror the kinetically equivalent source Rubiscos and can support tobacco growth. Plant Physiol. 2008, 146, 83–96. [Google Scholar] [CrossRef] [PubMed]
- Satagopan, S.; Scott, S.S.; Smith, T.G.; Tabita, F.R. A Rubisco mutant that confers growth under a normally “inhibitory” oxygen concentration. Biochemistry 2009, 48, 9076–9083. [Google Scholar] [CrossRef] [PubMed]
- Satagopan, S.; Tabita, F.R. Rubisco selection using the vigorously aerobic and metabolically versatile bacterium Ralstonia eutropha. FEBS J. 2016, 2869–2880. [Google Scholar] [CrossRef] [PubMed]
- Knight, S.; Andersson, I.; Brändén, C. Crystallographic analysis of ribulose 1,5-bisphosphate carboxylase from spinach at 2.4 A resolution. Subunit interactions and active site. J. Mol. Biol. 1990, 215, 113–160. [Google Scholar] [CrossRef]
- Ramage, R.T.; Read, B.A.; Tabita, F.R. Alteration of the alpha helix region of cyanobacterial ribulose 1,5-bisphosphate carboxylase/oxygenase to reflect sequences found in high substrate specificity enzymes. Arch. Biochem. Biophys. 1998, 349, 81–88. [Google Scholar] [CrossRef] [PubMed]
- Furbank, R.T.; Sage, R.F. Editorial overview: Physiology and metabolism: CO2 concentrating mechanisms in photosynthetic organisms: Evolution, efficiency and significance for crop improvement. Curr. Opin. Plant Biol. 2016, 31, iv–vii. [Google Scholar] [CrossRef]
- Hasse, D.; Larsson, A.M.; Andersson, I. Structure of Arabidopsis thaliana Rubisco activase. Acta Crystallogr. D Biol. Crystallogr. 2015, 71, 800–808. [Google Scholar] [CrossRef]
- Van Lun, M.; van der Spoel, D.; Andersson, I. Subunit interface dynamics in hexadecameric rubisco. J Mol. Biol. 2011, 411, 1083–1098. [Google Scholar] [CrossRef]
- Romero, P.A.; Arnold, F.H. Exploring protein fitness landscapes by directed evolution. Nat. Reviews. Mol. Cell Biol. 2009, 10, 866–876. [Google Scholar] [CrossRef]
- You, L.; Arnold, F.H. Directed evolution of subtilisin E in Bacillus subtilis to enhance total activity in aqueous dimethylformamide. Protein Eng. Des. Sel. 1996, 9, 77–83. [Google Scholar] [CrossRef]
- Vitlin Gruber, A.; Feiz, L. Rubisco Assembly in the Chloroplast. Front. Mol. Biosci. 2018, 5, 24. [Google Scholar] [CrossRef] [PubMed]
- Wilson, R.H.; Hayer-Hartl, M. Complex chaperone dependence of Rubisco biogenesis. Biochemistry 2018, 57, 3210–3216. [Google Scholar] [CrossRef] [PubMed]
- Whitney, S.M.; Birch, R.; Kelso, C.; Beck, J.L.; Kapralov, M.V. Improving recombinant Rubisco biogenesis, plant photosynthesis and growth by coexpressing its ancillary RAF1 chaperone. Proc. Natl. Acad. Sci. USA 2015, 112, 3564–3569. [Google Scholar] [CrossRef] [PubMed]
- Whitney, S.M.; Sharwood, R.E. Linked Rubisco subunits can assemble into functional oligomers without impeding catalytic performance. J Biol. Chem. 2007, 282, 3809–3818. [Google Scholar] [CrossRef] [PubMed]
- Conlan, B.; Whitney, S. Preparing Rubisco for a tune up. Nat. Plants 2018, 4, 12–13. [Google Scholar] [CrossRef]
- Wilson, R.H.; Thieulin-Pardo, G.; Hartl, F.-U.; Hayer-Hartl, M. Improved recombinant expression and purification of functional plant Rubisco. FEBS Lett. 2019, 593, 611–621. [Google Scholar] [CrossRef] [PubMed]
- Cai, Z.; Liu, G.; Zhang, J.; Li, Y. Development of an activity-directed selection system enabled significant improvement of the carboxylation efficiency of Rubisco. Prot. Cell 2014, 5, 552–562. [Google Scholar] [CrossRef]

| Amino Acid Mutation | aTrait Selected | Rubisco (Solubility) | kcatC | KC | KCair | kcatC/KC | kcatC/KCair | SC/O | b Selection Host | Reference | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| SeRubisco (Synechococcus sp. PCC 6301)—Expressed in E. coli at ~1% (w/w) of the Cell Soluble Protein. | |||||||||||
| F140I | solubility | c WT | c ↑ 46% | ↓ 48% | - | ↑ 288% | - | ↓ 9% | MM1 | [27] | |
| ↑ 4.9-fold | ↓ 12% | - | ↓ 16% | - | ↑ 6% | WT | RDE2 | [22] | |||
| V189I | d solubility | WT | ↑ 8% | ↓ 35% | - | ↑ 71% | - | ↓ 12% | MM1 | [27] | |
| ↑ 4.8-fold | ↓ 16% | - | ↓ 26% | - | ↑ 14% | ↓ 15% | RDE2 | [22] | |||
| - | ↓ 25% | ↓ 44% | ↓ 41% | ↑ 32% | ↑ 8% | - | R. capsulatus | [28] | |||
| M262T | d solubility | ↑ 6-fold | ↑ 13% | ↓ 13% | - | ↑ 29% | - | WT | RDE(K12) | [29] | |
| ↑ 4-fold | - | - | - | - | - | - | MM1 | [22] | |||
| - | WT | ↓ 23% | ↓ 15% | ↑ 23% | ↑ 18% | - | R. capsulatus | [28] | |||
| F345I | solubility | ↑ 8-fold | ↓ 14% | ↑ 4% | ↑ 10% | ↓ 17% | ↓ 22% | ↓ 13% | MM1 | [30] | |
| ↑ 6-fold | ↓ 15% | ↑ 10% | - | ↓ 21% | - | ↓7% | MM1 | [27] | |||
| ↑ 11.1-fold | ↓ 16% | - | ↑ 13% | - | ↓ 28% | ↓ 8% | RDE2 | [22] | |||
| F345L | solubility | ↑ 7-fold | ↓ 17% | ↑ 22% | - | ↓ 32% | - | ↓4% | MM1 | [30] | |
| F345V | solubility | - | ↓ 25% | ↑ 38% | - | ↓ 45% | - | ↓ 45% | R. capsulatus | [31] | |
| Te-Rubisco (Thermosynechococcus elongatus BP-1)—Expressed in E. coli at ~6% (w/w) of the Soluble Protein. | |||||||||||
| F345I | solubility | ↑ 2.2-fold | ↓ 27% | - | WT | - | ↓ 24% | ↓ 10% | RDE2 | [22] | |
| P415A | solubility | ↑ 1.6-fold | ↑ 5% | - | WT | - | WT | WT | RDE2 | [22] | |
| P415A | F345I | solubility/kinetic | ↑ 2.7-fold | ↓ 27% | - | WT | - | ↓ 24% | ↓ 8% | RDE2 | [22] |
| P415A | V98MS | solubility/kinetic | ↑ 3.1-fold | ↑ 44% | - | ↓ 13% | - | ↑ 44% | ↑ 6% | RDE2 | [22] |
| P415A | A48VS | kinetic | ↑ 1.5-fold | ↑ 33% | - | WT | - | ↑ 35% | ↓ 10% | RDE2 | [22] |
| P415A | H37LS | kinetic | ↑ 1.7-fold | ↑ 27% | - | WT | - | ↑ 28% | ↓ 7% | RDE2 | [22] |
| P415A | Y36NS/G112DS | kinetic | ↑ 1.5-fold | ↑ 41% | - | ↓ 12% | - | ↑ 60% | ↓ 10% | RDE2 | [22] |
| P415A | L74M/D397N | solubility/kinetic | ↑ 1.9-fold | ↑ 24% | - | ↓ 14% | - | ↑ 45% | = WT | RDE2 | [22] |
| RsRubisco Isoform | RbcL Residue Number | Rubisco | KC | kcatC | KcatC/KC | SC/O | KO | kcatO | kcatO/KO | |||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 11 | 83 | 252 | 345 | a (% CSP) | (μM) | (s−1) | (mM.s−1) | mol·mol−1 | (μM) | (s−1) | (mM·s−1) | |
| WT | Val | Lys | Arg | Tyr | 4.9 ± 0.4 a | 60 ± 1 a | 3.7 ± 0.2 a | 60 ± 1 | 58 ± 1 a | 1724 ± 280 | 1.8 | 1.0 |
| YZ1 | Ile | Gln | Leu | Phe | 2.9 ± 0.1 b | 66 ± 1 a | 4.7 ± 0.1c | 70 ± 1 | 57 ± 1 a | 1296 ± 207 | 1.6 | 1.3 |
| YZ1a | Val | Gln | Leu | Phe | 3.3 ± 0.4 b | 66 ± 1 a | 4.5 ± 0.1c | 68 ± 2 | - | - | - | - |
| YZ1b | Ile | Gln | Leu | Tyr | 3.0 ± 0.1 b | 61 ± 1 a | 3.8 ± 0.1 a | 63 ± 1 | - | - | - | - |
| YZ1c | Val | Gln | Leu | Tyr | 3.6 ± 0.a b | 63 ± 1 a | 3.9 ± 0.1 ab | 63 ± 1 | - | - | - | - |
| YZ1d | Ile | Gln | Arg | Phe | 4.6 ± 0.1 a | 62 ± 1 a | 4.6 ± 0.1c | 76 ± 1 | - | - | - | - |
| YZ1e | Ile | Lys | Leu | Phe | 4.5 ± 0.3 a | 63 ± 1 a | 4.3 ± 0.1bc | 70 ± 3 | - | - | - | - |
| YZ1f | Val | Gln | Arg | Tyr | 5.1 ± 0.4 a | 61 ± 2 a | 3.9 ± 0.2 ab | 65 ± 1 | 57 ± 1 a | 1220 ± 199 | 1.4 | 1.1 |
| YZ1g | Val | Lys | Leu | Tyr | 5.3 ± 0.2 a | 62 ± 4 a | 3.5 ± 0.1 a | 57 ± 3 | - | - | - | - |
| YZ1h | Ile | Lys | Arg | Phe | 4.6 ± 0.1 a | 61 ± 1 a | 4.4 ± 0.1a | 73 ± 1 | - | - | - | - |
| YZ1i | Val | Lys | Arg | Phe | 4.7 ± 0.2 a | 64 ± 1 a | 4.6 ± 0.1a | 71 ± 2 | 59 ± 1 a | 1551 ± 293 | 1.9 | 1.2 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhou, Y.; Whitney, S. Directed Evolution of an Improved Rubisco; In Vitro Analyses to Decipher Fact from Fiction. Int. J. Mol. Sci. 2019, 20, 5019. https://doi.org/10.3390/ijms20205019
Zhou Y, Whitney S. Directed Evolution of an Improved Rubisco; In Vitro Analyses to Decipher Fact from Fiction. International Journal of Molecular Sciences. 2019; 20(20):5019. https://doi.org/10.3390/ijms20205019
Chicago/Turabian StyleZhou, Yu, and Spencer Whitney. 2019. "Directed Evolution of an Improved Rubisco; In Vitro Analyses to Decipher Fact from Fiction" International Journal of Molecular Sciences 20, no. 20: 5019. https://doi.org/10.3390/ijms20205019
APA StyleZhou, Y., & Whitney, S. (2019). Directed Evolution of an Improved Rubisco; In Vitro Analyses to Decipher Fact from Fiction. International Journal of Molecular Sciences, 20(20), 5019. https://doi.org/10.3390/ijms20205019






