Identification of a 119-bp Promoter of the Maize Sulfite Oxidase Gene (ZmSO) That Confers High-Level Gene Expression and ABA or Drought Inducibility in Transgenic Plants
Abstract
1. Introduction
2. Results
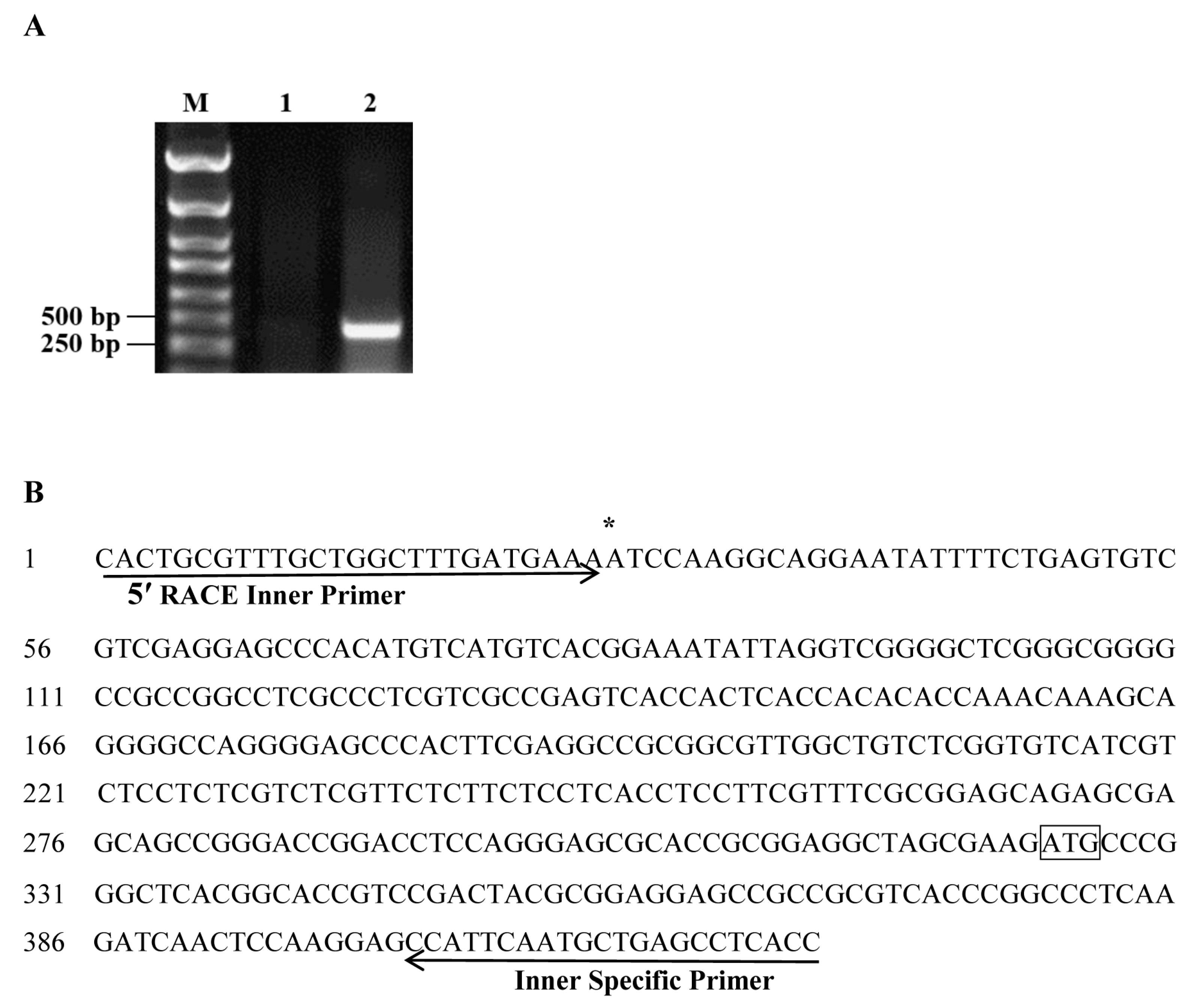
2.1. Isolation and Sequence Analysis of the ZmSO Promoter
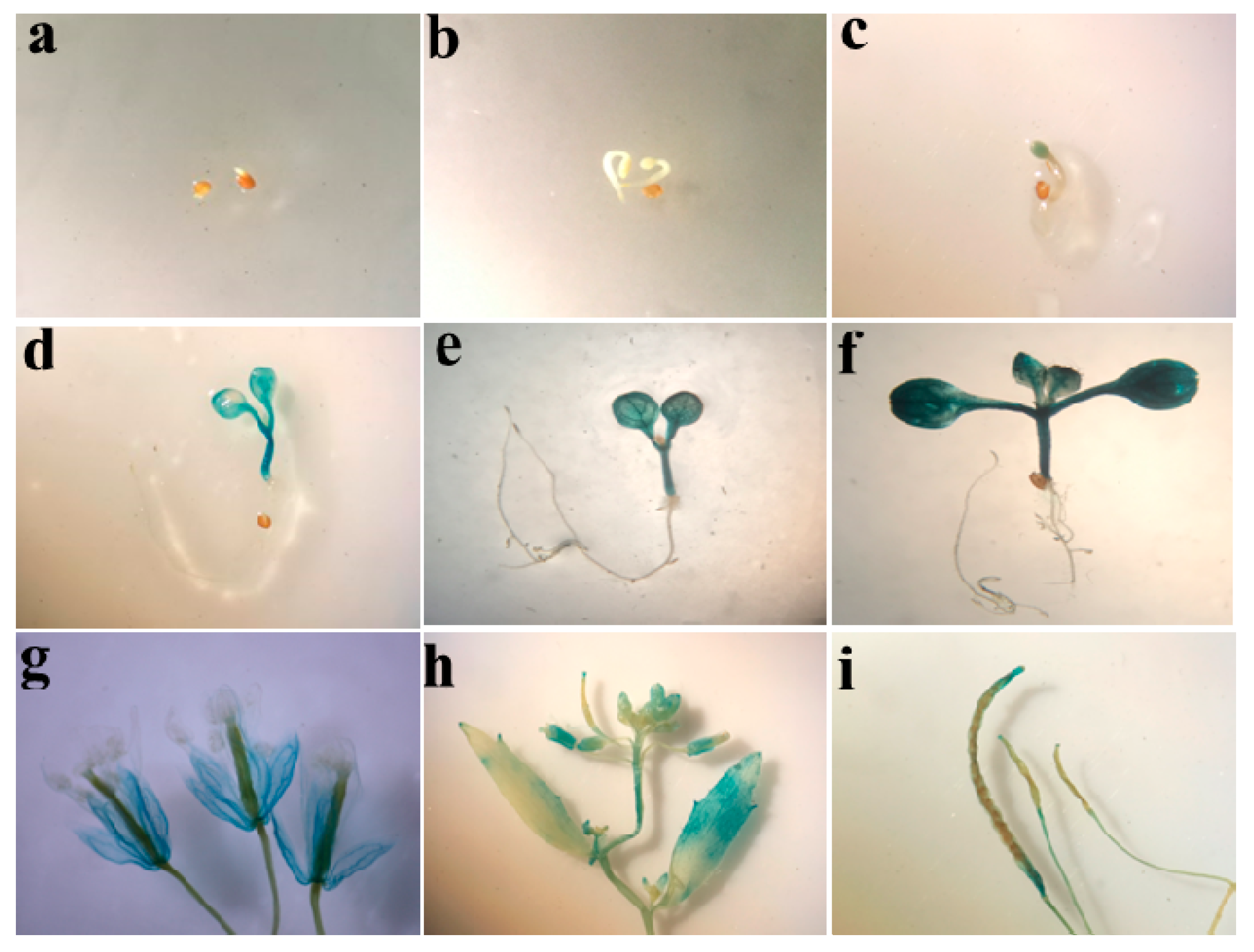
2.2. Promoter Activity of ZmSO in Transgenic Arabidopsis
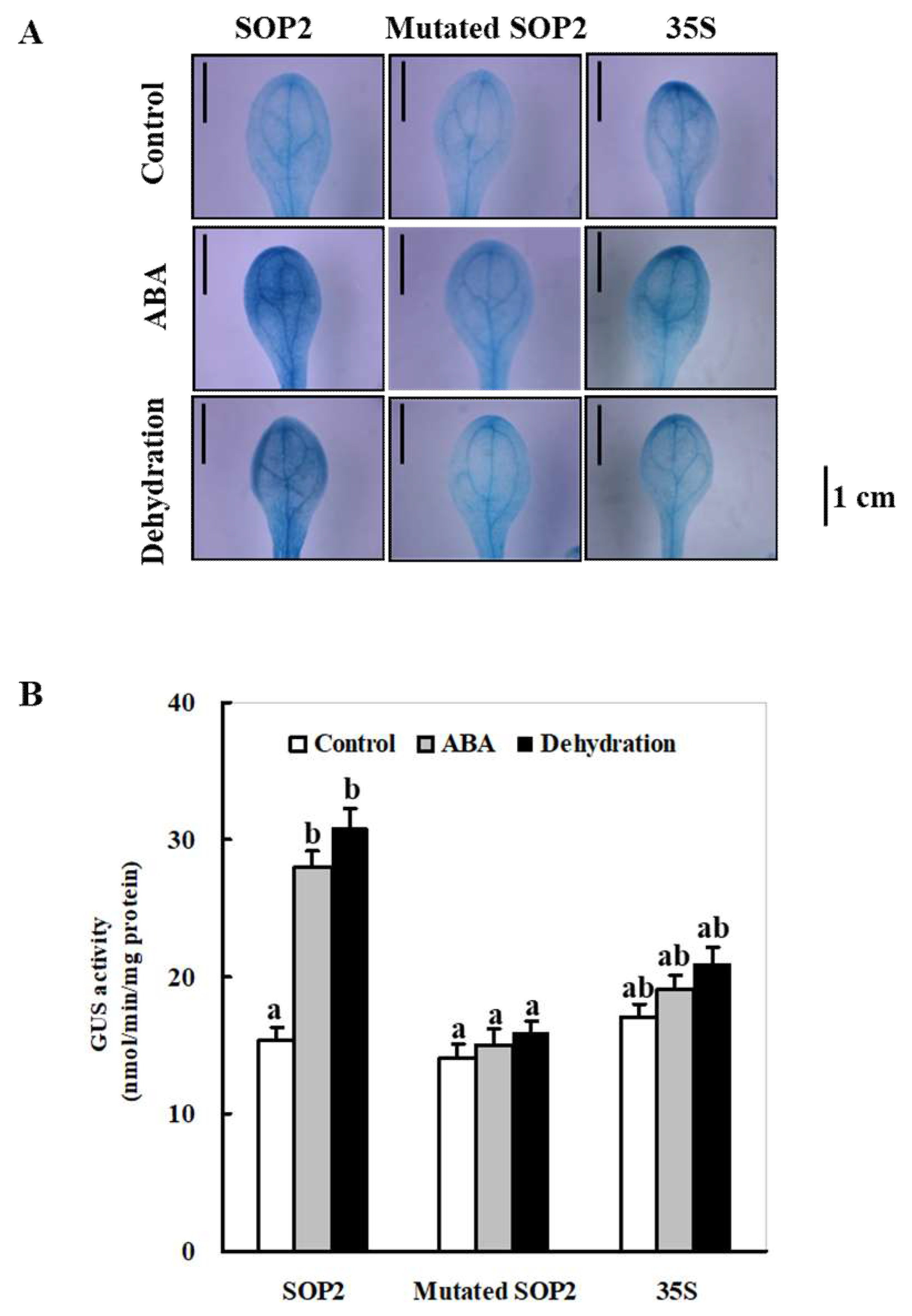
2.3. Changes of the ZmSO Promoter Activity in Arabidopsis in Response to ABA and Drought Stress
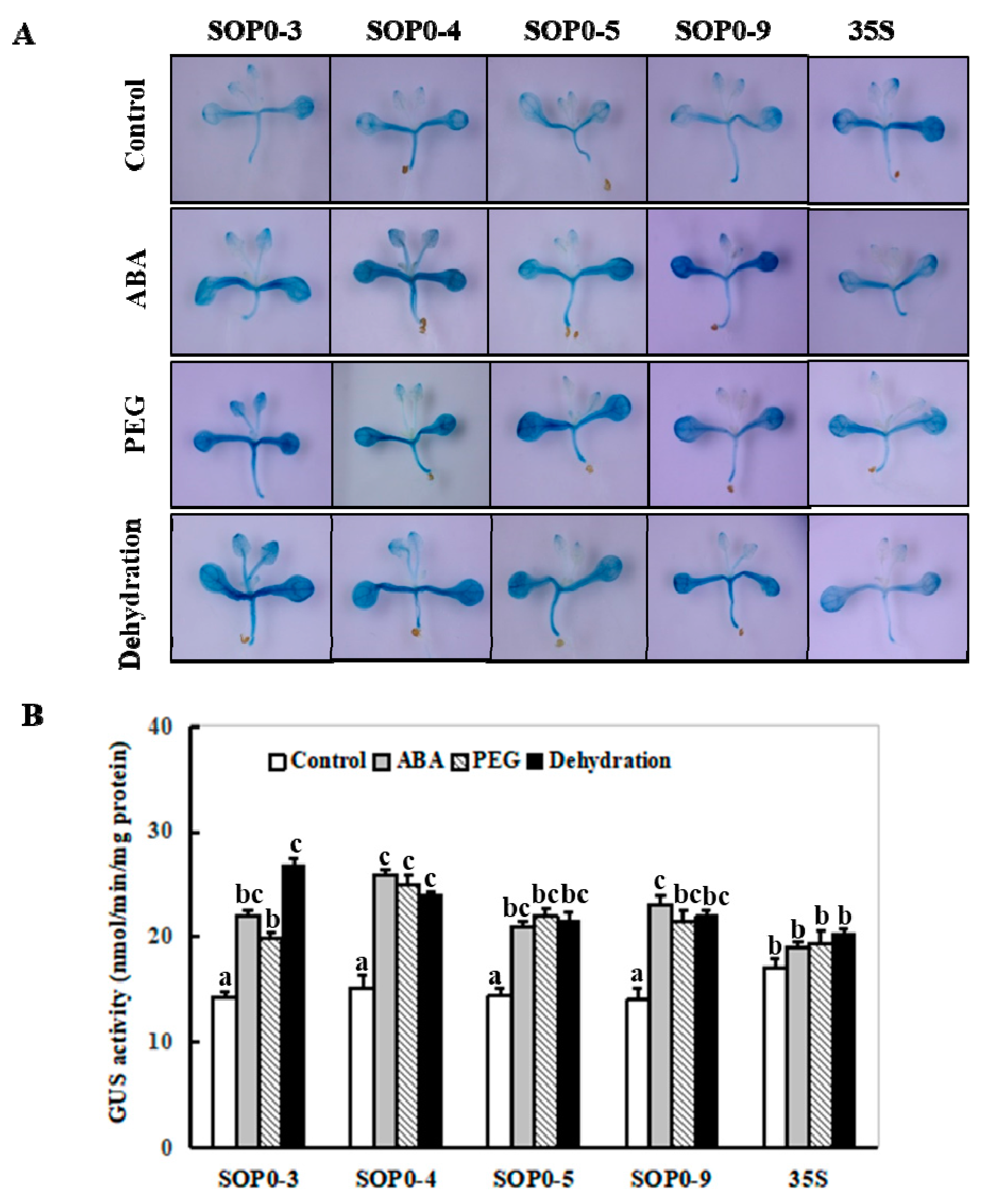
2.4. Identification of the Minimal Region of the ZmSO Promoter Required for Its High-Level Expression
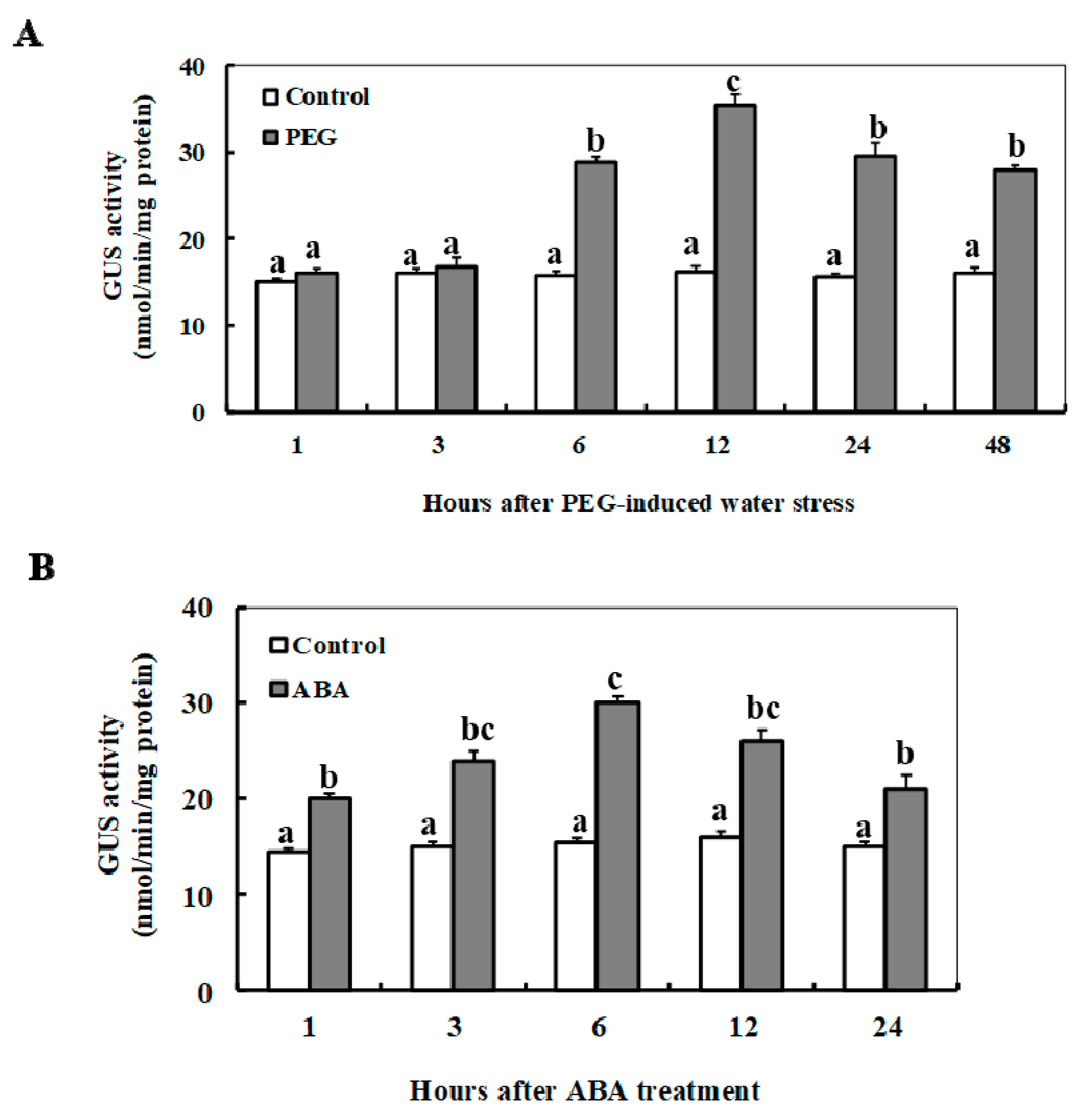
2.5. Dynamic Changes of the Minimal ZmSO Promoter Activity in Arabidopsis in Response to ABA or Drought Stress
2.6. The MBS Element in the ZmSO Promoter Region between −119 and −1 Is Responsible for ABA- and Drought-Stress-Induced Expression
3. Discussion
4. Materials and Methods
4.1. Cloning of the ZmSO Promoter
4.2. Identification of the ZmSO Gene Transcription Start Site
4.3. Bioinformatic Analysis of the ZmSO Promoter Sequence
4.4. Construction of Plasmids Harboring Full-Length and Mutant Promoters Fused to GUS
4.5. Agrobacterium-Mediated Transformation of Arabidopsis Plants
4.6. Histochemical and Quantitative GUS Assays
4.7. ABA, Dehydration, and Osmotic Stress Treatments
4.8. Transient GUS Expression Assays in Tobacco Seedlings
4.9. Quantitative Real-Time PCR
4.10. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Hu, H.; Xiong, L. Genetic Engineering and Breeding of Drought-Resistant Crops. Annu. Rev. Plant Boil. 2014, 65, 715–741. [Google Scholar] [CrossRef] [PubMed]
- Singh, B.; Bohra, A.; Mishra, S.; Joshi, R.; Pandey, S. Embracing new-generation ‘omics’ tools to improve drought tolerance in cereal and food-legume crops. Boil. Plant. 2015, 59, 413–428. [Google Scholar] [CrossRef]
- Tester, M.; Langridge, P. Breeding Technologies to Increase Crop Production in a Changing World. Science 2010, 327, 818–822. [Google Scholar] [CrossRef] [PubMed]
- Potenza, C.; Aleman, L.; Sengupta-Gopalan, C. Targeting transgene expression in research, agricultural, and environmental applications: Promoters used in plant transformation. In Vitro Cell. Dev. Boil. Anim. 2004, 40, 1–22. [Google Scholar] [CrossRef]
- Han, Y.J.; Kim, Y.M.; Hwang, O.J.; Kim, J.I. Characterization of a small constitutive promoter from Arabidopsis translationally controlled tumor protein (AtTCTP) gene for plant transformation. Plant Cell Rep. 2015, 34, 265–275. [Google Scholar] [CrossRef] [PubMed]
- Tao, Y.B.; He, L.L.; Niu, L.J.; Xu, Z.F. Isolation and characterization of an ubiquitin extension protein gene (JcUEP) promoter from Jatropha curcas. Planta 2015, 241, 823–836. [Google Scholar] [CrossRef] [PubMed]
- Yamaguchi-Shinozaki, K.; Shinozaki, K. Characterization of the expression of a desiccation-responsive rd29 gene of Arabidopsis thaliana and analysis of its promoter in transgenic plants. Mol. Genet. Genom. 1993, 236, 331–340. [Google Scholar] [CrossRef]
- Odell, J.T.; Nagy, F.; Chua, N.-H. Identification of DNA sequences required for activity of the cauliflower mosaic virus 35S promoter. Nature 1985, 313, 810–812. [Google Scholar] [CrossRef] [PubMed]
- Luth, D.; Blankenship, K.M.; Anderson, O.D.; Blechl, A.E. Activity of a maize ubiquitin promoter in transgenic rice. Plant Mol. Boil. 1993, 23, 567–581. [Google Scholar]
- Sinha, N.R.; Williams, R.E.; Hake, S. Overexpression of the maize homeo box gene, KNOTTED-I, causes a switch from determinate to indeterminate cell fates. Gene Dev. 1993, 7, 787–795. [Google Scholar] [CrossRef]
- Cheon, B.Y.; Kim, H.J.; Oh, K.H.; Bahn, S.C.; Ahn, J.H.; Choi, J.W.; Ok, S.H.; Bae, J.M.; Shin, J.S. Overexpression of human erythropoietin (EPO) affects plant morphologies: Retarded vegetative growth in tobacco and male sterility in tobacco and Arabidopsis. Transgenic Res. 2004, 13, 541–549. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Ren, W.; Zhi, D.; Wang, L.; Xia, G. Arabidopsis DREB1A/CBF3 bestowed transgenic tall fescue increased tolerance to drought stress. Plant Cell Rep. 2007, 26, 1521–1528. [Google Scholar] [CrossRef] [PubMed]
- Pan, Y.; Ma, X.; Liang, H.; Zhao, Q.; Zhu, D.; Yu, J. Spatial and temporal activity of the foxtail millet (Setaria italica) seed-specific promoter pF128. Planta 2015, 241, 57–67. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Yin, H.; Li, D.; Zhu, W.; Li, Q. Functional analysis of BADH gene promoter from Suaeda liaotungensis K. Plant Cell Rep. 2008, 27, 585–592. [Google Scholar] [CrossRef]
- Rai, M.; He, C.; Wu, R. Comparative functional analysis of three abiotic stress-inducible promoters in transgenic rice. Transgenic Res. 2009, 18, 787–799. [Google Scholar] [CrossRef] [PubMed]
- Tavakol, E.; Sardaro, M.L.S.; J, V.S.; Rossini, L.; Porceddu, E. Isolation, promoter analysis and expression profile of Dreb2 in response to drought stress in wheat ancestors. Gene 2014, 549, 24–32. [Google Scholar] [CrossRef]
- Alscher, R.G. Biosynthesis and antioxidant function of glutathione in plants. Physiol. Plant. 1989, 77, 457–464. [Google Scholar] [CrossRef]
- Chan, K.X.; Wirtz, M.; Phua, S.Y.; Estavillo, G.M.; Pogson, B.J. Balancing metabolites in drought: The sulfur assimilation conundrum. Trends Plant Sci. 2013, 18, 18–29. [Google Scholar] [CrossRef]
- Ahmad, N.; Malagoli, M.; Wirtz, M.; Hell, R. Drought stress in maize causes differential acclimation responses of glutathione and sulfur metabolism in leaves and roots. BMC Plant Boil. 2016, 16, 247. [Google Scholar] [CrossRef]
- Brychkova, G.; Xia, Z.; Yang, G.; Yesbergenova, Z.; Zhang, Z.; Davydov, O.; Fluhr, R.; Sagi, M. Sulfite oxidase protects plants against sulfur dioxide toxicity. Plant J. 2007, 50, 696–709. [Google Scholar] [CrossRef]
- Xia, Z.; Sun, K.; Wang, M.; Wu, K.; Zhang, H.; Wu, J. Overexpression of a Maize Sulfite Oxidase Gene in Tobacco Enhances Tolerance to Sulfite Stress via Sulfite Oxidation and CAT-Mediated H2O2 Scavenging. PLoS ONE 2012, 7, e37383. [Google Scholar] [CrossRef] [PubMed]
- Xia, Z.; Xu, Z.; Wei, Y.; Wang, M. Overexpression of the Maize Sulfite Oxidase Increases Sulfate and GSH Levels and Enhances Drought Tolerance in Transgenic Tobacco. Front. Plant Sci. 2018, 9, 298. [Google Scholar] [CrossRef] [PubMed]
- Joshi, C. An inspection of the domain between putative TATA box and translation start site in 79 plant genes. Nucleic Acids Res. 1987, 15, 6643–6653. [Google Scholar] [CrossRef] [PubMed]
- Shinozaki, K.; Yamaguchi-Shinozaki, K. Gene networks involved in drought stress response and tolerance. J. Exp. Bot. 2007, 58, 221–227. [Google Scholar] [CrossRef] [PubMed]
- Bhuria, M.; Goel, P.; Kumar, S.; Singh, A.K. The Promoter of AtUSP is Co-regulated by Phytohormones and Abiotic Stresses in Arabidopsis thaliana. Front. Plant Sci. 2016, 7, 1957. [Google Scholar] [CrossRef] [PubMed]
- Zavallo, D.; Bilbao, M.L.; Hopp, H.E.; Heinz, R. Isolation and functional characterization of two novel seed-specific promoters from sunflower (Helianthus annuus L.). Plant Cell Rep. 2010, 29, 239–248. [Google Scholar] [CrossRef] [PubMed]
- Koia, J.; Moyle, R.; Hendry, C.; Lim, L.; Botella, J.R. Pineapple translation factor SUI1 and ribosomal protein L36 promoters drive constitutive transgene expression patterns in Arabidopsis thaliana. Plant Mol. Biol. 2013, 81, 327–336. [Google Scholar] [CrossRef]
- Joung, Y.H.; Kamo, K. Expression of a polyubiquitin promoter isolated from Gladiolus. Plant Cell Rep. 2006, 25, 1081–1088. [Google Scholar] [CrossRef]
- Kamo, K.; Kim, A.-Y.; Park, S.H.; Joung, Y.H. The 5′UTR-intron of the Gladiolus polyubiquitin promoter GUBQ1 enhances translation efficiency in Gladiolus and Arabidopsis. BMC Plant Boil. 2012, 12, 79. [Google Scholar] [CrossRef]
- Schledzewski, K.; Mendel, R.R. Quantitative transient gene expression: Comparison of the promoters for maize polyubiquitin1, rice actin1, maize-derivedEmu andCaMV 35S in cells of barley, maize and tobacco. Transgenic Res. 1994, 3, 249–255. [Google Scholar] [CrossRef]
- Sun, L.R.; Li, Y.P.; Miao, W.W.; Piao, T.T.; Hao, Y.; Hao, F.S. NADK2 positively modulates abscisic acid-induced stomatal closure by affecting accumulation of H2O2, Ca2+ and nitric oxide in Arabidopsis guard cells. Plant Sci. 2017, 262, 81–90. [Google Scholar] [CrossRef] [PubMed]
- Lescot, M. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef] [PubMed]
- Clough, S.J.; Bent, A.F. Floral dip: A simplified method for Agrobaterium—Mediated transformation of Arabidopsis thaliana. Plant J. 1998, 16, 735–743. [Google Scholar] [CrossRef] [PubMed]
- Bai, L.; Ma, X.; Zhang, G.; Song, S.; Zhou, Y.; Gao, L.; Miao, Y.; Song, C.-P. A receptor-like kinase mediates ammonium homeostasis and is important for the polar growth of root hairs in Arabidopsis. Plant Cell 2014, 26, 1497–1511. [Google Scholar] [CrossRef] [PubMed]
- Jefferson, R.A.; Kavanagh, T.A.; Bevan, M.W. GUS fusions: Beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 1987, 6, 3901–3907. [Google Scholar] [CrossRef] [PubMed]
- Xia, Z.; Huo, Y.; Wei, Y.; Chen, Q.; Xu, Z.; Zhang, W. The Arabidopsis LYST INTERACTING PROTEIN 5 Acts in Regulating Abscisic Acid Signaling and Drought Response. Front. Plant Sci. 2016, 7, 758. [Google Scholar] [CrossRef] [PubMed]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Zheng, Y.; Chen, Z.; Ma, L.; Liao, C. The Ubiquitin E3 Ligase RHA2b Promotes Degradation of MYB30 in Abscisic Acid Signaling. Plant Physiol. 2018, 178, 428–440. [Google Scholar] [CrossRef]
- Zhang, Y.; Yang, C.; Li, Y.; Zheng, N.; Chen, H.; Zhao, Q.; Gao, T.; Guo, H.; Xie, Q. SDIR1 is a RING finger E3 ligase that positively regulates stress-responsive abscisic acid signaling in Arabidopsis. Plant Cell 2007, 19, 1912–1929. [Google Scholar] [CrossRef]
- Xia, Z.; Su, X.; Liu, J.; Wang, M. The RING-H2 finger gene 1 (RHF1) encodes an E3 ubiquitin ligase and participates in drought stress response in Nicotiana tabacum. Genetica 2013, 141, 11–21. [Google Scholar] [CrossRef]
- Xu, F.C.; Liu, H.L.; Xu, Y.Y.; Zhao, J.R.; Guo, Y.W.; Long, L.; Gao, W.; Song, C.P. Heterogeneous expression of the cotton R2R3-MYB transcription factor GbMYB60 increases salt sensitivity in transgenic Arabidopsis. Plant Cell Tissue Org. Cult. 2018, 133, 15–25. [Google Scholar] [CrossRef]
- Qi, J.; Song, C.-P.; Wang, B.; Zhou, J.; Kangasjärvi, J.; Zhu, J.-K.; Gong, Z. Reactive oxygen species signaling and stomatal movement in plant responses to drought stress and pathogen attack. J. Integr. Plant Boil. 2018, 60, 805–826. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; He, J.; Zhao, Y.; Wu, T.; Zhou, X.; Ding, Y.; Kong, L.; Wang, X.; Wang, Y.; Li, J.; et al. EAR1 Negatively Regulates ABA Signaling by Enhancing 2C Protein Phosphatase Activity. Plant Cell 2018, 30, 815–834. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]


© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xu, Z.; Wang, M.; Guo, Z.; Zhu, X.; Xia, Z. Identification of a 119-bp Promoter of the Maize Sulfite Oxidase Gene (ZmSO) That Confers High-Level Gene Expression and ABA or Drought Inducibility in Transgenic Plants. Int. J. Mol. Sci. 2019, 20, 3326. https://doi.org/10.3390/ijms20133326
Xu Z, Wang M, Guo Z, Zhu X, Xia Z. Identification of a 119-bp Promoter of the Maize Sulfite Oxidase Gene (ZmSO) That Confers High-Level Gene Expression and ABA or Drought Inducibility in Transgenic Plants. International Journal of Molecular Sciences. 2019; 20(13):3326. https://doi.org/10.3390/ijms20133326
Chicago/Turabian StyleXu, Ziwei, Meiping Wang, Ziting Guo, Xianfeng Zhu, and Zongliang Xia. 2019. "Identification of a 119-bp Promoter of the Maize Sulfite Oxidase Gene (ZmSO) That Confers High-Level Gene Expression and ABA or Drought Inducibility in Transgenic Plants" International Journal of Molecular Sciences 20, no. 13: 3326. https://doi.org/10.3390/ijms20133326
APA StyleXu, Z., Wang, M., Guo, Z., Zhu, X., & Xia, Z. (2019). Identification of a 119-bp Promoter of the Maize Sulfite Oxidase Gene (ZmSO) That Confers High-Level Gene Expression and ABA or Drought Inducibility in Transgenic Plants. International Journal of Molecular Sciences, 20(13), 3326. https://doi.org/10.3390/ijms20133326



