Physiological and Transcriptomic Changes during the Early Phases of Adventitious Root Formation in Mulberry Stem Hardwood Cuttings
Abstract
1. Introduction
2. Results
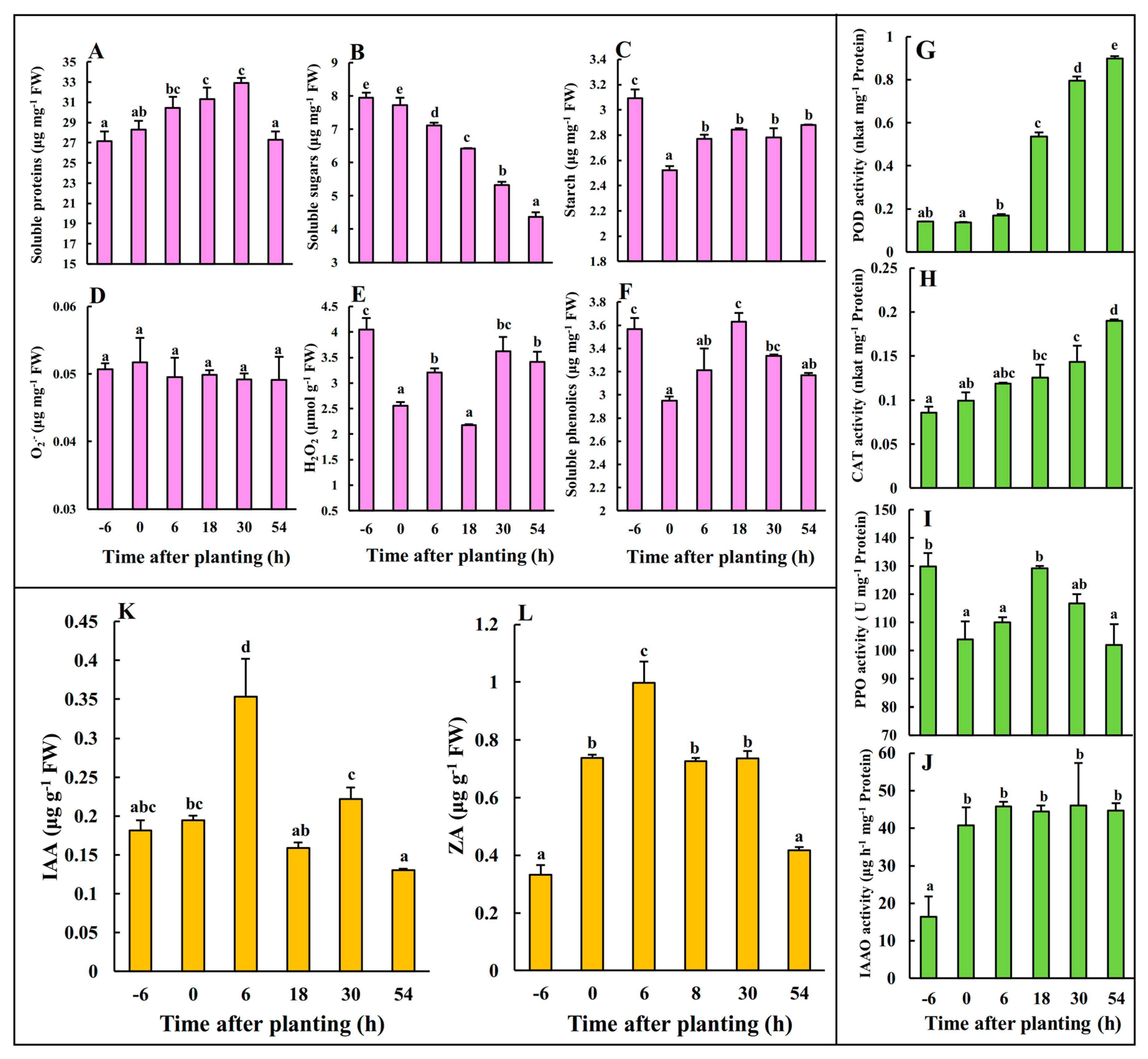
2.1. Physiological Changes during Early AR Phases
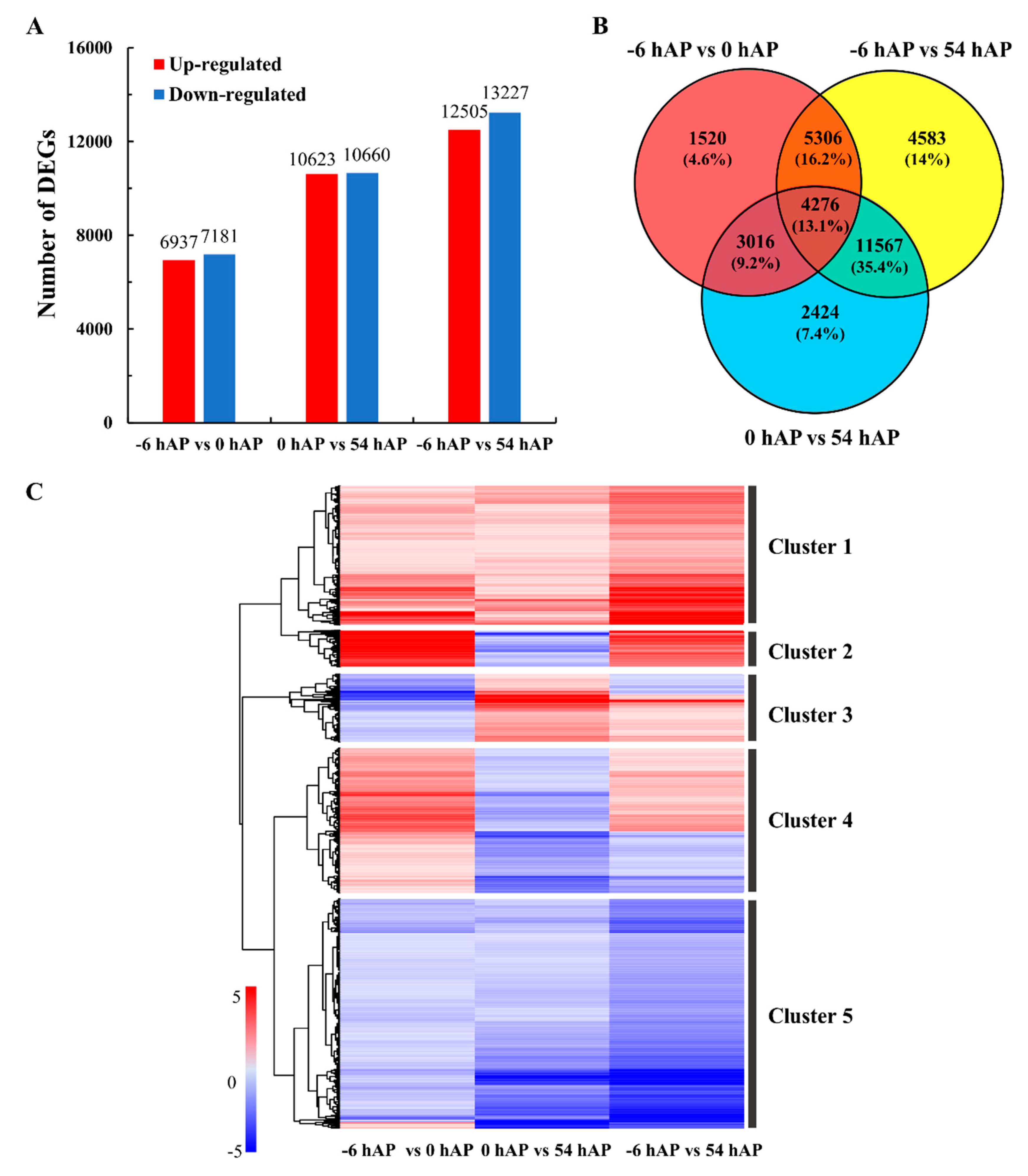
2.2. Comparative Transcriptomes Uncover Sets of Important Common Genes during Early AR Phases
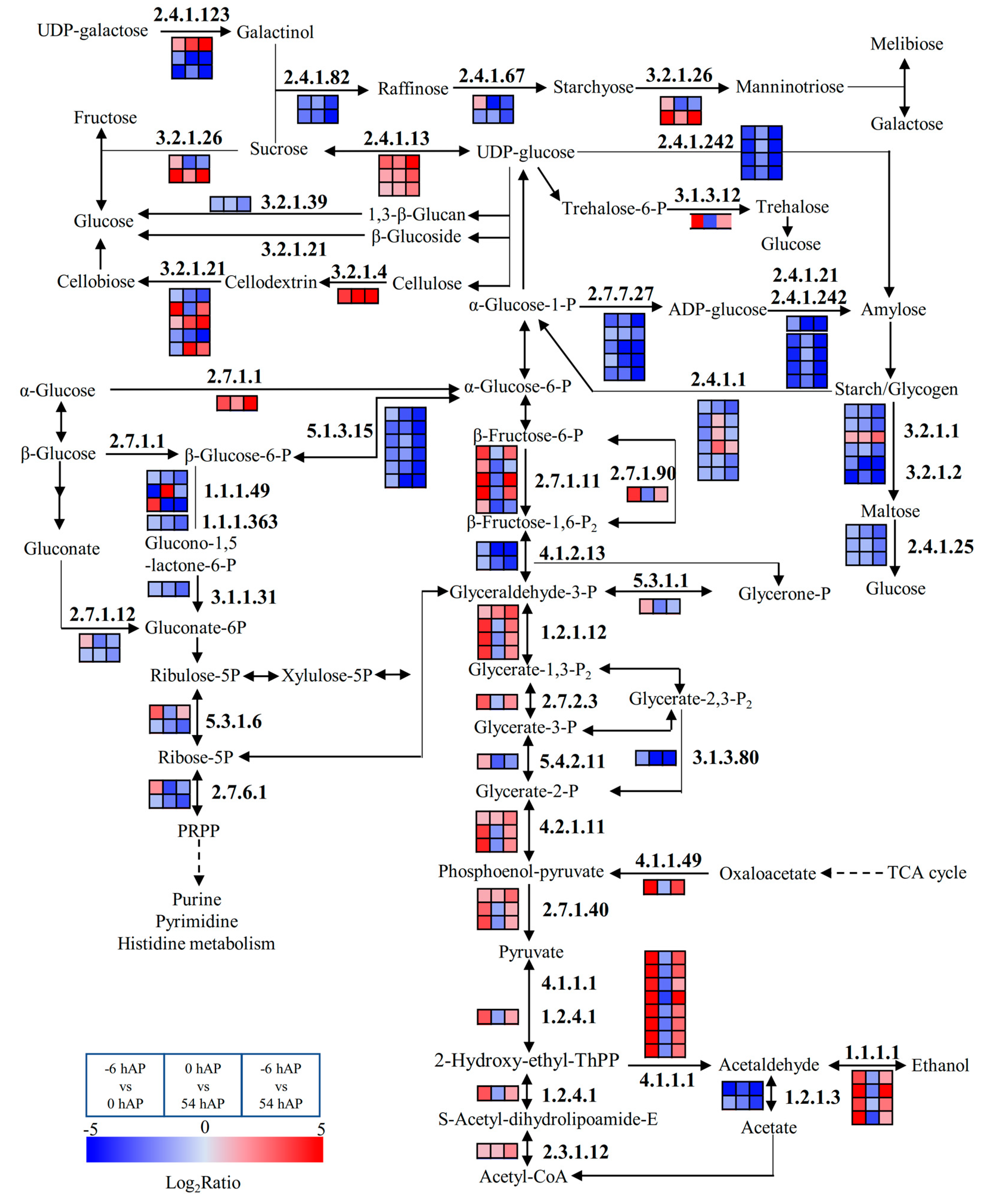
2.3. Several Pathways were Significantly Modified during Early AR Phases
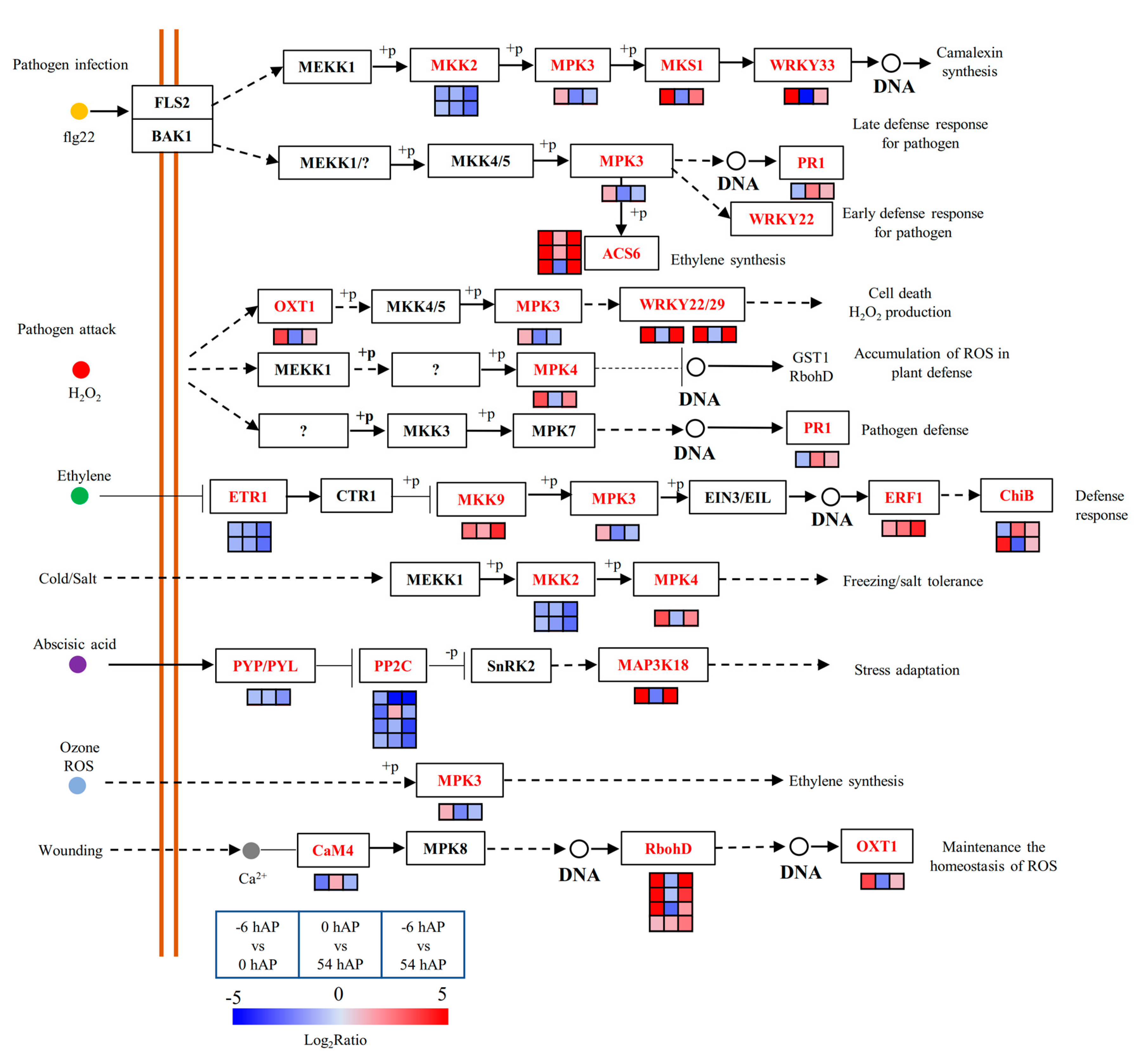
2.4. TFs and Hormone-Related Genes Play Essential Roles during Early AR Phases
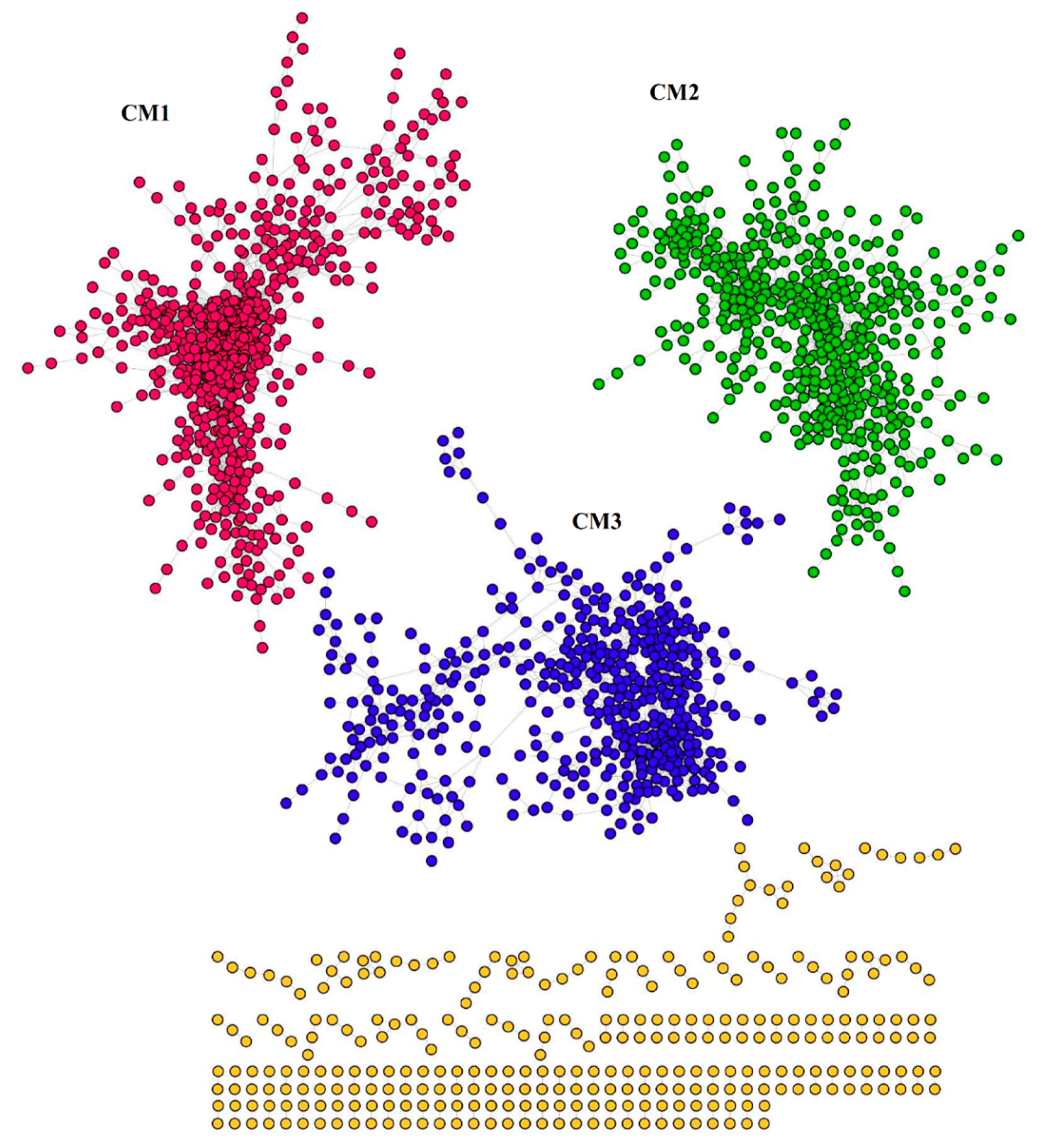
2.5. The Common DEGs were Highly Co-Expressed
3. Discussion
4. Materials and Methods
4.1. Cutting Cultivation and Harvest
4.2. Analysis of Rooting-Related Metabolites
4.3. Assay of Rooting-Related Enzyme Activities
4.4. Quantification of Phytohormones
4.5. Library Preparation and Sequencing
4.6. Raw Data Analysis, Annotation, Mapping and Selection of Differential Expressed Genes (DEGs)
4.7. Bioinformation Analysis
4.8. Validation of RNA-Seq by qRT-PCR
4.9. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Monteuuis, O. In vitro grafting of woody species. Propag. Ornam. Plants 2012, 12, 11–24. [Google Scholar]
- Druege, U.; Franken, P.; Hajirezaei, M.R. Plant hormone homeostasis, signaling, and function during adventitious root formation in cuttings. Front. Plant Sci. 2016, 7, 381. [Google Scholar] [CrossRef] [PubMed]
- Steffens, B.; Rasmussen, A. The physiology of adventitious roots. Plant Physiol. 2016, 170, 603–617. [Google Scholar] [CrossRef] [PubMed]
- Oinam, G.; Yeung, E.; Kurepin, L.; Haslam, T.; Lopez-Villalobos, A. Adventitious root formation in ornamental plants: I. General overview and recent successes. Propag. Ornam. Plants 2011, 11, 78–90. [Google Scholar]
- Nakhooda, M.; Jain, S.M. A review of eucalyptus propagation and conservation. Propag. Ornam. Plants 2016, 16, 101–119. [Google Scholar]
- Sandhu, M.; Wani, S.H.; Jimenez, V.M. In vitro propagation of bamboo species through axillary shoot proliferation: A review. Plant Cell Tiss. Org. 2018, 132, 27–53. [Google Scholar] [CrossRef]
- De Klerk, G.J.; Van der Krieken, W.; de Jong, J.C. Review the formation of adventitious roots: New concepts, new possibilities. In Vitro Cell. Dev.-Pl. 1999, 35, 189–199. [Google Scholar] [CrossRef]
- Zhao, X.; Zheng, H.; Li, S.; Yang, C.; Jiang, J.; Liu, G. The rooting of poplar cuttings: A review. New Forest. 2014, 45, 21–34. [Google Scholar] [CrossRef]
- Damiano, C.; Padro, M.D.A.; Frattarelli, A. Propagation and establishment in vitro of myrtle (Myrtus communis L.), pomegranate (Punica granatum L.) and mulberry (Morus alba L.). Propag. Ornam. Plants 2008, 8, 3–8. [Google Scholar]
- Ragonezi, C.; Klimaszewska, K.; Castro, M.R.; Lima, M.; de Oliveira, P.; Zavattieri, M.A. Adventitious rooting of conifers: Influence of physical and chemical factors. Trees 2010, 24, 975–992. [Google Scholar] [CrossRef]
- Lee, Y.; Lee, D.E.; Lee, H.S.; Kim, S.K.; Lee, W.S.; Kim, S.H.; Kim, M.W. Influence of auxins, cytokinins, and nitrogen on production of rutin from callus and adventitious roots of the white mulberry tree (Morus alba L.). Plant Cell Tiss. Org. 2011, 105, 9–19. [Google Scholar] [CrossRef]
- Werner, T.; Motyka, V.; Laucou, V.; Smets, R.; Onckelen, H.V. Cytokinin-deficient transgenic arabidopsis plants show multiple developmental alterations indicating opposite functions of cytokinins in the regulation of shoot and root meristem activity. Plant Cell 2003, 15, 2532–2550. [Google Scholar] [CrossRef] [PubMed]
- Gutierrez, L.; Mongelard, G.; Flokova, K.; Pacurar, D.I.; Novak, O.; Staswick, P.; Kowalczyk, M.; Pacurar, M.; Demailly, H.; Geiss, G.; et al. Auxin controls arabidopsis adventitious root initiation by regulating jasmonic acid homeostasis. Plant Cell 2012, 24, 2515–2527. [Google Scholar] [CrossRef] [PubMed]
- Pacurar, D.I.; Perrone, I.; Bellini, C. Auxin is a central player in the hormone cross-talks that control adventitious rooting. Physiol. Plantarum 2014, 151, 83–96. [Google Scholar] [CrossRef] [PubMed]
- Stevens, M.E.; Woeste, K.E.; Pijut, P.M. Localized gene expression changes during adventitious root formation in black walnut (Juglans nigra L.). Tree Physiol. 2018, 38, 877–894. [Google Scholar] [CrossRef] [PubMed]
- Li, K.; Liu, Z.; Xing, L.; Wei, Y.; Mao, J.; Meng, Y.; Bao, L.; Han, M.; Zhao, C.; Zhang, D. miRNAs associated with auxin signaling, stress response, and cellular activities mediate adventitious root formation in apple rootstocks. Plant Physiol. Biochem. 2019, 139, 66–81. [Google Scholar] [CrossRef] [PubMed]
- Minibayeva, F.; Kolesnikov, O.; Chasov, A.; Beckett, R.P.; Luthje, S.; Vylegzhanina, N.; Buck, F.; Bottger, M. Wound-induced apoplastic peroxidase activities: Their roles in the production and detoxification of reactive oxygen species. Plant Cell Environ. 2009, 32, 497–508. [Google Scholar] [CrossRef] [PubMed]
- De Klerk, G.J.; Guan, H.Y.; Huisman, P.; Marinova, S. Effects of phenolic compounds on adventitious root formation and oxidative decarboxylation of applied indoleacetic acid in Malus ‘jork 9’. Plant Growth Regul. 2011, 63, 175–185. [Google Scholar] [CrossRef]
- Klopotek, Y.; Haensch, K.T.; Hause, B.; Hajirezaei, M.R.; Druege, U. Dark exposure of petunia cuttings strongly improves adventitious root formation and enhances carbohydrate availability during rooting in the light. J. Plant Physiol. 2010, 167, 547–554. [Google Scholar] [CrossRef]
- Iwase, A.; Ohme-Takagi, M.; Sugimoto, K. WIND1: A key molecular switch for plant cell dedifferentiation. Plant Signal. Behav. 2011, 6, 1943–1945. [Google Scholar] [CrossRef]
- Chen, X.; Cheng, J.; Chen, L.; Zhang, G.; Huang, H.; Zhang, Y.; Xu, L. Auxin-independent nac pathway acts in response to explant-specific wounding and promotes root tip emergence during de novo root organogenesis in Arabidopsis. Plant Physiol. 2016, 170, 2136–2145. [Google Scholar] [CrossRef]
- Liu, J.; Sheng, L.; Xu, Y.; Li, J.; Yang, Z.; Huang, H.; Xu, L. WOX 11 and 12 are involved in the first-step cell fate transition during de novo root organogenesis in Arabidopsis. Plant Cell 2014, 26, 1081–1093. [Google Scholar] [CrossRef]
- Lee, H.W.; Cho, C.; Pandey, S.K.; Park, Y.; Kim, M.-J.; Kim, J. LBD16 and LBD18 acting downstream of ARF7 and ARF19 are involved in adventitious root formation in Arabidopsis. BMC Plant Biol. 2019, 19, 46. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Yu, J.; Ge, Y.C.; Qin, P.; Xu, L. Pivotal role of LBD16 in root and root-like organ initiation. Cell. Mol. Life Sci. 2018, 75, 3329–3338. [Google Scholar] [CrossRef]
- Gutierrez, L.; Bussell, J.D.; Păcurar, D.I.; Schwambach, J.; Păcurar, M.; Bellini, C. Phenotypic plasticity of adventitious rooting in Arabidopsis is controlled by complex regulation of AUXIN RESPONSE FACTOR transcripts and microRNA abundance. Plant Cell 2009, 21, 3119–3132. [Google Scholar] [CrossRef] [PubMed]
- Lewis, D.R.; Negi, S.; Sukumar, P.; Muday, G.K. Ethylene inhibits lateral root development, increases IAA transport and expression of PIN3 and PIN7 auxin efflux carriers. Development 2011, 138, 3485–3495. [Google Scholar] [CrossRef] [PubMed]
- Trupiano, D.; Yordanov, Y.; Regan, S.; Meilan, R.; Tschaplinski, T.; Scippa, G.S.; Busov, V. Identification, characterization of an AP2/ERF transcription factor that promotes adventitious, lateral root formation in Populus. Planta 2013, 238, 271–282. [Google Scholar] [CrossRef]
- Voss, U.; Wilson, M.H.; Kenobi, K.; Gould, P.D.; Robertson, F.C.; Peer, W.A.; Lucas, M.; Swarup, K.; Casimiro, I.; Holman, T.J.; et al. The circadian clock rephases during lateral root organ initiation in Arabidopsis thaliana. Nat. Commun. 2015, 6, 7641. [Google Scholar] [CrossRef]
- Bellini, C.; Pacurar, D.I.; Perrone, I. Adventitious roots and lateral roots: Similarities and differences. Annu. Rev. Plant Biol. 2014, 65, 639–666. [Google Scholar] [CrossRef]
- Cao, X.; Shen, Q.; Shang, C.; Yang, H.; Liu, L.; Cheng, J. Determinants of shoot biomass production in mulberry: Combined selection with leaf morphological and physiological traits. Plants 2019, 8, 118. [Google Scholar] [CrossRef]
- Cheng, J.; Huang, P.; Du, W.; Zhang, G.; Fang, R.; Liu, Y.; Chai, J. A Method of Breeding Seedlings Using Multilayer Heating Boxes. Patent No. CN201510061596.0, 10 June 2015. [Google Scholar]
- Du, W.; Ban, Y.; Nie, H.; Tang, Z.; Du, X.; Cheng, J. A comparative transcriptome analysis leads to new insights into the molecular events governing root formation in mulberry softwood cuttings. Plant Mol. Biol. Rep. 2016, 34, 365–373. [Google Scholar] [CrossRef]
- Cao, X.; Du, W.; Shang, C.; Shen, Q.; Liu, L.; Cheng, J. Comparative transcriptome reveals circadian and hormonal control of adventitious rooting in mulberry hardwood cuttings. Acta Physiol. Plant. 2018, 40, 197. [Google Scholar] [CrossRef]
- Du, X.L.; Zhang, X.F.; Nie, H.; Liu, M.L.; Cheng, J.L. Transcript profiling analysis reveals crucial genes regulating main metabolism during adventitious root formation in cuttings of Morus alba L. Plant Growth Regul. 2016, 79, 251–262. [Google Scholar] [CrossRef]
- Tang, Z.; Du, W.; Du, X.; Ban, Y.; Cheng, J. iTRAQ protein profiling of adventitious root formation in mulberry hardwood cuttings. J. Plant Growth Regul. 2016, 35, 618–631. [Google Scholar] [CrossRef]
- Du, X.L.; Cao, X.; Yin, C.R.; Tang, Z.; Du, W.; Ban, Y.Y.; Cheng, J.L. Comprehensive analysis of R2R3-MYB genes during adventitious root formation in cuttings of Morus alba. J. Plant Growth Regul. 2017, 36, 290–299. [Google Scholar] [CrossRef]
- Liu, M.; Sun, L.; Zhang, X.; Nie, H.; Cheng, J. Physiological and biochemical analysis of mulberry hardwood cuttings rooting process. Chin. Seric. 2011, 32, 9–14. [Google Scholar]
- Naija, S.; Elloumi, N.; Jbir, N.; Ammar, S.; Kevers, C. Anatomical and biochemical changes during adventitious rooting of apple rootstocks MM 106 cultured in vitro. CR Biol. 2008, 331, 518–525. [Google Scholar] [CrossRef] [PubMed]
- Da costa, C.T.; de Almeida, M.R.; Ruedell, C.M.; Schwambach, J.; Maraschin, F.S.; Fett-Neto, A.G. When stress and development go hand in hand: Main hormonal controls of adventitious rooting in cuttings. Front. Plant Sci. 2013, 4, 133. [Google Scholar] [CrossRef]
- Ribeiro, C.L.; Silva, C.M.; Drost, D.R.; Novaes, E.; Novaes, C.; Dervinis, C.; Kirst, M. Integration of genetic, genomic and transcriptomic information identifies putative regulators of adventitious root formation in Populus. BMC Plant Biol. 2016, 16, 66. [Google Scholar] [CrossRef]
- Agullo-Anton, M.A.; Ferrandez-Ayela, A.; Fernandez-Garcia, N.; Nicolas, C.; Albacete, A.; Perez-Alfocea, F.; Sanchez-Bravo, J.; Perez-Perez, J.M.; Acosta, M. Early steps of adventitious rooting: Morphology, hormonal profiling and carbohydrate turnover in carnation stem cuttings. Physiol. Plantarum 2014, 150, 446–462. [Google Scholar] [CrossRef]
- Ren, H.; Hu, H.; Luo, X.; Zhang, C.; Li, X.; Li, P.; Li, W.; Khawar, A.; Sun, X.; Ren, Z.; et al. Dynamic changes of phytohormone signaling in the base of Taxus media stem cuttings during adventitious root formation. Sci. Hortic. 2019, 246, 338–346. [Google Scholar] [CrossRef]
- Li, S.W.; Xue, L. The interaction between H2O2 and NO, Ca2+, cGMP, and MAPKs during adventitious rooting in mung bean seedlings. In Vitro Cell. Dev. Biol. Plant 2010, 46, 142–148. [Google Scholar] [CrossRef]
- Druege, U.; Zerche, S.; Kadner, R.; Ernst, M. Relation between nitrogen status, carbohydrate distribution and subsequent rooting of Chrysanthemum cuttings as affected by pre-harvest nitrogen supply and cold-storage. Ann. Bot. 2000, 85, 687–701. [Google Scholar] [CrossRef]
- Druege, U.; Zerche, S.; Kadner, R. Nitrogen- and storage-affected carbohydrate partitioning in high-light-adapted Pelargonium cuttings in relation to survival and adventitious root formation under low light. Ann. Bot. 2004, 94, 831–842. [Google Scholar] [CrossRef] [PubMed]
- Druege, U.; Hilo, A.; Perez-Perez, J.M.; Klopotek, Y.; Acosta, M.; Shahinnia, F.; Zerche, S.; Franken, P.; Hajirezaei, M.R. Molecular and physiological control of adventitious rooting in cuttings: Phytohormone action meets resource allocation. Ann. Bot. 2019, 123, 929–949. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Hua, J.; Yin, Y.; Gu, C.; Yu, C.; Shi, Q.; Guo, J.; Xuan, L.; Yu, F. An integrated transcriptome and proteome analysis reveals putative regulators of adventitious root formation in Taxodium ‘zhongshanshan’. Int. J. Mol. Sci. 2019, 20, 1225. [Google Scholar] [CrossRef] [PubMed]
- Mu, X.; Luo, J. Evolutionary analyses of NIN-like proteins in plants and their roles in nitrate signaling. Cell. Mol. Life Sci. 2019, 1–12. [Google Scholar] [CrossRef]
- Luo, J.; Li, H.; Liu, T.; Polle, A.; Peng, C.; Luo, Z.B. Nitrogen metabolism of two contrasting poplar species during acclimation to limiting nitrogen availability. J. Exp. Bot. 2013, 64, 4207–4224. [Google Scholar] [CrossRef]
- Luo, J.; Qin, J.; He, F.; Li, H.; Liu, T.; Polle, A.; Peng, C.; Luo, Z.B. Net fluxes of ammonium and nitrate in association with H+ fluxes in fine roots of Populus popularis. Planta 2013, 237, 919–931. [Google Scholar] [CrossRef]
- Benkova, E.; Michniewicz, M.; Sauer, M.; Teichmann, T.; Seifertova, D.; Jurgens, G.; Friml, J. Local efflux-dependent auxin gradients as a common module for plant organ formation. Cell 2003, 115, 591–602. [Google Scholar] [CrossRef]
- Ahkami, A.H.; Melzer, M.; Ghaffari, M.R.; Pollmann, S.; Ghorbani Javid, M.; Shahinnia, F.; Hajirezaei, M.R.; Druege, U. Distribution of indole-3-acetic acid in petunia hybrida shoot tip cuttings and relationship between auxin transport, carbohydrate metabolism and adventitious root formation. Planta 2013, 238, 499–517. [Google Scholar] [CrossRef] [PubMed]
- Rasmussen, A.; Hosseini, S.A.; Hajirezaei, M.-R.; Druege, U.; Geelen, D. Adventitious rooting declines with the vegetative to reproductive switch and involves a changed auxin homeostasis. J. Exp. Bot. 2015, 66, 1437–1452. [Google Scholar] [CrossRef] [PubMed]
- Della Rovere, F.; Fattorini, L.; D’Angeli, S.; Veloccia, A.; Falasca, G.; Altamura, M.M. Auxin and cytokinin control formation of the quiescent centre in the adventitious root apex of Arabidopsis. Ann. Bot. 2013, 112, 1395–1407. [Google Scholar] [CrossRef] [PubMed]
- Luo, J.; Zhou, J.J.; Zhang, J.Z. Aux/IAA gene family in plants: Molecular structure, regulation, and function. Int. J. Mol. Sci. 2018, 19, 259. [Google Scholar] [CrossRef] [PubMed]
- Garrido, G.; Ramón Guerrero, J.; Angel Cano, E.; Acosta, M.; Sánchez-Bravo, J. Origin and basipetal transport of the IAA responsible for rooting of carnation cuttings. Physiol. Plantarum 2002, 114, 303–312. [Google Scholar] [CrossRef]
- Sukumar, P.; Maloney, G.S.; Muday, G.K. Localized induction of the ATP-Binding Cassette B19 auxin transporter enhances adventitious root formation in Arabidopsis. Plant Physiol. 2013, 162, 1392–1405. [Google Scholar] [CrossRef] [PubMed]
- Balzan, S.; Johal, G.S.; Carraro, N. The role of auxin transporters in monocots development. Front. Plant Sci. 2014, 5, 393. [Google Scholar] [CrossRef]
- Armengot, L.; Marquès-Bueno, M.M.; Jaillais, Y. Regulation of polar auxin transport by protein and lipid kinases. J. Exp. Bot. 2016, 67, 4015–4037. [Google Scholar] [CrossRef]
- Zhou, J.J.; Luo, J. The PIN-formed auxin efflux carriers in plants. Int. J. Mol. Sci. 2018, 19, 2759. [Google Scholar] [CrossRef]
- Simon, S.; Skupa, P.; Viaene, T.; Zwiewka, M.; Tejos, R.; Klima, P.; Carna, M.; Rolcik, J.; De Rycke, R.; Moreno, I.; et al. PIN6 auxin transporter at endoplasmic reticulum and plasma membrane mediates auxin homeostasis and organogenesis in Arabidopsis. New Phytol. 2016, 211, 65–74. [Google Scholar] [CrossRef]
- Mao, J.; Zhang, D.; Meng, Y.; Li, K.; Wang, H.; Han, M. Inhibition of adventitious root development in apple rootstocks by cytokinin is based on its suppression of adventitious root primordia formation. Physiol. Plantarum 2019, 166, 663–676. [Google Scholar] [CrossRef]
- Jones, B.; Gunnerås, S.A.; Petersson, S.V.; Tarkowski, P.; Graham, N.; May, S.; Dolezal, K.; Sandberg, G.; Ljung, K. Cytokinin regulation of auxin synthesis in Arabidopsis involves a homeostatic feedback loop regulated via auxin and cytokinin signal transduction. Plant Cell 2010, 22, 2956–2969. [Google Scholar] [CrossRef]
- Aloni, R.; Aloni, E.; Langhans, M.; Ullrich, C.I. Role of cytokinin and auxin in shaping root architecture: Regulating vascular differentiation, lateral root initiation, root apical dominance and root gravitropism. Ann. Bot. 2006, 97, 883–893. [Google Scholar] [CrossRef]
- Ahkami, A.H.; Lischewski, S.; Haensch, K.-T.; Porfirova, S.; Hofmann, J.; Rolletschek, H.; Melzer, M.; Franken, P.; Hause, B.; Druege, U.; et al. Molecular physiology of adventitious root formation in Petunia hybrida cuttings: Involvement of wound response and primary metabolism. New Phytol. 2009, 181, 613–625. [Google Scholar] [CrossRef]
- Ding, Z.; Friml, J. Auxin regulates distal stem cell differentiation in Arabidopsis roots. Proc. Natl. Acad. Sci. USA 2010, 107, 12046–12051. [Google Scholar] [CrossRef]
- Vielba, J.M.; Díaz-Sala, C.; Ferro, E.; Rico, S.; Lamprecht, M.; Abarca, D.; Ballester, A.; Sánchez, C. CsSCL1 is differentially regulated upon maturation in chestnut microshoots and is specifically expressed in rooting-competent cells. Tree Physiol. 2011, 31, 1152–1160. [Google Scholar] [CrossRef]
- Che, P.; Lall, S.; Nettleton, D.; Howell, S.H. Gene expression programs during shoot, root, and callus development in Arabidopsis tissue culture. Plant Physiol. 2006, 141, 620–637. [Google Scholar] [CrossRef]
- Raya-González, J.; Oropeza-Aburto, A.; López-Bucio, J.S.; Guevara-García, Á.A.; de Veylder, L.; López-Bucio, J.; Herrera-Estrella, L. MEDIATOR18 influences Arabidopsis root architecture, represses auxin signaling and is a critical factor for cell viability in root meristems. Plant J. 2018, 96, 895–909. [Google Scholar] [CrossRef] [PubMed]
- Zhang, K.; Zhao, L.; Yang, X.; Li, M.; Sun, J.; Wang, K.; Li, Y.; Zheng, Y.; Yao, Y.; Li, W. GmRAV1 regulates regeneration of roots and adventitious buds by the cytokinin signaling pathway in Arabidopsis and soybean. Physiol. Plantarum 2019, 165, 814–829. [Google Scholar] [CrossRef]
- Asahina, M.; Azuma, K.; Pitaksaringkarn, W.; Yamazaki, T.; Mitsuda, N.; Ohme-Takagi, M.; Yamaguchi, S.; Kamiya, Y.; Okada, K.; Nishimura, T.; et al. Spatially selective hormonal control of RAP2.6L and ANAC071 transcription factors involved in tissue reunion in Arabidopsis. Proc. Natl. Acad. Sci. USA 2011, 108, 16128–16132. [Google Scholar] [CrossRef]
- Deveaux, Y.; Toffano-Nioche, C.; Claisse, G.; Thareau, V.; Morin, H.; Laufs, P.; Moreau, H.; Kreis, M.; Lecharny, A. Genes of the most conserved WOX clade in plants affect root and flower development in Arabidopsis. BMC Evol. Biol. 2008, 8, 291. [Google Scholar] [CrossRef] [PubMed]
- Kong, D.; Hao, Y.; Cui, H. The WUSCHEL related homeobox protein WOX7 regulates the sugar response of lateral root development in Arabidopsis thaliana. Mol. Plant 2016, 9, 261–270. [Google Scholar] [CrossRef] [PubMed]
- Luo, J.; Xia, W.; Cao, P.; Xiao, Z.; Zhang, Y.; Liu, M.; Zhan, C.; Wang, N. Integrated transcriptome analysis reveals plant hormones jasmonic acid and salicylic acid coordinate growth and defense responses upon fungal infection in poplar. Biomolecules 2019, 9, 12. [Google Scholar] [CrossRef] [PubMed]
- Luo, J.; Liang, Z.; Wu, M.; Mei, L. Genome-wide identification of BOR genes in poplar and their roles in response to various environmental stimuli. Environ. Exp. Bot. 2019, 164, 101–113. [Google Scholar] [CrossRef]
- Gibon, Y.; Bläsing, O.E.; Palacios-Rojas, N.; Pankovic, D.; Hendriks, J.H.; Fisahn, J.; Höhne, M.; Günther, M.; Stitt, M. Adjustment of diurnal starch turnover to short days: Depletion of sugar during the night leads to a temporary inhibition of carbohydrate utilization, accumulation of sugars and post-translational activation of ADP-glucose pyrophosphorylase in the following light period. Plant J. 2004, 39, 847–862. [Google Scholar] [PubMed]
- Graf, A.; Smith, A.M. Starch and the clock: The dark side of plant productivity. Trends Plant Sci. 2011, 16, 169–175. [Google Scholar] [CrossRef] [PubMed]
- Izumi, M. Roles of the clock in controlling starch metabolism. Plant Physiol. 2019, 179, 1441–1443. [Google Scholar] [CrossRef]
- Yemm, E.W.; Willis, A.J. The estimation of carbohydrates in plant extracts by anthrone. Biochem. J. 1954, 57, 508–514. [Google Scholar] [CrossRef]
- Luo, Z.B.; Calfapietra, C.; Scarascia-Mugnozza, G.; Liberloo, M.; Polle, A. Carbon-based secondary metabolites and internal nitrogen pools in Populus nigra under free air CO2 enrichment (FACE) and nitrogen fertilisation. Plant Soil 2008, 304, 45–57. [Google Scholar] [CrossRef]
- Cao, X.; Jia, J.; Zhang, C.; Li, H.; Liu, T.; Jiang, X.; Polle, A.; Peng, C.; Luo, Z.B. Anatomical, physiological and transcriptional responses of two contrasting poplar genotypes to drought and re-watering. Physiol. Plantarum 2014, 151, 480–494. [Google Scholar] [CrossRef]
- Jia, J.; Li, S.; Cao, X.; Li, H.; Shi, W.; Polle, A.; Liu, T.X.; Peng, C.; Luo, Z.B. Physiological and transcriptional regulation in poplar roots and leaves during acclimation to high temperature and drought. Physiol. Plantarum 2016, 157, 38–53. [Google Scholar] [CrossRef] [PubMed]
- Concellón, A.a.; Añón, M.a.C.; Chaves, A.R. Characterization and changes in polyphenol oxidase from eggplant fruit (Solanum melongena L.) during storage at low temperature. Food Chem. 2004, 88, 17–24. [Google Scholar] [CrossRef]
- Güneș, T. Peroxidase and IAA oxidase activities during rooting in cuttings of three poplar species. Turk. J. Bot. 2000, 24, 97–101. [Google Scholar]
- Loffler, B.M.; Kunze, H. Refinement of the coomassie brilliant blue g assay for quantitative protein determination. Anal. Biochem. 1989, 177, 100–102. [Google Scholar] [CrossRef]
- Shi, W.G.; Li, H.; Liu, T.X.; Polle, A.; Peng, C.H.; Luo, Z.B. Exogenous abscisic acid alleviates zinc uptake and accumulation in Populus × canescens exposed to excess zinc. Plant Cell Environ. 2015, 38, 207–223. [Google Scholar] [CrossRef] [PubMed]
- Luo, J.; Zhou, J.J.; Masclaux-Daubresse, C.; Wang, N.; Wang, H.; Zheng, B. Morphological and physiological responses to contrasting nitrogen regimes in Populus cathayana is linked to resources allocation and carbon/nitrogen partition. Environ. Exp. Bot. 2019, 162, 247–255. [Google Scholar] [CrossRef]
- Luo, J.; Zhou, J.; Li, H.; Shi, W.; Polle, A.; Lu, M.; Sun, X.; Luo, Z.B. Global poplar root and leaf transcriptomes reveal links between growth and stress responses under nitrogen starvation and excess. Tree Physiol. 2015, 35, 1283–1302. [Google Scholar] [CrossRef] [PubMed]
- Langmead, B.; Trapnell, C.; Pop, M.; Salzberg, S.L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009, 10, 1–10. [Google Scholar] [CrossRef]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q.; et al. Full-length transcriptome assembly from RNA-seq data without a reference genome. Nat. Biotechnol. 2011, 29, 644–652. [Google Scholar] [CrossRef]
- Pertea, G.; Huang, X.; Liang, F.; Antonescu, V.; Sultana, R.; Karamycheva, S.; Lee, Y.; White, J.; Cheung, F.; Parvizi, B.; et al. TIGR gene indices clustering tools (TGICL): A software system for fast clustering of large est datasets. Bioinformatics 2003, 19, 651–652. [Google Scholar] [CrossRef]
- Luo, J.; Shi, W.G.; Li, H.G.; Janz, D.; Luo, Z.B. The conserved salt-responsive genes in the roots of Populus × canescens and Arabidopsis thaliana. Environ. Exp. Bot. 2016, 129, 48–56. [Google Scholar] [CrossRef]
- Oliveros, J.C. An Interactive Tool for Comparing Lists with Venn’s Diagrams 2007–2015. Available online: https://bioinfogp.cnb.csic.es/tools/venny/index.html (accessed on 20 June 2019).
- Kolde, R.; Kolde, M.R. Package ‘pheatmap’ 2015. Available online: https://cran.r-project.org/web/packages/pheatmap/index.html (accessed on 20 June 2019).
- Yu, G.C.; Wang, L.G.; Han, Y.Y.; He, Q.Y. Clusterprofiler: An R package for comparing biological themes among gene clusters. Omics 2012, 16, 284–287. [Google Scholar] [CrossRef] [PubMed]
- Luo, W.; Brouwer, C. Pathview: An R/Bioconductor package for pathway-based data integration and visualization. Bioinformatics 2013, 29, 1830–1831. [Google Scholar] [CrossRef]
- Zhu, D.; Hero, A.O.; Qin, Z.S.; Swaroop, A. High throughput screening of co-expressed gene pairs with controlled false discovery rate (FDR) and minimum acceptable strength (MAS). J. Comput. Biol. 2005, 12, 1029–1045. [Google Scholar] [CrossRef] [PubMed]
- Luo, J.; Zhou, J.J. Growth performance, photosynthesis, and root characteristics are associated with nitrogen use efficiency in six poplar species. Environ. Exp. Bot. 2019, 164, 40–51. [Google Scholar] [CrossRef]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Cao, X.; Jia, J.B.; Li, H.; Li, M.C.; Luo, J.; Liang, Z.S.; Liu, T.X.; Liu, W.G.; Peng, C.H.; Luo, Z.B. Photosynthesis, water use efficiency and stable carbon isotope composition are associated with anatomical properties of leaf and xylem in six poplar species. Plant Biol. 2012, 14, 612–620. [Google Scholar] [CrossRef]

© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shang, C.; Yang, H.; Ma, S.; Shen, Q.; Liu, L.; Hou, C.; Cao, X.; Cheng, J. Physiological and Transcriptomic Changes during the Early Phases of Adventitious Root Formation in Mulberry Stem Hardwood Cuttings. Int. J. Mol. Sci. 2019, 20, 3707. https://doi.org/10.3390/ijms20153707
Shang C, Yang H, Ma S, Shen Q, Liu L, Hou C, Cao X, Cheng J. Physiological and Transcriptomic Changes during the Early Phases of Adventitious Root Formation in Mulberry Stem Hardwood Cuttings. International Journal of Molecular Sciences. 2019; 20(15):3707. https://doi.org/10.3390/ijms20153707
Chicago/Turabian StyleShang, Chunqiong, Honglei Yang, Sang Ma, Qiudi Shen, Li Liu, Chengxiang Hou, Xu Cao, and Jialing Cheng. 2019. "Physiological and Transcriptomic Changes during the Early Phases of Adventitious Root Formation in Mulberry Stem Hardwood Cuttings" International Journal of Molecular Sciences 20, no. 15: 3707. https://doi.org/10.3390/ijms20153707
APA StyleShang, C., Yang, H., Ma, S., Shen, Q., Liu, L., Hou, C., Cao, X., & Cheng, J. (2019). Physiological and Transcriptomic Changes during the Early Phases of Adventitious Root Formation in Mulberry Stem Hardwood Cuttings. International Journal of Molecular Sciences, 20(15), 3707. https://doi.org/10.3390/ijms20153707





