Evolutionary and Comparative Expression Analyses of TCP Transcription Factor Gene Family in Land Plants
Abstract
:1. Introduction
2. Results
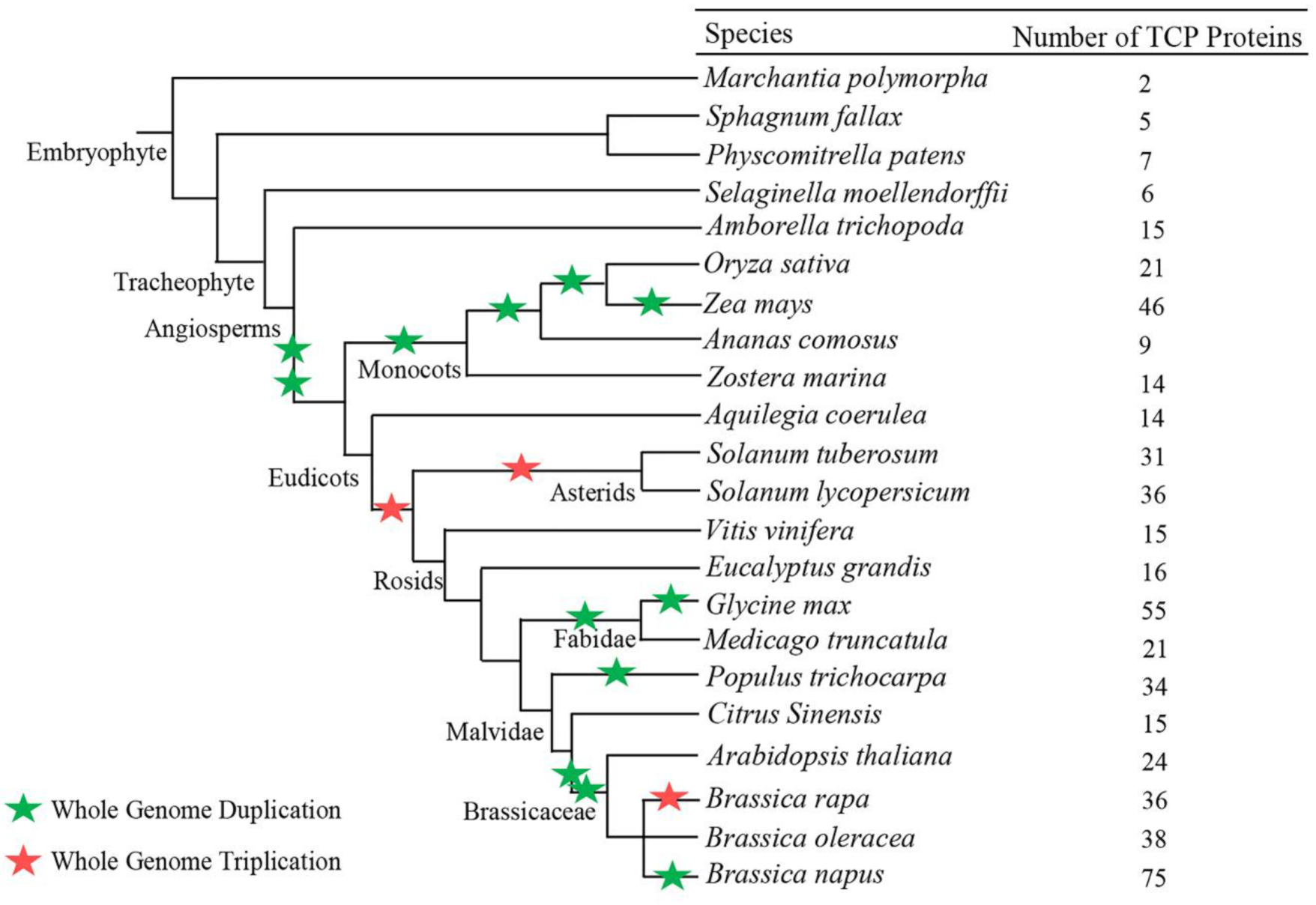
2.1. TCP (Teosinte-Branched 1/Cycloidea/Proliferating) Proteins are Widely Represented in Land Plant Genomes
2.2. Classification and Distribution of TCP Proteins in Land Plants
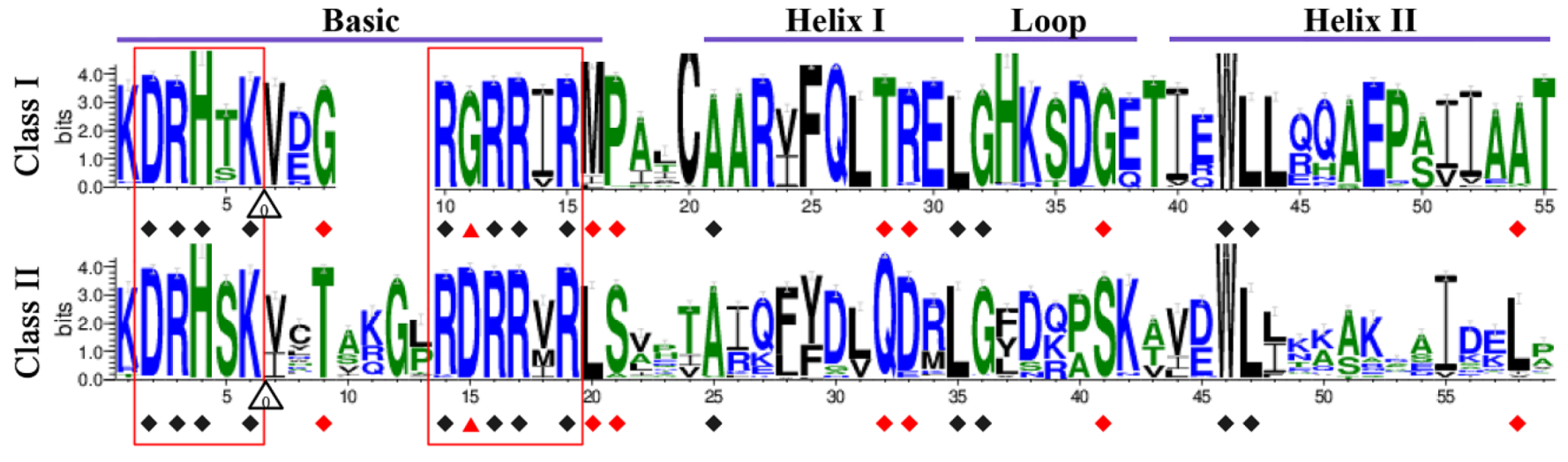
2.3. Conserved Sequence Characteristics within TCP Domains
2.4. Distribution of Non-TCP Motifs Supports the Classification of TCP Genes in Land Plants
2.5. Rapid Expansion of TCP Genes in Land Plants
2.6. MicroRNA319 Targets of TCP Genes were Conserved in Angiosperms
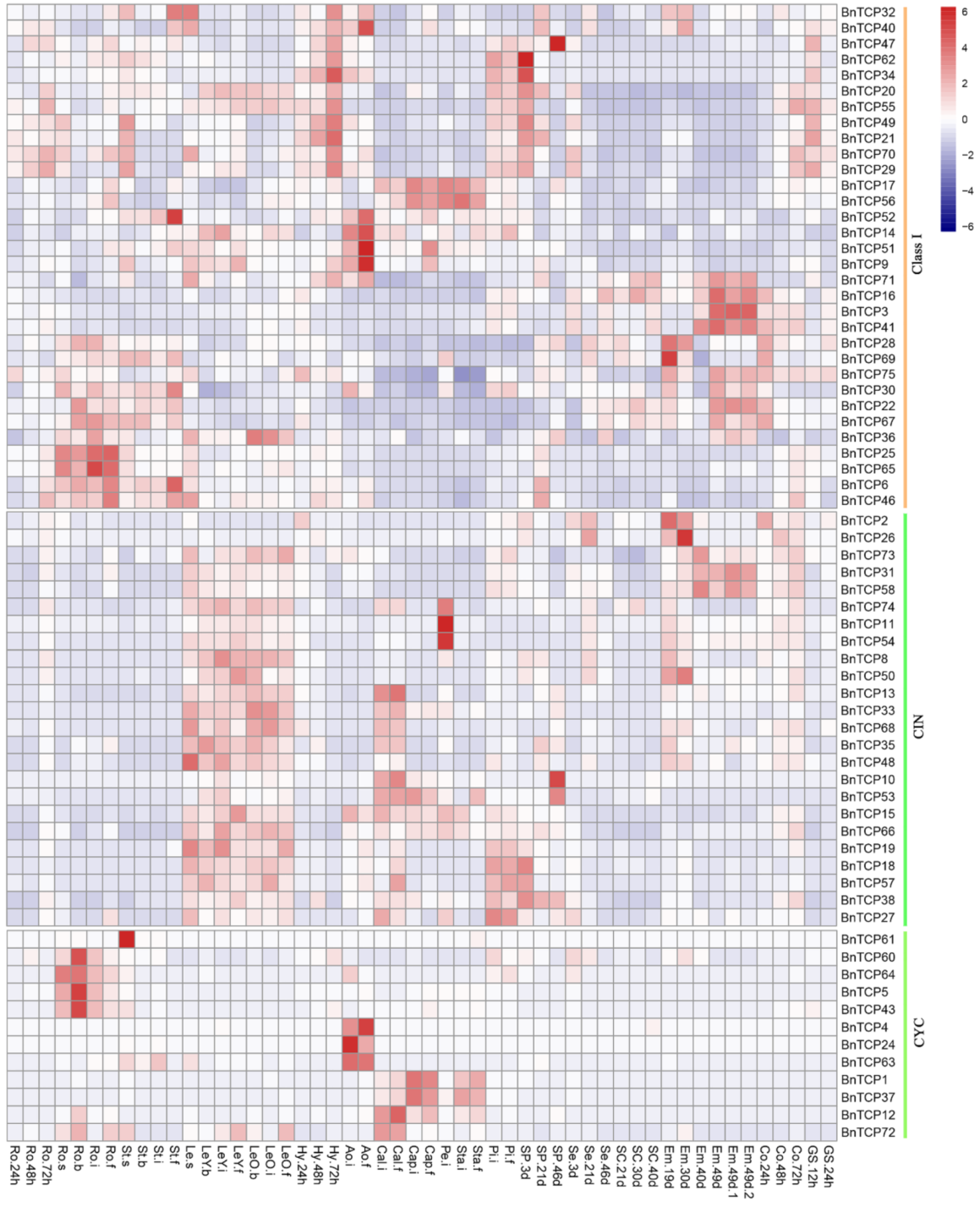
2.7. The Expressions of TCP Genes in Land Plants Suggest Function Diversification and Conservation
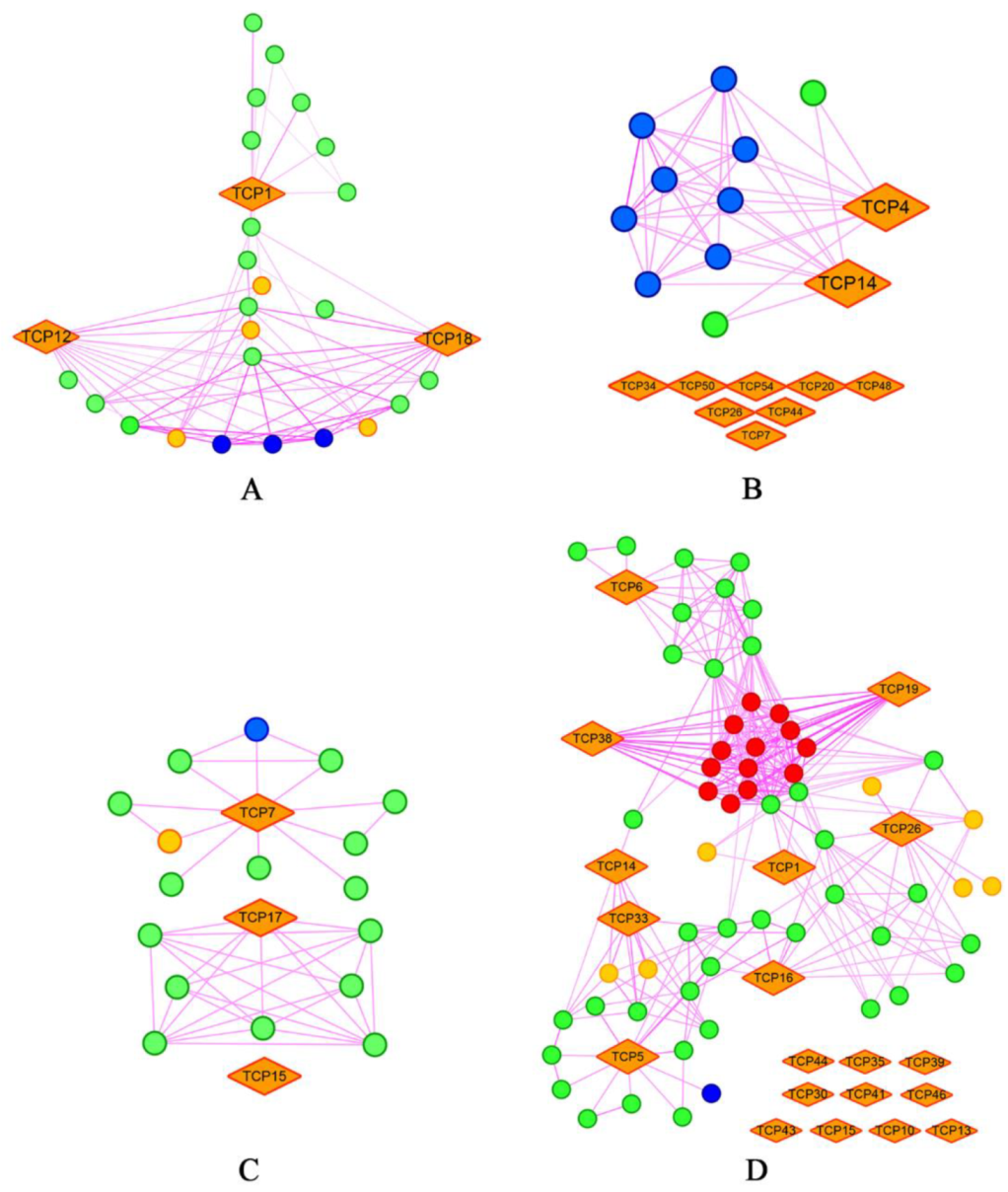
2.8. Diversified and Conserved Protein Interaction Network of TCP Proteins in Land Plants
3. Discussion
3.1. Similar, Antagonistic, and Evolutionary Functions of TCP Proteins in Plants
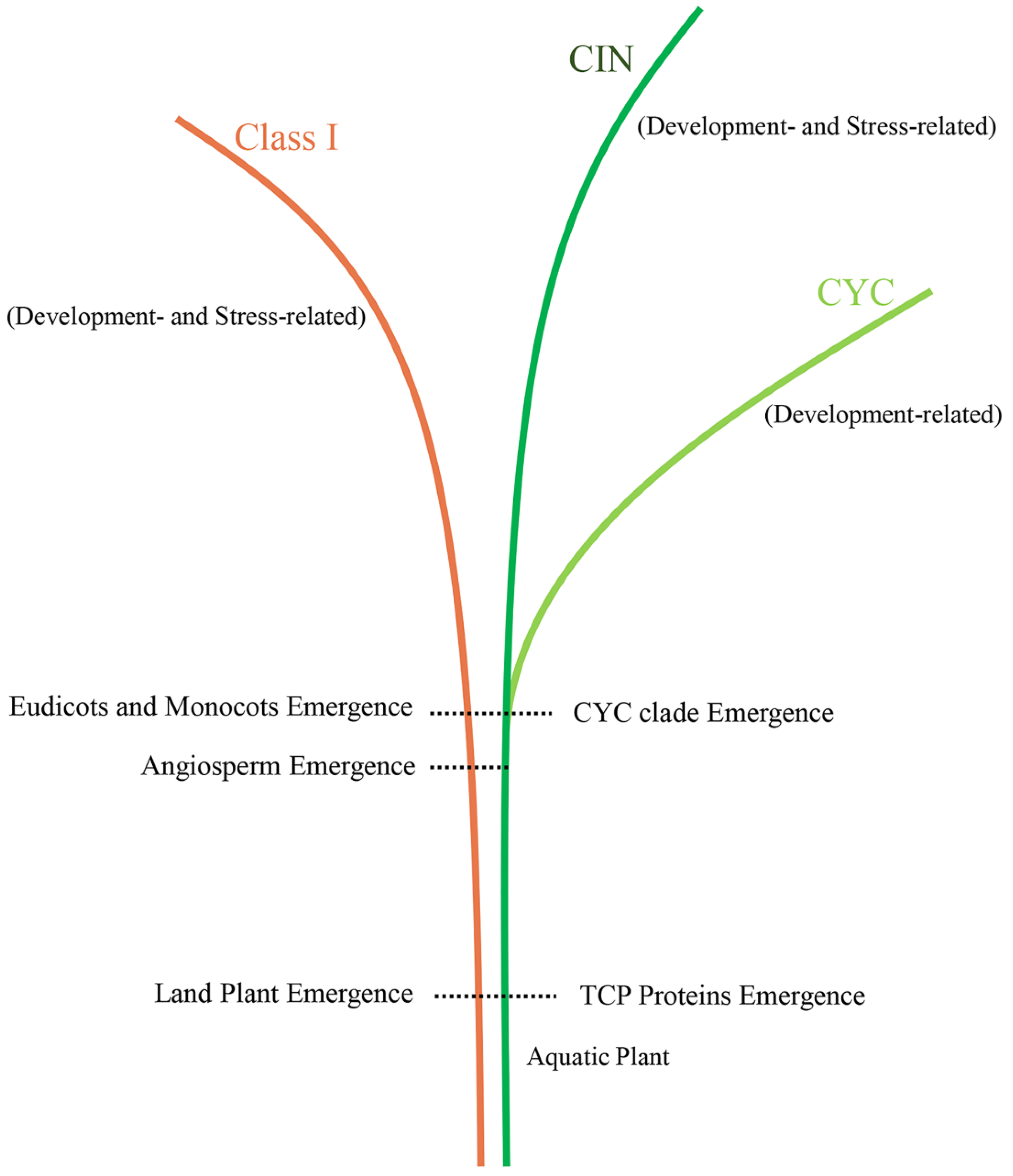
3.2. Origin and Evolution of the TCP Gene Family in Land Plants
4. Materials and Methods
4.1. Sequence Retrieval
4.2. Multiple Sequence Alignments and Gene Structure Analysis
4.3. Phylogenetic Analysis
4.4. Genome Synteny and Gene Duplication Analyses
4.5. Gene Expression Analysis
4.6. Interaction Network and miR319-Target Analysis
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Takeda, T.; Suwa, Y.; Suzuki, M. The OsTB1 gene negatively regulates lateral branching in rice. Plant J. 2003, 33, 513–520. [Google Scholar] [CrossRef]
- Aguilar-Martínez, J.A.; Poza-Carrión, C.; Cubas, P. Arabidopsis BRANCHED1 acts as an integrator of branching signals within axillary buds. Plant Cell 2007, 19, 458–472. [Google Scholar]
- Kieffer, M.; Master, V.; Waites, R.; Davies, B. TCP14 and TCP15 affect internode length and leaf shape in Arabidopsis. Plant J. 2011, 68, 147–158. [Google Scholar] [CrossRef] [Green Version]
- Lucero, L.E.; Uberti-Manassero, N.G.; Arce, A.L.; Colombatti, F.; Alemano, S.G.; Gonzalez, D.H. TCP15 modulates cytokinin and auxin responses during gynoecium development in Arabidopsis. Plant J. 2015, 84, 267–282. [Google Scholar] [CrossRef]
- Nicolas, M.; Cubas, P. TCP factors: New kids on the signaling block. Curr Opin Plant Biol. 2016, 33, 33–41. [Google Scholar] [CrossRef]
- Tatematsu, K.; Nakabayashi, K.; Kamiya, Y.; Nambara, E. Transcription factor AtTCP14 regulates embryonic growth potential during seed germination in Arabidopsis thaliana. Plant J. 2008, 53, 42–52. [Google Scholar] [CrossRef]
- Pruneda-Paz, J.L.; Breton, G.; Para, A.; Kay, S.A. A functional genomics approach reveals CHE as a component of the Arabidopsis circadian clock. Science 2009, 323, 1481–1485. [Google Scholar] [CrossRef]
- Kosugi, S.; Ohashi, Y. PCF1 and PCF2 specifically bind to cis-elements in the rice proliferating cell nuclear antigen gene. Plant Cell 1997, 9, 1607–1619. [Google Scholar]
- Doebley, J.; Stec, A.; Hubbard, L. The evolution of apical dominance in maize. Nature 1997, 386, 485–488. [Google Scholar] [CrossRef]
- Luo, D.; Carpenter, R.; Vincent, C.; Copsey, L.; Coen, E. Origin of floral asymmetry in Antirrhinum. Nature 1996, 383, 794–799. [Google Scholar] [CrossRef]
- Cubas, P.; Lauter, N.; Doebley, J.; Coen, E. The TCP domain: A motif found in proteins regulating plant growth and development. Plant J. 1999, 18, 215–222. [Google Scholar] [CrossRef]
- Navaud, O.; Dabos, P.; Carnus, E.; Tremousaygue, D.; Hervé, C. TCP transcription factors predate the emergence of land plants. J. Mol. Evol. 2007, 65, 23–33. [Google Scholar] [CrossRef]
- Howarth, D.G.; Donoghue, M.J. Phylogenetic analysis of the ‘ECE’ (CYC/TB1) clade reveals duplications predating the core eudicots. Proc. Natl. Acad. Sci. USA 2006, 103, 9101–9106. [Google Scholar] [CrossRef]
- Martín-Trillo, M.; Cubas, P. TCP genes: A family snapshot ten years later. Trends Plant Sci. 2010, 15, 31–39. [Google Scholar]
- Crawford, B.C.; Nath, U.; Carpenter, R.; Coen, E.S. CINCINNATA controls both cell differentiation and growth in petal lobes and leaves of Antirrhinum. Plant Physiol. 2004, 135, 244–253. [Google Scholar] [CrossRef]
- Li, S. The Arabidopsis thaliana TCP transcription factors: A broadening horizon beyond development. Plant Signal. Behav. 2015, 10, e1044192. [Google Scholar]
- Moreno-Pachon, N.M.; Mutimawurugo, M.C.; Heynen, E.; Sergeeva, L.; Benders, A.; Blilou, I.; Hilhorst, H.W.M.; Immink, R.G.H. Role of Tulipa gesneriana TEOSINTE BRANCHED1 (TgTB1) in the control of axillary bud outgrowth in bulbs. Plant Reprod. 2017, 31, 145–157. [Google Scholar] [CrossRef]
- Viola, I.L.; Uberti Manassero, N.G.; Ripoll, R.; Gonzalez, D.H. The Arabidopsis class I TCP transcription factor AtTCP11 is a developmental regulator with distinct DNA-binding properties due to the presence of a threonine residue at position 15 of the TCP domain. Biochem. J. 2011, 435, 143–155. [Google Scholar] [CrossRef]
- Danisman, S.; van der Wal, F.; Dhondt, S.; Waites, R.; de Folter, S.; Bimbo, A.; van Dijk, A.D.; Muino, J.M.; Cutri, L.; Dornelas, M.C.; et al. Arabidopsis class I and class II TCP transcription factors regulate jasmonic acid metabolism and leaf development antagonistically. Plant Physiol. 2012, 159, 1511–1523. [Google Scholar] [CrossRef]
- Li, Z.Y.; Li, B.; Dong, A.W. The Arabidopsis transcription factor AtTCP15 regulates endoreduplication by modulating expression of key cell-cycle genes. Mol. Plant 2012, 5, 270–280. [Google Scholar] [CrossRef]
- Balsemão-Pires, E.; Andrade, L.R.; Sachetto-Martins, G. Functional study of TCP23 in Arabidopsis thaliana during plant development. Plant Physiol. Biochem. 2013, 67, 120–125. [Google Scholar]
- Chai, W.; Jiang, P.; Huang, G.; Jiang, H.; Li, X. Identification and expression profiling analysis of TCP family genes involved in growth and development in maize. Physiol. Mol. Biol. Plants 2017, 23, 779–791. [Google Scholar] [CrossRef]
- Zhao, J.; Zhai, Z.; Li, Y.; Geng, S.; Song, G.; Guan, J.; Jia, M.; Wang, F.; Sun, G.; Feng, N.; et al. Genome-Wide identification and expression profiling of the TCP family genes in spike and grain development of wheat (Triticum aestivum L.). Front. Plant Sci. 2018, 9, 1282. [Google Scholar] [CrossRef]
- Cubas, P. Role of TCP genes in the evolution of key morphological characters in angiosperms. In Developmental Genetics and Plant Evolution; Cronk, Q.C.B., Hawkins, J., Bateman, R.M., Eds.; CRC Press: London, UK, 2002; pp. 247–266. [Google Scholar]
- Ma, X.; Ma, J.; Fan, D.; Li, C.; Jiang, Y.; Luo, K. Genome-wide Identification of TCP family transcription factors from Populus euphratica and their involvement in leaf shape regulation. Sci. Rep. 2016, 6, 32795. [Google Scholar] [CrossRef]
- Ma, J.; Wang, Q.; Sun, R.; Xie, F.; Jones, D.C.; Zhang, B. Genome-wide identification and expression analysis of TCP transcription factors in Gossypium raimondii. Sci. Rep. 2014, 4, 6645. [Google Scholar] [CrossRef]
- Camacho, C.; Coulouris, G.; Avagyan, V.; Ma, N.; Papadopoulos, J.; Bealer, K.; Madden, T.L. BLAST+: Architecture and applications. BMC Bioinform. 2009, 10, 421. [Google Scholar] [CrossRef]
- Goodstein, D.M.; Shu, S.; Howson, R.; Neupane, R.; Hayes, R.D.; Fazo, J.; Mitros, T.; Dirks, W.; Hellsten, U.; Putnam, N.; et al. Phytozome: A comparative platform for green plant genomics. Nucleic Acids Res. 2012, 40, D1178–D1186. [Google Scholar] [CrossRef]
- Cheng, F.; Liu, S.; Wu, J.; Fang, L.; Sun, S.; Liu, B.; Li, P.; Hua, W.; Wang, X. BRAD, the genetics and genomics database for Brassica plants. BMC Plant Biol. 2011, 11, 136. [Google Scholar] [CrossRef]
- Chalhoub, B.; Denoeud, F.; Liu, S.; Parkin, I.A.; Tang, H.; Wang, X.; Chiquet, J.; Belcram, H.; Tong, C.; Samans, B.; et al. Early allopolyploid evolution in the post-Neolithic Brassica napus oilseed genome. Science 2014, 345, 950–953. [Google Scholar] [CrossRef]
- Yao, X.; Ma, H.; Wang, J.; Zhang, D. Genome-wide comparative analysis and expression pattern of TCP gene families in Arabidopsis thaliana and Oryza sativa. J. Integr. Plant Biol. 2007, 49, 885–897. [Google Scholar] [CrossRef]
- Mondragón-Palomino, M.; Trontin, C. High time for a roll call: Gene duplication and phylogenetic relationships of TCP-like genes in monocots. Ann. Bot. 2011, 107, 1533–1544. [Google Scholar] [CrossRef]
- Parapunova, V.; Busscher, M.; Busscher-Lange, J. Identification, cloning and characterization of the tomato TCP transcription factor family. BMC Plant Biol. 2014, 14, 157. [Google Scholar] [CrossRef]
- Tamura, K.; Peterson, D.; Peterson, N.; Stecher, G.; Nei, M.; Kumar, S. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 2011, 28, 2731–2739. [Google Scholar] [CrossRef]
- Bailey, T.L.; Williams, N.; Misleh, C.; Li, W.W. MEME: Discovering and analyzing DNA and protein sequence motifs. Nucleic Acids Res. 2006, 34, W369–W373. [Google Scholar] [CrossRef]
- Viola, I.L.; Reinheimer, R.; Ripoll, R.; Manassero, N.G.; Gonzalez, D.H. Determinants of the DNA binding specificity of class I and class II TCP transcription factors. J. Biol. Chem. 2012, 287, 347–356. [Google Scholar] [CrossRef]
- Kim, S.H.; Son, G.H.; Bhattacharjee, S.; Kim, H.J.; Nam, J.C.; Nguyen, P.D.; Hong, J.C.; Gassmann, W. The Arabidopsis immune adaptor SRFR1 interacts with TCP transcription factors that redundantly contribute to effector-triggered immunity. Plant J. 2014, 78, 978–989. [Google Scholar] [CrossRef]
- Li, S.; Gutsche, N.; Zachgo, S. The ROXY1 C-terminal L**LL motif is essential for the interaction with TGA transcription factors. Plant Physiol. 2011, 157, 2056–2068. [Google Scholar] [CrossRef]
- Du, H.; Liang, Z.; Zhao, S.; Nan, M.G.; Tran, L.S.; Lu, K.; Huang, Y.B.; Li, J.N. The evolutionary history of R2R3-MYB proteins across 50 eukaryotes: New insights into subfamily classification and expansion. Sci. Rep. 2015, 5, 11037. [Google Scholar] [CrossRef]
- Crooks, G.E.; Hon, G.; Chandonia, J.M.; Brenner, S.E. WebLogo: A sequence logo generator. Genome Res. 2004, 14, 1188–1190. [Google Scholar] [CrossRef]
- Bresso, E.G.; Chorostecki, U.; Rodriguez, R.E.; Palatnik, J.F.; Schommer, C. Spatial control of gene expression by miR319-regulated TCP transcription factors in leaf development. Plant Physiol. 2017, 176, 1694–1708. [Google Scholar] [CrossRef]
- Lyons, E.; Freeling, M. How to usefully compare homologous plant genes and chromosomes as DNA sequences. Plant J. 2008, 53, 661–673. [Google Scholar] [CrossRef] [PubMed]
- Tang, H.; Wang, X.; Bowers, J.E.; Ming, R.; Alam, M.; Paterson, A.H. Unraveling ancient hexaploidy through multiply-aligned angiosperm gene maps. Genome Res. 2008, 18, 1944–1954. [Google Scholar] [CrossRef] [Green Version]
- Axtell, M.J. Classification and Comparison of Small RNAs from Plants. Annu. Rev. Plant Biol. 2013, 64, 137–159. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ori, N.; Cohen, A.R.; Etzioni, A.; Brand, A.; Yanai, O.; Shleizer, S.; Menda, N.; Amsellem, Z.; Efroni, I.; Pekker, I.; et al. Regulation of LANCEOLATE by miR319 is required for compound-leaf development in tomato. Nat. Genet. 2007, 39, 787–791. [Google Scholar] [CrossRef] [PubMed]
- Cuperus, J.T.; Fahlgren, N.; Carrington, J.C. Evolution and functional diversification of MIRNA genes. Plant Cell 2011, 23, 431–442. [Google Scholar] [CrossRef]
- Miskiewicz, J.; Szachniuk, M. Discovering structural motifs in miRNA precursors from the Viridiplantae kingdom. Molecules 2018, 23, E1367. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.D.; Ling, L.Z.; Yi, T.S. Evolution and divergence of SBP-box genes in land plants. BMC Genom. 2015, 16, 787. [Google Scholar] [CrossRef]
- Du, J.; Hu, S.; Yu, Q.; Wang, C.; Yang, Y.; Sun, H.; Yang, Y.; Sun, X. Genome-wide identification and characterization of BrrTCP transcription factors in Brassica rapa ssp. rapa. Front. Plant Sci. 2017, 8, 1588. [Google Scholar] [CrossRef]
- Shi, P.; Guy, K.M.; Wu, W.; Fang, B.; Yang, J.; Zhang, M.; Hu, Z. Genome-wide identification and expression analysis of the ClTCP transcription factors in Citrullus lanatus. BMC Plant Biol. 2016, 16, 85. [Google Scholar] [CrossRef]
- Manassero, N.G.; Viola, I.L.; Welchen, E.; Gonzalez, D.H. TCP transcription factors: Architectures of plant form. Biomol. Concepts 2013, 4, 111–127. [Google Scholar] [CrossRef]
- Galili, T.; O’Callaghan, A.; Sidi, J.; Sievert, C. Heatmaply: An R package for creating interactive cluster heatmaps for online publishing. Bioinformatics 2018, 34, 1600–1602. [Google Scholar] [CrossRef] [PubMed]
- Mukhtar, M.S.; Carvunis, A.R.; Dreze, M.; Epple, P.; Steinbrenner, J.; Moore, J.; Tasan, M.; Galli, M.; Hao, T.; Nishimura, M.T. Independently evolved virulence effectors converge onto hubs in a plant immune system network. Science 2011, 333, 596–601. [Google Scholar] [CrossRef] [PubMed]
- Niwa, M.; Daimon, Y.; Kurotani, K.; Higo, A.; Pruneda-Paz, J.L.; Breton, G.; Mitsuda, N.; Kay, S.A.; Ohme-Takagi, M.; Endo, M.; et al. BRANCHED1 interacts with FLOWERING LOCUS T to repress the floral transition of the axillary meristems in Arabidopsis. Plant Cell 2013, 25, 1228–1242. [Google Scholar] [CrossRef] [PubMed]
- Szklarczyk, D.; Morris, J.H.; Cook, H.; Kuhn, M.; Wyder, S.; Simonovic, M.; Santos, A.; Doncheva, N.T.; Roth, A.; Bork, P.; et al. The STRING database in 2017: Quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res. 2017, 45, D362–D368. [Google Scholar] [CrossRef] [PubMed]
- Koyama, T.; Ohme-Takagi, M.; Sato, F. Generation of serrated and wavy petals by inhibition of the activity of TCP transcription factors in Arabidopsis thaliana. Plant Signal. Behav. 2011, 6, 697–699. [Google Scholar] [CrossRef] [PubMed]
- Braun, N.; de Saint Germain, A.; Pillot, J.P.; Boutet-Mercey, S.; Dalmais, M.; Antoniadi, I.; Li, X.; Maia-Grondard, A.; Le Signor, C.; Bouteiller, N.; et al. The pea TCP transcription factor PsBRC1 acts downstream of strigolactones to control shoot branching. Plant Physiol. 2012, 158, 225–238. [Google Scholar] [CrossRef]
- Martín-Trillo, M.; Grandío, E.G.; Serra, F.; Marcel, F.; Rodríguez-Buey, M.L.; Schmitz, G.; Theres, K.; Bendahmane, A.; Dopazo, H.; Cubas, P. Role of tomato BRANCHED1-like genes in the control of shoot branching. Plant J. 2011, 67, 701–714. [Google Scholar]
- Guan, J.C.; Koch, K.E.; Suzuki, M.; Wu, S.; Latshaw, S.; Petruff, T.; Goulet, C.; Klee, H.J.; McCarty, D.R. Diverse roles of strigolactone signaling in maize architecture and the uncoupling of a branching-specific subnetwork. Plant Physiol. 2012, 160, 1303–1317. [Google Scholar] [CrossRef] [PubMed]
- Dong, Z.; Li, W.; Unger-Wallace, E.; Yang, J.; Vollbrecht, E.; Chuck, G. Ideal crop plant architecture is mediated by tassels replace upper ears1, a BTB/POZ ankyrin repeat gene directly targeted by TEOSINTE BRANCHED1. Proc. Natl. Acad. Sci. USA 2017, 114, E8656–E8664. [Google Scholar] [CrossRef]
- González-Grandío, E.; Pajoro, A.; Franco-Zorrilla, J.M.; Tarancón, C.; Immink, R.G.; Cubas, P. Abscisic acid signaling is controlled by a BRANCHED1/HD-ZIP I cascade in Arabidopsis axillary buds. Proc. Natl. Acad. Sci. USA 2017, 114, E245–E254. [Google Scholar]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef] [PubMed]
- Vadde, B.V.L.; Challa, K.R.; Nath, U. The TCP4 transcription factor regulates trichome cell differentiation by directly activating GLABROUS INFLORESCENCE STEMS in Arabidopsis thaliana. Plant J. 2017, 93, 259–269. [Google Scholar] [CrossRef] [PubMed]
- Kubota, A.; Ito, S.; Shim, J.S.; Johnson, R.S.; Song, Y.H.; Breton, G.; Goralogia, G.S.; Kwon, M.S.; Laboy Cintrón, D.; Koyama, T.; et al. TCP4-dependent induction of CONSTANS transcription requires GIGANTEA in photoperiodic flowering in Arabidopsis. PLoS Genet. 2017, 13, e1006856. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Mao, Y.; Yang, J.; He, Y. TCP24 modulates secondary cell wall thickening and anther endothecium development. Front. Plant Sci. 2015, 6, 436. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, S.T.; Sun, X.L.; Hoshino, Y.; Yu, Y.; Jia, B.; Sun, Z.W.; Sun, M.Z.; Duan, X.B.; Zhu, Y.M. MicroRNA319 positively regulates cold tolerance by targeting OsPCF6 and OsTCP21 in rice (Oryza sativa L.). PLoS ONE 2014, 9, e91357. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Ding, Z.; Wu, K.; Yang, L.; Li, Y.; Yang, Z.; Shi, S.; Liu, X.; Zhao, S.; Yang, Z.; et al. Suppression of jasmonic acid-mediated defense by viral-inducible MicroRNA319 facilitates virus infection in rice. Mol. Plant 2016, 9, 1302–1314. [Google Scholar] [CrossRef] [PubMed]
- Broholm, S.K.; Tähtiharju, S.; Laitinen, R.A.; Albert, V.A.; Teeri, T.H.; Elomaa, P. A TCP domain transcription factor controls flower type specification along the radial axis of the Gerbera (Asteraceae) inflorescence. Proc. Natl. Acad. Sci. USA 2008, 105, 9117–9122. [Google Scholar] [CrossRef] [PubMed]
- Yuan, Z.; Gao, S.; Xue, D.W.; Luo, D.; Li, L.T.; Ding, S.Y.; Yao, X.; Wilson, Z.A.; Qian, Q.; Zhang, D.B. RETARDED PALEA1 controls palea development and floral zygomorphy in rice. Plant Physiol. 2009, 149, 235–244. [Google Scholar] [CrossRef] [PubMed]
- Feng, X.; Zhao, Z.; Tian, Z. Control of petal shape and floral zygomorphy in Lotus japonicus. Proc. Natl. Acad. Sci. USA 2006, 103, 4970–4975. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Potuschak, T.; Colón-Carmona, A.; Gutiérrez, R.A.; Doerner, P. Arabidopsis TCP20 links regulation of growth and cell division control pathways. Proc. Natl. Acad. Sci. USA 2005, 102, 12978–12983. [Google Scholar] [CrossRef] [PubMed]
- Aguilar-Martínez, J.A.; Sinha, N. Analysis of the role of Arabidopsis class I TCP genes AtTCP7, AtTCP8, AtTCP22, and AtTCP23 in leaf development. Front. Plant Sci. 2013, 4, 406. [Google Scholar]
- Zhang, T.; Qu, Y.; Wang, H.; Wang, J.; Song, A.; Hu, Y.; Chen, S.; Jiang, J.; Chen, F. The heterologous expression of a chrysanthemum TCP-P transcription factor CmTCP14 suppresses organ size and delays senescence in Arabidopsis thaliana. Plant Physiol. Biochem. 2017, 115, 239–248. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.F.; Chen, Y.Y.; Hsiao, Y.Y.; Shen, C.Y.; Hsu, J.L.; Yeh, C.M.; Mitsuda, N.; Ohme-Takagi, M.; Liu, Z.J.; Tsai, W.C. Genome-wide identification and characterization of TCP genes involved in ovule development of Phalaenopsis equestris. J. Exp. Bot. 2016, 67, 5051–5066. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.Y.; Zhao, P.M.; Cheng, H.Q.; Han, L.B.; Wu, X.M.; Gao, P.; Wang, H.Y.; Yang, C.L.; Zhong, N.Q.; Zuo, J.R.; et al. The cotton transcription factor TCP14 functions in auxin-mediated epidermal cell differentiation and elongation. Plant Physiol. 2013, 162, 1669–1680. [Google Scholar] [CrossRef] [PubMed]
- Guan, P.; Ripoll, J.J.; Wang, R.; Vuong, L.; Bailey-Steinitz, L.J.; Ye, D.; Crawford, N.M. Interacting TCP and NLP transcription factors control plant responses to nitrate availability. Proc. Natl. Acad. Sci. USA 2017, 114, 2419–2424. [Google Scholar] [CrossRef] [Green Version]
- Mukhopadhyay, P.; Tyagi, A.K. OsTCP19 influences developmental and abiotic stress signaling by modulating ABI4-mediated pathways. Sci. Rep. 2015, 5, 9998. [Google Scholar] [CrossRef] [PubMed]
- Almeida, D.M.; Gregorio, G.B.; Oliveira, M.M.; Saibo, N.J. Five novel transcription factors as potential regulators of OsNHX1 gene expression in a salt tolerant rice genotype. Plant Mol. Biol. 2017, 93, 61–77. [Google Scholar] [CrossRef] [PubMed]
- Krouk, G.; Lacombe, B.; Bielach, A.; Perrine-Walker, F.; Malinska, K.; Mounier, E.; Hoyerova, K.; Tillard, P.; Leon, S.; Ljung, K.; et al. Nitrate-regulated auxin transport by NRT1.1 defines a mechanism for nutrient sensing in plants. Dev. Cell 2010, 18, 927–937. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Zachgo, S. TCP3 interacts with R2R3-MYB proteins, promotes flavonoid biosynthesis and negatively regulates the auxin response in Arabidopsis thaliana. Plant J. 2013, 76, 901–913. [Google Scholar] [CrossRef] [PubMed]
- Schommer, C.; Palatnik, J.F.; Aggarwal, P.; Chételat, A.; Cubas, P.; Farmer, E.E.; Nath, U.; Weigel, D. Control of jasmonate biosynthesis and senescence by miR319 targets. PLoS Biol. 2008, 6, e230. [Google Scholar] [CrossRef] [PubMed]
- Lopez, J.A.; Sun, Y.; Blair, P.B.; Mukhtar, M.S. TCP three-way handshake: Linking developmental processes with plant immunity. Trends Plant Sci. 2015, 20, 238–245. [Google Scholar] [CrossRef] [PubMed]
- Stam, R.; Motion, G.; Boevink, P.C.; Huitema, E. A conserved oomycete CRN effector targets and modulates tomato TCP14-2 to enhance virulence. BioRxiv 2013, 001248. [Google Scholar]
- Sugio, A.; Kingdom, H.N.; MacLean, A.M.; Grieve, V.M.; Hogenhout, S.A. Phytoplasma protein effector SAP11 enhances insect vector reproduction by manipulating plant development and defense hormone biosynthesis. Proc. Natl. Acad. Sci. USA 2011, 108, E1254–E1263. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tan, C.M.; Li, C.H.; Tsao, N.W.; Su, L.W.; Lu, Y.T.; Chang, S.H.; Lin, Y.Y.; Liou, J.C.; Hsieh, L.C.; Yu, J.Z.; et al. Phytoplasma SAP11 alters 3-isobutyl-2-methoxypyrazine biosynthesis in Nicotiana benthamiana by suppressing NbOMT1. J. Exp. Bot. 2016, 67, 4415–4425. [Google Scholar] [CrossRef] [PubMed]
- Janik, K.; Mithöfer, A.; Raffeiner, M.; Stellmach, H.; Hause, B.; Schlink, K. An effector of apple proliferation phytoplasma targets TCP transcription factors-a generalized virulence strategy of phytoplasma? Mol. Plant Pathol. 2017, 18, 435–442. [Google Scholar] [CrossRef] [PubMed]
- Sugio, A.; MacLean, A.M.; Hogenhout, S.A. The small phytoplasma virulence effector SAP11 contains distinct domains required for nuclear targeting and CIN-TCP binding and destabilization. New Phytol. 2014, 202, 838–848. [Google Scholar] [CrossRef] [PubMed]
- Chang, S.H.; Tan, C.M.; Wu, C.T.; Lin, T.H.; Jiang, S.Y.; Liu, R.C.; Tsai, M.C.; Su, L.W.; Yang, J.Y. Alterations of plant architecture and phase transition by the phytoplasma virulence factor SAP11. J. Exp. Bot. 2018, 69, 5389–5401. [Google Scholar] [CrossRef] [Green Version]
- Wang, N.; Yang, H.; Yin, Z.; Liu, W.; Sun, L.; Wu, Y. Phytoplasma effector SWP1 induces witches’ broom symptom by destabilizing the TCP transcription factor BRANCHED1. Mol. Plant Pathol. 2018, 19, 2623–2634. [Google Scholar] [CrossRef]
- Mao, Y.; Wu, F.; Yu, X.; Bai, J.; Zhong, W.; He, Y. MicroRNA319a-targeted Brassica rapa ssp. pekinensis TCP genes modulate head shape in Chinese cabbage by differential cell division arrest in leaf regions. Plant Physiol. 2014, 164, 710–720. [Google Scholar] [CrossRef]
- Uberti-Manassero, N.G.; Coscueta, E.R.; Gonzalez, D.H. Expression of a repressor form of the Arabidopsis thaliana transcription factor TCP16 induces the formation of ectopic meristems. Plant Physiol. Biochem. 2016, 108, 57–62. [Google Scholar] [CrossRef]
- Takeda, T.; Amano, K.; Ohto, M.A.; Nakamura, K.; Sato, S.; Kato, T.; Tabata, S.; Ueguchi, C. RNA interference of the Arabidopsis putative transcription factor TCP16 gene results in abortion of early pollen development. Plant Mol. Biol. 2006, 61, 165–177. [Google Scholar] [CrossRef] [PubMed]
- Hervé, C.; Dabos, P.; Bardet, C.; Jauneau, A.; Auriac, M.C.; Ramboer, A.; Lacout, F.; Tremousaygue, D. In vivo interference with AtTCP20 function induces severe plant growth alterations and deregulates the expression of many genes important for development. Plant Physiol. 2009, 149, 1462–1477. [Google Scholar] [CrossRef] [PubMed]
- Guan, P.; Wang, R.; Nacry, P.; Breton, G.; Kay, S.A.; Pruneda-Paz, J.L.; Davani, A.; Crawford, N.M. Nitrate foraging by Arabidopsis roots is mediated by the transcription factor TCP20 through the systemic signaling pathway. Proc. Natl. Acad. Sci. USA 2014, 111, 15267–15272. [Google Scholar] [CrossRef] [PubMed]
- Danisman, S.; van Dijk, A.D.; Bimbo, A.; van der Wal, F.; Hennig, L.; de Folter, S.; Angenent, G.C.; Immink, R.G. Analysis of functional redundancies within the Arabidopsis TCP transcription factor family. J. Exp. Bot. 2013, 64, 5673–5685. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, X.; Gao, J.; Zhu, Z.; Dong, X.; Wang, X.; Ren, G.; Zhou, X.; Kuai, B. TCP transcription factors are critical for the coordinated regulation of ISOCHORISMATE SYNTHASE 1 expression in Arabidopsis thaliana. Plant J. 2015, 82, 151–162. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Zhang, D.; An, J.; Yin, H.; Fang, S.; Chu, J.; Zhao, Y.; Li, J. TCP transcription factors regulate shade avoidance via directly mediating the expression of both PHYTOCHROME INTERACTING FACTORs and auxin biosynthetic genes. Plant Physiol. 2017, 176, 1850–1861. [Google Scholar] [CrossRef] [PubMed]
- Baba, K.; Nakano, T.; Yamagishi, K.; Yoshida, S. Involvement of a nuclear-encoded basic helix-loop-helix protein in transcription of the light-responsive promoter of psbD. Plant Physiol. 2001, 125, 595–603. [Google Scholar] [CrossRef] [PubMed]
- Ortiz-Ramírez, C.; Hernandez-Coronado, M.; Thamm, A.; Catarino, B.; Wang, M.; Dolan, L.; Feijó, J.A.; Becker, J.D. A transcriptome atlas of Physcomitrella patens provides insights into the evolution and development of land plants. Mol. Plant 2016, 9, 205–220. [Google Scholar]
- Guo, Z.; Fujioka, S.; Blancaflor, E.B.; Miao, S.; Gou, X.; Li, J. TCP1 modulates brassinosteroid biosynthesis by regulating the expression of the key biosynthetic gene DWARF4 in Arabidopsis thaliana. Plant Cell 2010, 22, 1161–1173. [Google Scholar] [CrossRef]
- Mizuno, S.; Sonoda, M.; Tamura, Y.; Nishino, E.; Suzuki, H.; Sato, T.; Oizumi, T. Chiba Tendril-Less locus determines tendril organ identity in melon (Cucumis melo L.) and potentially encodes a tendril-specific TCP homolog. J. Plant Res. 2015, 128, 941–951. [Google Scholar] [CrossRef]
- Wang, S.; Yang, X.; Xu, M.; Lin, X.; Lin, T.; Qi, J.; Shao, G.; Tian, N.; Yang, Q.; Zhang, Z.; et al. A rare SNP identified a TCP transcription factor essential for tendril development in cucumber. Mol. Plant 2015, 8, 1795–1808. [Google Scholar] [CrossRef] [PubMed]
- Nicolas, M.; Rodríguez-Buey, M.L.; Franco-Zorrilla, J.M.; Cubas, P. A recently evolved alternative splice site in the BRANCHED1a gene controls potato plant architecture. Curr. Biol. 2015, 25, 1799–1809. [Google Scholar] [CrossRef] [PubMed]
- Ramsay, L.; Comadran, J.; Druka, A.; Marshall, D.F.; Thomas, W.T.; Macaulay, M.; MacKenzie, K.; Simpson, C.; Fuller, J.; Bonar, N.; et al. INTERMEDIUM-C, a modifier of lateral spikelet fertility in barley, is an ortholog of the maize domestication gene TEOSINTE BRANCHED 1. Nat. Genet. 2011, 43, 169–172. [Google Scholar] [CrossRef] [PubMed]
- Kebrom, T.H.; Burson, B.L.; Finlayson, S.A. Phytochrome B represses TEOSINTE BRANCHED 1 expression and induces sorghum axillary bud outgrowth in response to light signals. Plant Physiol. 2006, 140, 1109–1117. [Google Scholar] [CrossRef] [PubMed]
- Xu, K.; Wang, Y.; Shi, L.; Sun, F.; Liu, S.; Xi, Y. PvTB1, a Teosinte Branched1 gene homolog, negatively regulates tillering in switchgrass. J. Plant Growth Regul. 2016, 35, 44–53. [Google Scholar] [CrossRef]
- Bai, F.; Reinheimer, R.; Durantini, D.; Kellogg, E.A.; Schmidt, R.J. TCP transcription factor, BRANCH ANGLE DEFECTIVE 1 (BAD1), is required for normal tassel branch angle formation in maize. Proc. Natl. Acad. Sci. USA 2012, 109, 12225–12230. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, D.; Li, X.; Sun, M.; Zhang, T.; Pan, H.; Cheng, T.; Wang, J.; Zhang, Q. Identification and characterization of CYC-like genes in regulation of ray floret development in Chrysanthemum morifolium. Front. Plant Sci. 2016, 7, 1633. [Google Scholar] [CrossRef] [PubMed]
- McCourt, R.M.; Delwiche, C.F.; Karol, K.G. Charophyte algae and land plant origins. Trends Ecol. Evol. 2004, 19, 661–666. [Google Scholar] [CrossRef]
- Bowman, J.L.; Floyd, S.K.; Sakakibara, K. Green genes-comparative genomics of the green branch of life. Cell 2007, 129, 229–234. [Google Scholar] [CrossRef]
- Xu, R.; Sun, P.; Jia, F.; Lu, L.; Li, Y.; Zhang, S.; Huang, J. Genomewide analysis of TCP transcription factor gene family in Malus domestica. J. Genet. 2014, 93, 733–746. [Google Scholar] [CrossRef]
- De Paolo, S.; Gaudio, L.; Aceto, S. Analysis of the TCP genes expressed in the inflorescence of the orchid Orchis italica. Sci. Rep. 2015, 5, 16265. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zheng, K.; Ni, Z.; Qu, Y.; Cai, Y.; Yang, Z.; Sun, G.; Chen, Q. Genome-wide identification and expression analyses of TCP transcription factor genes in Gossypium barbadense. Sci. Rep. 2018, 8, 14526. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Li, D.D.; Han, L.H.; Tao, M.; Hu, Q.Q.; Wu, W.Y.; Zhang, J.B.; Li, X.B.; Huang, G.Q. Genome-wide identification and characterization of TCP transcription factor genes in upland cotton (Gossypium hirsutum). Sci. Rep. 2017, 7, 10118. [Google Scholar] [CrossRef] [PubMed]
- Citerne, H.L.; Le Guilloux, M.; Sannier, J.; Nadot, S.; Damerval, C. Combining phylogenetic and syntenic analyses for understanding the evolution of TCP ECE genes in eudicots. PLoS ONE 2013, 8, e74803. [Google Scholar] [CrossRef] [PubMed]
- Blanc, G.; Wolfe, K.H. Functional divergence of duplicated genes formed by polyploidy during Arabidopsis evolution. Plant Cell 2004, 16, 1679–1691. [Google Scholar] [CrossRef] [PubMed]
- Artimo, P.; Jonnalagedda, M.; Arnold, K.; Baratin, D.; Csardi, G.; de Castro, E.; Duvaud, S.; Flegel, V.; Fortier, A.; Gasteiger, E.; et al. ExPASy: SIB bioinformatics resource portal. Nucleic Acids Res. 2012, W597–W603. [Google Scholar] [CrossRef]
- Lee, T.H.; Tang, H.; Wang, X.; Paterson, A.H. PGDD: A database of gene and genome duplication in plants. Nucleic Acids Res. 2013, 41, D1152–D1158. [Google Scholar] [CrossRef] [PubMed]
- Bowman, J.L.; Kohchi, T.; Yamato, K.T.; Jenkins, J.; Shu, S.; Ishizaki, K.; Yamaoka, S.; Nishihama, R.; Nakamura, Y.; Berger, F.; et al. Insights into land plant evolution garnered from the Marchantia polymorpha genome. Cell 2017, 171, 287–304. [Google Scholar] [CrossRef]
- Dai, X.; Zhuang, Z.; Zhao, P.X. psRNATarget: A plant small RNA target analysis server (2017 release). Nucleic Acids Res. 2018, 46, W49–W54. [Google Scholar] [CrossRef]


| Class (Clade) | Species | Name | Biological Function |
|---|---|---|---|
| Class I | Brassica rapa | BrpTCP4 | Head shape from round to cylindrical [90]. Plant immunity [84]. |
| Arabidopsis thaliana | AtTCP15 | Regulate the expression of the key cell cycle genes [20]; regulate the cytokinin and auxin response [4]. | |
| AtTCP14, 15 | Influence the internode length and leaf shape [3]. Related to the defense of Hpa pathogen [53] and DC3000 [37]. Plant immunity [82]. | ||
| AtTCP14 | Involved in seed germination [6]. | ||
| AtTCP16 | Regulate meristem induction and differentiation [91]. Involved in Pollen development [92]. | ||
| AtTCP20 | Involved in cell proliferation, division and differentiation [93]. Regulate leaf pavement cell sizes and senescence [19]. Mediate nitrate foraging by Arabidopsis roots [94]. Influence the absorption of nitrate [74]. | ||
| AtTCP19, 20 | Control leaf senescence [95]. | ||
| AtTCP21 | Regulate circadian clock activity [7]. Plant immunity [82]. | ||
| AtTCP23 | Related to flowering time and plant development [21]. | ||
| AtTCP8 | Regulate SA biosynthesis and signal transduction [96]. | ||
| Gossypium hirsutum | GhTCP14 | Regulate cotton fiber cells development [73]. | |
| Oryza sativa | OsTCP19 | Involved in salinity and drought tolerance [75]. | |
| PCF2 | Involved in salt stress tolerance [78]. | ||
| Phalaenopsis equestris | PePCF10 | Involved in leaf and ovule development [72]. | |
| Chrysanthemum morifolium | CmTCP14 | Inhibited organ size and delayed senescence [71]. | |
| Class II (CIN) | Arabidopsis thaliana | AtTCP17 | Up-regulate phytochrome interaction factors and auxin biosynthesis genes to avoid shade [97]. |
| AtTCP4 | Involved in trichome differentiation and photoperiodic flowering [63,64]. Promote JA biosynthesis involving in leaf development and senescence [81]. | ||
| AtTCP24 | Regulate anther wall development [65]. | ||
| AtTCP3 | Promote flavonoid biosynthesis and negatively regulate auxin response [80]. Regulate flower morphology [56]. | ||
| AtTCP13 | Involved in light response [98]. | ||
| AtTCP2, 3, 4, 10 | Involved in leaf development [41]. Plant immunity [84]. | ||
| Oryza sativa | OsTCP21, PCF6 | Involved in cold stress tolerance and plant defense [66,67]. | |
| Solanum lycopersicum | LA | Involved in leaf development [45]. | |
| Phalaenopsis equestris | PeCIN8 | Regulate ovule, leaf, and petal development [72]. | |
| Physcomitrella patens | PpTCP5 | Negatively regulate sporophyte branching [99]. | |
| Class II (CYC) | Arabidopsis thaliana | AtTCP18 | Suppresses the growth of axillary bud [2]. Repress the floral transition [54]. Plant immunity [89]. |
| AtTCP1 | Influence petioles, rosette leaves, and inflorescent stems development [100]. | ||
| Cucumis melon | CmTCP1 | Involved in development of tendrils from lateral shoots [101]. | |
| Cucumis sativus | TEN | Regulate tendril-less phenotype [102]. | |
| Gerbera hybrida | GhCYC2 | Partake in flower symmetry [66]. | |
| Pisum sativum | PsBRC1 | Regulate shoot branching [57]. | |
| Solanum lycopersicum | SlBRC1b | Suppress shoot branching [58]. | |
| Solanum tuberosum | BRC1a | Regulate lateral branching [103]. | |
| Hordeum vulgare | INT-C | Regulate tillering and fertility of lateral spikelets [104]. | |
| Oryza sativa | FC1 | Influence plant height tillering [1]. | |
| REP1 | Control palea development and floral zygomorphy [67]. | ||
| Sorghum bicolor | SbTB1 | Negatively regulate tillering [105]. | |
| Switchgrass | PvTB1 | Negatively regulate tillering [106]. | |
| Zea mays | BAD1 | Regulate inflorescence architecture [107]. | |
| TB1 | Negatively regulates tillering [9]. | ||
| Tulipa gesneriana | TgTB1 | Suppress the growth of axillary bud [17]. | |
| Chrysanthemum morifolium | CmCYC2c | Regulate petal development [108]. | |
| Lotus japonicus | LjCYC5 | Regulate the symmetry of the inflorescence [68]. |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, M.-M.; Wang, M.-M.; Yang, J.; Wen, J.; Guo, P.-C.; Wu, Y.-W.; Ke, Y.-Z.; Li, P.-F.; Li, J.-N.; Du, H. Evolutionary and Comparative Expression Analyses of TCP Transcription Factor Gene Family in Land Plants. Int. J. Mol. Sci. 2019, 20, 3591. https://doi.org/10.3390/ijms20143591
Liu M-M, Wang M-M, Yang J, Wen J, Guo P-C, Wu Y-W, Ke Y-Z, Li P-F, Li J-N, Du H. Evolutionary and Comparative Expression Analyses of TCP Transcription Factor Gene Family in Land Plants. International Journal of Molecular Sciences. 2019; 20(14):3591. https://doi.org/10.3390/ijms20143591
Chicago/Turabian StyleLiu, Ming-Ming, Mang-Mang Wang, Jin Yang, Jing Wen, Peng-Cheng Guo, Yun-Wen Wu, Yun-Zhuo Ke, Peng-Feng Li, Jia-Na Li, and Hai Du. 2019. "Evolutionary and Comparative Expression Analyses of TCP Transcription Factor Gene Family in Land Plants" International Journal of Molecular Sciences 20, no. 14: 3591. https://doi.org/10.3390/ijms20143591
APA StyleLiu, M.-M., Wang, M.-M., Yang, J., Wen, J., Guo, P.-C., Wu, Y.-W., Ke, Y.-Z., Li, P.-F., Li, J.-N., & Du, H. (2019). Evolutionary and Comparative Expression Analyses of TCP Transcription Factor Gene Family in Land Plants. International Journal of Molecular Sciences, 20(14), 3591. https://doi.org/10.3390/ijms20143591





