Quantitative Biology of Human Shelterin and Telomerase: Searching for the Weakest Point
Abstract
1. Introduction
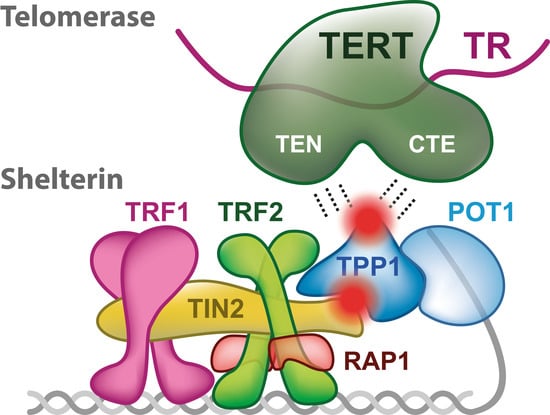
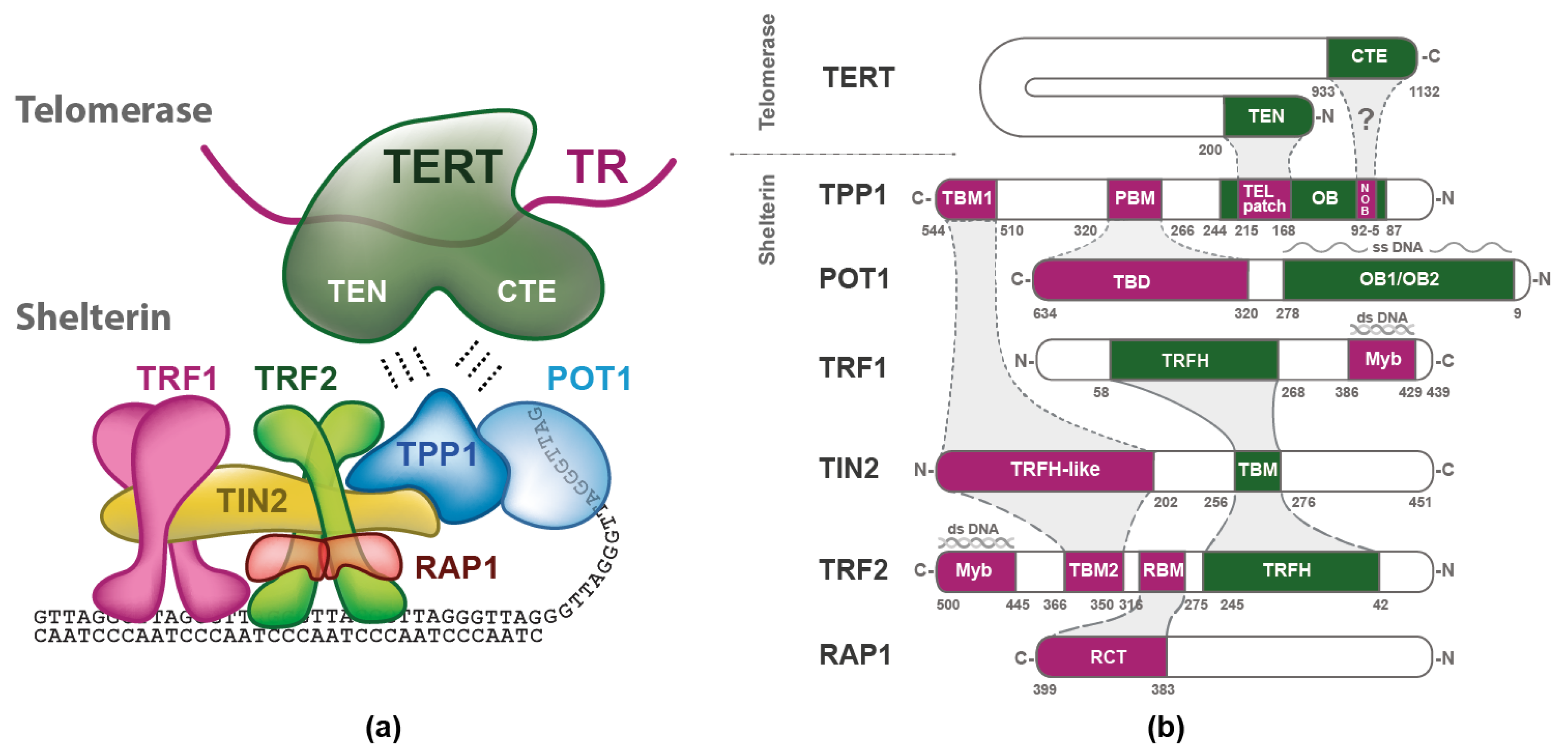
2. Shelterin Structure and Binding Features
3. Telomerase
3.1. Structure of TERT and TR
3.2. The Function of TERT and TR
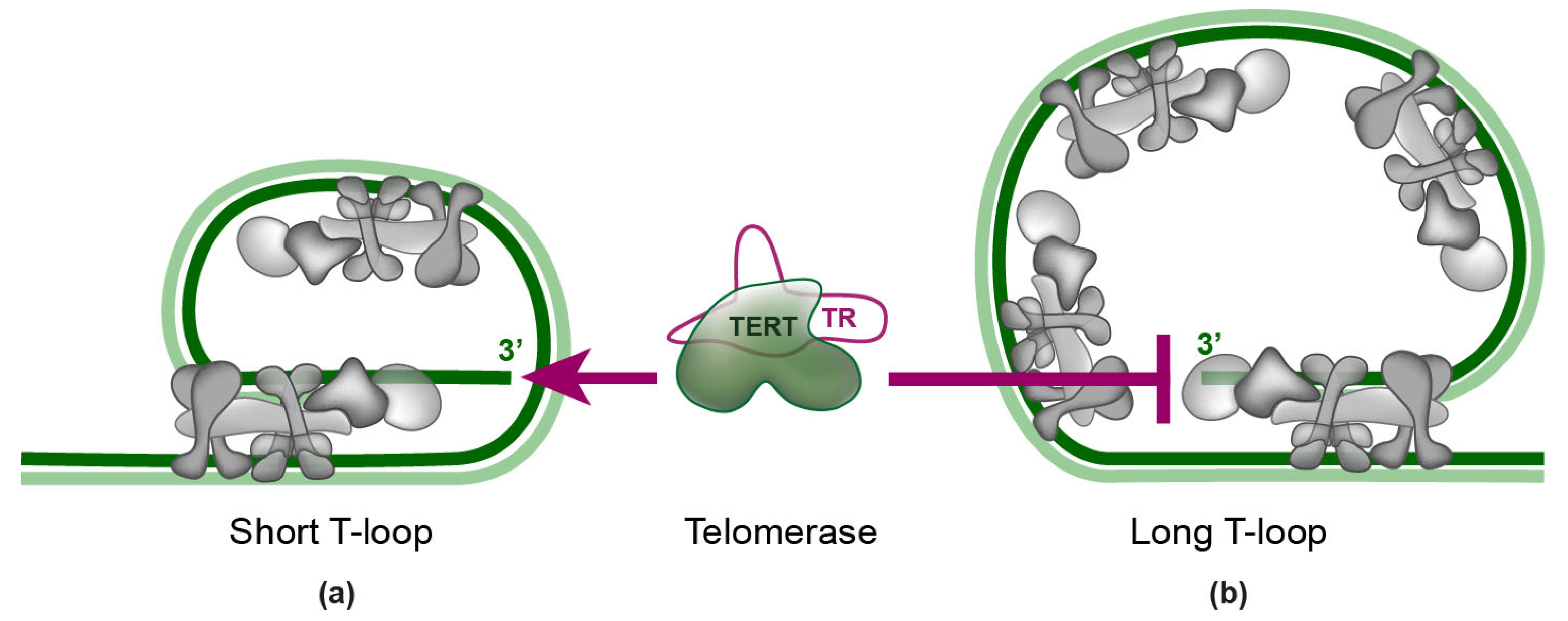
4. Telomerase Recruitment via Shelterin
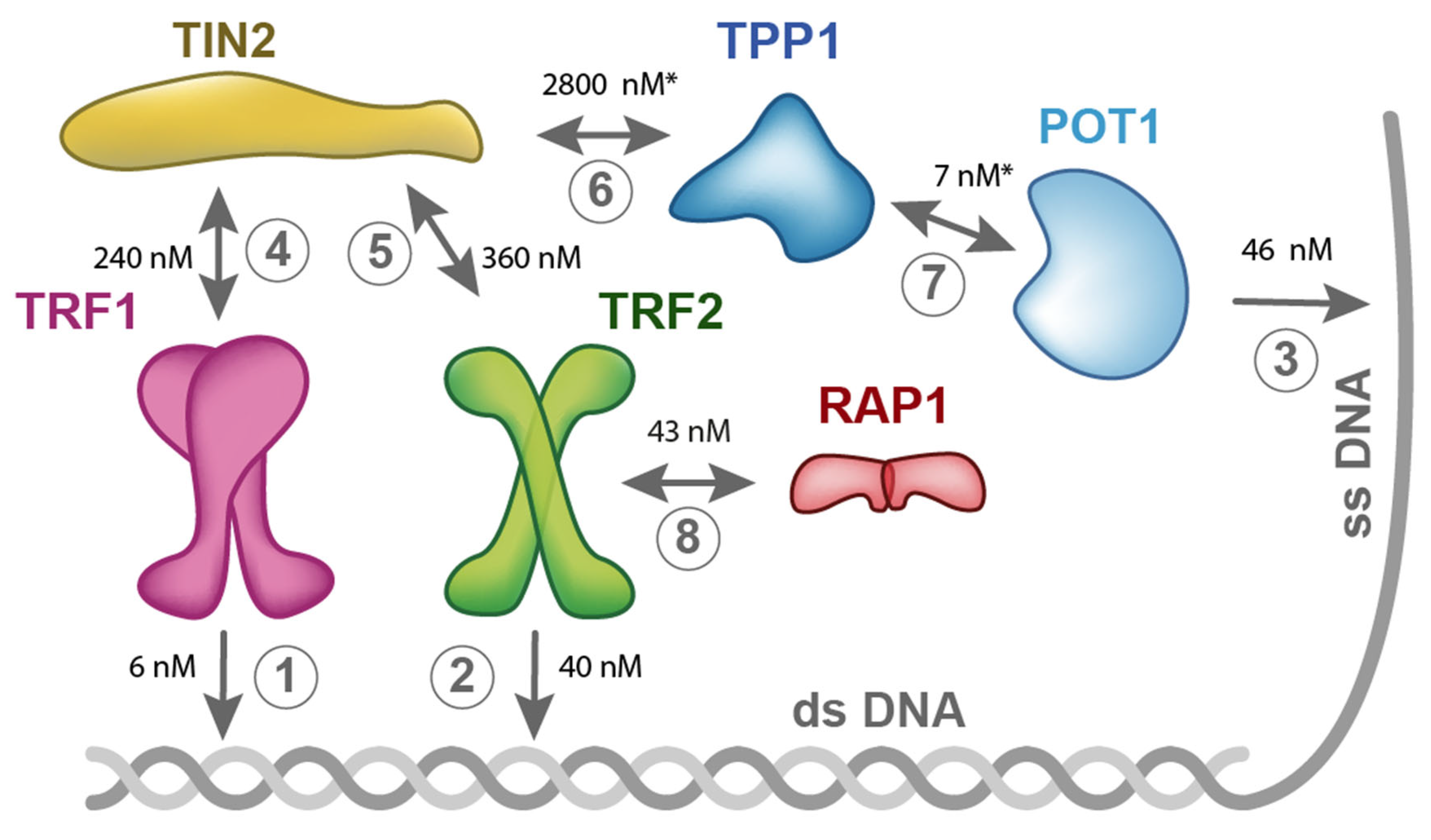
5. Quantitative Description of Interactions within Shelterin
6. Implication for Anticancer Drug Design
6.1. GRN163L (Imetelstat)
6.2. Clinical Studies of GRN163L
6.3. T-oligo Cancer Therapy
7. Perspectives
Funding
Acknowledgments
Conflicts of Interest
References
- Levy, M.Z.; Allsopp, R.C.; Futcher, A.B.; Greider, C.W.; Harley, C.B. Telomere end-replication problem and cell aging. J. Mol. Biol. 1992, 225, 951–960. [Google Scholar] [CrossRef]
- Erdel, F.; Kratz, K.; Willcox, S.; Griffith, J.D.; Greene, E.C.; de Lange, T. Telomere recognition and assembly mechanism of mammalian shelterin. Cell Rep. 2017, 18, 41–53. [Google Scholar] [CrossRef] [PubMed]
- Janovič, T.; Stojaspal, M.; Veverka, P.; Horáková, D.; Hofr, C. Human telomere repeat binding factor TRF1 replaces TRF2 bound to shelterin core hub TIN2 when TPP1 is absent. J. Mol. Biol. 2019. [Google Scholar] [CrossRef] [PubMed]
- De Lange, T. Shelterin-mediated telomere protection. Annu. Rev. Genet. 2018, 52, 223–247. [Google Scholar] [CrossRef] [PubMed]
- Broccoli, D.; Smogorzewska, A.; Chong, L.; de Lange, T. Human telomeres contain two distinct Myb-related proteins, TRF1 and TRF2. Nat. Genet. 1997, 17, 231–235. [Google Scholar] [CrossRef] [PubMed]
- Janouskova, E.; Necasova, I.; Pavlouskova, J.; Zimmermann, M.; Hluchy, M.; Marini, V.; Novakova, M.; Hofr, C. Human RAP1 modulates TRF2 attraction to telomeric DNA. Nucleic. Acids Res. 2015, 43, 2691–2700. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Yang, Y.; van Overbeek, M.; Donigian, J.; Baciu, P.; de Lange, T.; Lei, M. A shared docking motif in TRF1 and TRF2 used for differential recruitment of telomeric proteins. Science 2008, 319, 1092–1096. [Google Scholar] [CrossRef] [PubMed]
- Ye, J.Z.S.; Donigian, J.R.; van Overbeek, M.; Loayza, D.; Luo, Y.; Krutchinsky, A.N.; Chait, B.T.; de Lange, T. TIN2 binds TRF1 and TRF2 simultaneously and stabilizes the TRF2 complex on telomeres. J. Biol. Chem. 2004, 279, 47264–47271. [Google Scholar] [CrossRef] [PubMed]
- Grill, S.; Tesmer, V.M.; Nandakumar, J. The N terminus of the OB domain of telomere protein TPP1 is critical for telomerase action. Cell Rep. 2018, 22, 1132–1140. [Google Scholar] [CrossRef]
- Hu, C.; Rai, R.; Huang, C.; Broton, C.; Long, J.; Xu, Y.; Xue, J.; Lei, M.; Chang, S.; Chen, Y. Structural and functional analyses of the mammalian TIN2-TPP1-TRF2 telomeric complex. Cell Res. 2017, 27, 1485–1502. [Google Scholar] [CrossRef]
- Lei, M.; Podell, E.R.; Cech, T.R. Structure of human POT1 bound to telomeric single-stranded DNA provides a model for chromosome end-protection. Nat. Struct. Mol. Biol. 2004, 11, 1223–1229. [Google Scholar] [CrossRef] [PubMed]
- Baumann, P.; Cech, T.R. POT1, the putative telomere end-binding protein in fission yeast and humans. Science 2001, 292, 1171–1175. [Google Scholar] [CrossRef] [PubMed]
- Loayza, D.; Parsons, H.; Donigian, J.; Hoke, K.; de Lange, T. DNA binding features of human POT1—A nonamer 5′-TAGGGTTAG-3′ minimal binding site, sequence specificity, and internal binding to multimeric sites. J. Biol. Chem. 2004, 279, 13241–13248. [Google Scholar] [CrossRef] [PubMed]
- Lim, C.J.; Zaug, A.J.; Kim, H.J.; Cech, T.R. Reconstitution of human shelterin complexes reveals unexpected stoichiometry and dual pathways to enhance telomerase processivity. Nat. Commun. 2017, 8, 1075. [Google Scholar] [CrossRef] [PubMed]
- Revzin, A. The Biology of Nonspecific DNA–Protein Interactions; CRC Press: Boca Raton, FL, USA, 1990. [Google Scholar]
- Griffith, J.D.; Comeau, L.; Rosenfield, S.; Stansel, R.M.; Bianchi, A.; Moss, H.; de Lange, T. Mammalian telomeres end in a large duplex loop. Cell 1999, 97, 503–514. [Google Scholar] [CrossRef]
- Doksani, Y.; Wu, J.Y.; de Lange, T.; Zhuang, X. Super-resolution fluorescence imaging of telomeres reveals TRF2-dependent t-loop formation. Cell 2013, 155, 345–356. [Google Scholar] [CrossRef] [PubMed]
- Nandakumar, J.; Cech, T.R. Finding the end: Recruitment of telomerase to telomeres. Nat. Rev. Mol. Cell Biol. 2013, 14, 69–82. [Google Scholar] [CrossRef]
- He, Q.; Zeng, P.; Tan, J.-H.; Ou, T.-M.; Gu, L.-Q.; Huang, Z.-S.; Li, D. G-quadruplex-mediated regulation of telomere binding protein POT1 gene expression. BBA 2014, 1840, 2222–2233. [Google Scholar] [CrossRef]
- De Lange, T. Shelterin: The protein complex that shapes and safeguards human telomeres. Genes Dev. 2005, 19, 2100–2110. [Google Scholar] [CrossRef]
- Chen, J.L.; Blasco, M.A.; Greider, C.W. Secondary structure of vertebrate telomerase RNA. Cell 2000, 100, 503–514. [Google Scholar] [CrossRef]
- Mitchell, J.R.; Collins, K. Human telomerase activation requires two independent interactions between telomerase RNA and telomerase reverse transcriptase. Mol. Cell 2000, 6, 361–371. [Google Scholar] [CrossRef]
- Chen, J.-L.; Opperman, K.K.; Greider, C.W. A critical stem-loop structure in the CR4-CR5 domain of mammalian telomerase RNA. Nucleic. Acids Res. 2002, 30, 592–597. [Google Scholar] [CrossRef] [PubMed]
- Bley, C.J.; Qi, X.; Rand, D.P.; Borges, C.R.; Nelson, R.W.; Chen, J.J.-L. RNA-protein binding interface in the telomerase ribonucleoprotein. PNAS 2011, 108, 20333–20338. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.-L.; Greider, C.W. Determinants in mammalian telomerase RNA that mediate enzyme processivity and cross-species incompatibility. EMBO J. 2003, 22, 304–314. [Google Scholar] [CrossRef] [PubMed]
- Lai, C.K.; Miller, M.C.; Collins, K. Roles for RNA in telomerase nucleotide and repeat addition processivity. Mol. Cell 2003, 11, 1673–1683. [Google Scholar] [CrossRef]
- Kim, N.W.; Piatyszek, M.A.; Prowse, K.R.; Harley, C.B.; West, M.D.; Ho, P.L.C.; Coviello, G.M.; Wright, W.E.; Weinrich, S.L.; Shay, J.W. Specific association of human telomerase activity with immortal cells and cancer. Science 1994, 266, 2011–2015. [Google Scholar] [CrossRef]
- Horn, S.; Figl, A.; Rachakonda, P.S.; Fischer, C.; Sucker, A.; Gast, A.; Kadel, S.; Moll, I.; Nagore, E.; Hemminki, K.; et al. TERT promoter mutations in familial and sporadic melanoma. Science 2013, 339, 959–961. [Google Scholar] [CrossRef]
- Huang, F.W.; Hodis, E.; Xu, M.J.; Kryukov, G.V.; Chin, L.; Garraway, L.A. Highly recurrent TERT promoter mutations in human melanoma. Science 2013, 339, 957–959. [Google Scholar] [CrossRef]
- Dokal, I. Dyskeratosis congenita. Hematol. Am. Soc. Hematol. Educ. Program 2011, 2011, 480–486. [Google Scholar] [CrossRef]
- Lingner, J.; Hughes, T.R.; Shevchenko, A.; Mann, M.; Lundblad, V.; Cech, T.R. Reverse transcriptase motifs in the catalytic subunit of telomerase. Science 1997, 276, 561–567. [Google Scholar] [CrossRef]
- Meyerson, M.; Counter, C.M.; Eaton, E.N.; Ellisen, L.W.; Steiner, P.; Caddle, S.D.; Ziaugra, L.; Beijersbergen, R.L.; Davidoff, M.J.; Liu, Q.; et al. hEST2, the putative human telomerase catalytic subunit gene, is up-regulated in tumor cells and during immortalization. Cell 1997, 90, 785–795. [Google Scholar] [CrossRef]
- Wu, R.A.; Tam, J.; Collins, K. DNA-binding determinants and cellular thresholds for human telomerase repeat addition processivity. EMBO J. 2017, 36, 1908–1927. [Google Scholar] [CrossRef] [PubMed]
- Mitchell, J.R.; Wood, E.; Collins, K. A telomerase component is defective in the human disease dyskeratosis congenita. Nature 1999, 402, 551–555. [Google Scholar] [CrossRef] [PubMed]
- Egan, E.D.; Collins, K. Biogenesis of telomerase ribonucleoproteins. RNA 2012, 18, 1747–1759. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, T.H.D.; Tam, J.; Wu, R.A.; Greber, B.J.; Toso, D.; Nogales, E.; Collins, K. Cryo-em structure of substrate-bound human telomerase holoenzyme. Nature 2018, 557, 190–195. [Google Scholar] [CrossRef] [PubMed]
- Alves, D.; Li, H.; Codrington, R.; Orte, A.; Ren, X.; Klenerman, D.; Balasubramanian, S. Single-molecule analysis of human telomerase monomer. Nat. Chem. Biol. 2008, 4, 287. [Google Scholar] [CrossRef] [PubMed]
- Jiang, J.; Miracco, E.J.; Hong, K.; Eckert, B.; Chan, H.; Cash, D.D.; Min, B.; Zhou, Z.H.; Collins, K.; Feigon, J. The architecture of tetrahymena telomerase holoenzyme. Nature 2013, 496, 187–192. [Google Scholar] [CrossRef]
- Egan, E.D.; Collins, K. Specificity and stoichiometry of subunit interactions in the human telomerase holoenzyme assembled in vivo. Mol. Cell. Biol. 2010, 30, 2775–2786. [Google Scholar] [CrossRef]
- Wu, R.A.; Dagdas, Y.S.; Yilmaz, S.T.; Yildiz, A.; Collins, K. Single-molecule imaging of telomerase reverse transcriptase in human telomerase holoenzyme and minimal RNP complexes. Elife 2015, 4. [Google Scholar] [CrossRef] [PubMed]
- Lue, N.F. A physical and functional constituent of telomerase anchor site. J. Biol. Chem. 2005, 280, 26586–26591. [Google Scholar] [CrossRef]
- Moriarty, T.J.; Ward, R.J.; Taboski, M.A.S.; Autexier, C. An anchor site-type defect in human telomerase that disrupts telomere length maintenance and cellular immortalization. Mol. Biol. Cell 2005, 16, 3152–3161. [Google Scholar] [CrossRef] [PubMed]
- Jurczyluk, J.; Nouwens, A.S.; Holien, J.K.; Adams, T.E.; Lovrecz, G.O.; Parker, M.W.; Cohen, S.B.; Bryan, T.M. Direct involvement of the TEN domain at the active site of human telomerase. Nucleic Acids Res. 2011, 39, 1774–1788. [Google Scholar] [CrossRef] [PubMed]
- Armbruster, B.N.; Banik, S.S.; Guo, C.; Smith, A.C.; Counter, C.M. N-terminal domains of the human telomerase catalytic subunit required for enzyme activity in vivo. Mol. Cell. Biol. 2001, 21, 7775–7786. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, J.C.; Zaug, A.J.; Kufer, R.; Cech, T.R. Dynamics of human telomerase recruitment depend on template-telomere base pairing. Mol. Biol. Cell 2018, 29, 869–880. [Google Scholar] [CrossRef] [PubMed]
- Podlevsky, J.D.; Chen, J.J.-L. Evolutionary perspectives of telomerase RNA structure and function. RNA Biol. 2016, 13, 720–732. [Google Scholar] [CrossRef] [PubMed]
- Chan, H.; Wang, Y.; Feigon, J. Progress in human and tetrahymena telomerase structure determination. Annu. Rev. Biophys. 2017, 46, 199–225. [Google Scholar] [CrossRef] [PubMed]
- Theimer, C.A.; Blois, C.A.; Feigon, J. Structure of the human telomerase RNA pseudoknot reveals conserved tertiary interactions essential for function. Mol. Cell 2005, 17, 671–682. [Google Scholar] [CrossRef]
- Shefer, K.; Brown, Y.; Gorkovoy, V.; Nussbaum, T.; Ulyanov, N.B.; Tzfati, Y. A triple helix within a pseudoknot is a conserved and essential element of telomerase RNA. Mol. Cell. Biol. 2007, 27, 2130–2143. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Qiao, F.; Cech, T.R. Triple-helix structure in telomerase RNA contributes to catalysis. Nat. Struct. Mol. Biol. 2008, 15, 634–640. [Google Scholar] [CrossRef]
- Schmidt, J.C.; Zaug, A.J.; Cech, T.R. Live cell imaging reveals the dynamics of telomerase recruitment to telomeres. Cell 2016. [Google Scholar] [CrossRef]
- Schmidt, J.C.; Dalby, A.B.; Cech, T.R. Identification of human TERT elements necessary for telomerase recruitment to telomeres. Elife 2014, 3. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Sfeir, A.J.; Zou, Y.; Buseman, C.M.; Chow, T.T.; Shay, J.W.; Wright, W.E. Telomere extension occurs at most chromosome ends and is uncoupled from fill-in in human cancer cells. Cell 2009, 138, 463–475. [Google Scholar] [CrossRef]
- Cristofari, G.; Lingner, J. Telomere length homeostasis requires that telomerase levels are limiting. EMBO J. 2006, 25, 565–574. [Google Scholar] [CrossRef] [PubMed]
- Xi, L.H.; Schmidt, J.C.; Zaug, A.J.; Ascarrunz, D.R.; Cech, T.R. A novel two-step genome editing strategy with CRISPR-Cas9 provides new insights into telomerase action and TERT gene expression. Genome Biol. 2015, 16. [Google Scholar] [CrossRef] [PubMed]
- Venteicher, A.S.; Abreu, E.B.; Meng, Z.; McCann, K.E.; Terns, R.M.; Veenstra, T.D.; Terns, M.P.; Artandi, S.E. A human telomerase holoenzyme protein required for Cajal body localization and telomere synthesis. Science 2009, 323, 644–648. [Google Scholar] [CrossRef] [PubMed]
- Xin, H.; Liu, D.; Wan, M.; Safari, A.; Kim, H.; Sun, W.; O’Connor, M.S.; Songyang, Z. TPP1 is a homologue of ciliate TEBP-β and interacts with POT1 to recruit telomerase. Nature 2007, 445, 559. [Google Scholar] [CrossRef]
- Zhong, F.L.; Batista, L.F.Z.; Freund, A.; Pech, M.F.; Venteicher, A.S.; Artandi, S.E. TPP1 OB-fold domain controls telomere maintenance by recruiting telomerase to chromosome ends. Cell 2012, 150, 481–494. [Google Scholar] [CrossRef]
- Sexton, A.N.; Youmans, D.T.; Collins, K. Specificity requirements for human telomere protein interaction with telomerase holoenzyme. J. Biol. Chem. 2012, 287, 34455–34464. [Google Scholar] [CrossRef]
- Abreu, E.; Aritonovska, E.; Reichenbach, P.; Cristofari, G.; Culp, B.; Terns, R.M.; Lingner, J.; Terns, M.P. TIN2-tethered TPP1 recruits human telomerase to telomeres in vivo. Mol. Cell. Biol. 2010, 30, 2971–2982. [Google Scholar] [CrossRef]
- Smogorzewska, A.; van Steensel, B.; Bianchi, A.; Oelmann, S.; Schaefer, M.R.; Schnapp, G.; de Lange, T. Control of human telomere length by TRF1 and TRF2. Mol. Cell. Biol. 2000, 20, 1659–1668. [Google Scholar] [CrossRef]
- Marcand, S.; Gilson, E.; Shore, D. A protein-counting mechanism for telomere length regulation in yeast. Science 1997, 275, 986–990. [Google Scholar] [CrossRef] [PubMed]
- O’Connor, M.S.; Safari, A.; Xin, H.W.; Liu, D.; Songyang, Z. A critical role for TPP1 and TIN2 interaction in high-order telomeric complex assembly. Proc. Natl. Acad. Sci. USA 2006, 103, 11874–11879. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.; O’Connor, M.S.; Qin, J.; Songyang, Z. Telosome, a mammalian telomere-associated complex formed by multiple telomeric proteins. J. Biol. Chem. 2004, 279, 51338–51342. [Google Scholar] [CrossRef] [PubMed]
- Rice, C.; Shastrula, P.K.; Kossenkov, A.V.; Hills, R.; Baird, D.M.; Showe, L.C.; Doukov, T.; Janicki, S.; Skordalakes, E. Structural and functional analysis of the human POT1-TPP1 telomeric complex. Nat. Commun. 2017, 8, 14928. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Podell, E.R.; Zaug, A.J.; Yang, Y.T.; Baciu, P.; Cech, T.R.; Lei, M. The POT1-TPP1 telomere complex is a telomerase processivity factor. Nature 2007, 445, 506–510. [Google Scholar] [CrossRef] [PubMed]
- Nandakumar, J.; Bell, C.F.; Weidenfeld, I.; Zaug, A.J.; Leinwand, L.A.; Cech, T.R. The TEL patch of telomere protein TPP1 mediates telomerase recruitment and processivity. Nature 2012, 492, 285–289. [Google Scholar] [CrossRef] [PubMed]
- Dalby, A.B.; Hofr, C.; Cech, T.R. Contributions of the TEL-patch amino acid cluster on TPP1 to telomeric DNA synthesis by human telomerase. J. Mol. Biol. 2015, 427, 1291–1303. [Google Scholar] [CrossRef] [PubMed]
- Necasova, I.; Janouskova, E.; Klumpler, T.; Hofr, C. Basic domain of telomere guardian TRF2 reduces d-loop unwinding whereas RAP1 restores it. Nucleic Acids Res. 2017, 45, 12170–12180. [Google Scholar] [CrossRef] [PubMed]
- Berardinelli, F.; Coluzzi, E.; Sgura, A.; Antoccia, A. Targeting telomerase and telomeres to enhance ionizing radiation effects in in vitro and in vivo cancer models. Mutat. Res. 2017, 773, 204–219. [Google Scholar] [CrossRef] [PubMed]
- Schrank, Z.; Khan, N.; Osude, C.; Singh, S.; Miller, R.J.; Merrick, C.; Mabel, A.; Kuckovic, A.; Puri, N. Oligonucleotides targeting telomeres and telomerase in cancer. Molecules 2018, 23, 2267. [Google Scholar] [CrossRef] [PubMed]
- Allsopp, R.C.; Vaziri, H.; Patterson, C.; Goldstein, S.; Younglai, E.V.; Futcher, A.B.; Greider, C.W.; Harley, C.B. Telomere length predicts replicative capacity of human fibroblasts. PNAS 1992, 89, 10114–10118. [Google Scholar] [CrossRef] [PubMed]
- Dikmen, Z.G.; Gellert, G.C.; Jackson, S.; Gryaznov, S.; Tressler, R.; Dogan, P.; Wright, W.E.; Shay, J.W. In vivo inhibition of lung cancer by GRN163L: A novel human telomerase inhibitor. Cancer Res. 2005, 65, 7866–7873. [Google Scholar] [CrossRef] [PubMed]
- Ruden, M.; Puri, N. Novel anticancer therapeutics targeting telomerase. Cancer Treat. Rev. 2013, 39, 444–456. [Google Scholar] [CrossRef] [PubMed]
- Gryaznov, S.M.; Jackson, S.; Dikmen, G.; Harley, C.; Herbert, B.-S.; Wright, W.E.; Shay, J.W. Oligonucleotide conjugate GRN163L targeting human telomerase as potential anticancer and antimetastatic agent. Nucleosides Nucleotides Nucleic Acids 2007, 26, 1577–1579. [Google Scholar] [CrossRef] [PubMed]
- Jackson, S.R.; Zhu, C.-H.; Paulson, V.; Watkins, L.; Dikmen, Z.G.; Gryaznov, S.M.; Wright, W.E.; Shay, J.W. Antiadhesive effects of GRN163L-an oligonucleotide N3′->P5′ thio-phosphoramidate targeting telomerase. Cancer Res. 2007, 67, 1121–1129. [Google Scholar] [CrossRef] [PubMed]
- Shea-Herbert, B.; Pongracz, K.; Shay, J.W.; Gryaznov, S.M. Oligonucleotide N3′→P5′ phosphoramidates as efficient telomerase inhibitors. Oncogene 2002, 21, 638. [Google Scholar] [CrossRef] [PubMed]
- Asai, A.; Oshima, Y.; Yamamoto, Y.; Uochi, T.-A.; Kusaka, H.; Akinaga, S.; Yamashita, Y.; Pongracz, K.; Pruzan, R.; Wunder, E.; et al. A novel telomerase template antagonist (GRN163) as a potential anticancer agent. Cancer Res. 2003, 63, 3931–3939. [Google Scholar] [PubMed]
- Gryaznov, S.; Pongracz, K.; Matray, T.; Schultz, R.; Pruzan, R.; Aimi, J.; Chin, A.; Harley, C.; Shea-Herbert, B.; Shay, J.; et al. Telomerase inhibitors—Oligonucleotide phosphoramidates as potential therapeutic agents. Nucleosides Nucleotides Nucleic Acids 2001, 20, 401–410. [Google Scholar] [CrossRef]
- Jafri, M.A.; Ansari, S.A.; Alqahtani, M.H.; Shay, J.W. Roles of telomeres and telomerase in cancer, and advances in telomerase-targeted therapies. Genome Med. 2016, 8, 69. [Google Scholar] [CrossRef]
- Tokcaer-Keskin, Z.; Dikmen, Z.G.; Ayaloglu-Butun, F.; Gultekin, S.; Gryaznov, S.M.; Akcali, K.C. The effect of telomerase template antagonist GRN163L on bone-marrow-derived rat mesenchymal stem cells is reversible and associated with altered expression of cyclin D1, CDK4 and CDK6. Stem Cell Rev. 2010, 6, 224–233. [Google Scholar] [CrossRef]
- Salloum, R.; Hummel, T.R.; Kumar, S.S.; Dorris, K.; Li, S.; Lin, T.; Daryani, V.M.; Stewart, C.F.; Miles, L.; Poussaint, T.Y.; et al. A molecular biology and phase II study of Imetelstat (GRN163L) in children with recurrent or refractory central nervous system malignancies: A pediatric brain tumor consortium study. J. Neurooncol. 2016, 129, 443–451. [Google Scholar] [CrossRef] [PubMed]
- Chiappori, A.A.; Kolevska, T.; Spigel, D.R.; Hager, S.; Rarick, M.; Gadgeel, S.; Blais, N.; Von Pawel, J.; Hart, L.; Reck, M.; et al. A randomized phase II study of the telomerase inhibitor imetelstat as maintenance therapy for advanced non-small-cell lung cancer. Ann. Oncol. 2015, 26, 354–362. [Google Scholar] [CrossRef] [PubMed]
- Gomez-Millan, J.; Goldblatt, E.M.; Gryaznov, S.M.; Mendonca, M.S.; Herbert, B.-S. Specific telomere dysfunction induced by GRN163L increases radiation sensitivity in breast cancer cells. Int. J. Radiat. Oncol. Biol. Phys. 2007, 67, 897–905. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Zhang, J.; Yang, S.; Kuang, Z.; Tan, G.; Yang, G.; Wei, Q.; Guo, Z. Telomerase antagonist Imetelstat increases radiation sensitivity in esophageal squamous cell carcinoma. Oncotarget 2017, 8, 13600–13619. [Google Scholar] [CrossRef] [PubMed]
- Ferrandon, S.; Malleval, C.; El Hamdani, B.; Battiston-Montagne, P.; Bolbos, R.; Langlois, J.-B.; Manas, P.; Gryaznov, S.M.; Alphonse, G.; Honnorat, J.; et al. Telomerase inhibition improves tumor response to radiotherapy in a murine orthotopic model of human glioblastoma. Mol. Cancer 2015, 14, 134. [Google Scholar] [CrossRef]
- Puri, N.; Eller, M.S.; Byers, H.R.; Dykstra, S.; Kubera, J.; Gilchrest, B.A. Telomere-based DNA damage responses: A new approach to melanoma. FASEB J. 2004, 18, 1373–1381. [Google Scholar] [CrossRef] [PubMed]
- Ohashi, N.; Yaar, M.; Eller, M.S.; Truzzi, F.; Gilchrest, B.A. Features that determine telomere homolog oligonucleotide-induced therapeutic DNA damage-like responses in cancer cells. J. Cell. Physiol. 2007, 210, 582–595. [Google Scholar] [CrossRef] [PubMed]
- Pitman, R.T.; Wojdyla, L.; Puri, N. Mechanism of DNA damage responses induced by exposure to an oligonucleotide homologous to the telomere overhang in melanoma. Oncotarget 2013, 4, 761–771. [Google Scholar] [CrossRef]
- Puri, N.; Pitman, R.T.; Mulnix, R.E.; Erickson, T.; Iness, A.N.; Vitali, C.; Zhao, Y.; Salgia, R. Non-small cell lung cancer is susceptible to induction of DNA damage responses and inhibition of angiogenesis by telomere overhang oligonucleotides. Cancer Lett. 2014, 343, 14–23. [Google Scholar] [CrossRef]
- Chhabra, G.; Wojdyla, L.; Frakes, M.; Schrank, Z.; Leviskas, B.; Ivancich, M.; Vinay, P.; Ganapathy, R.; Ramirez, B.E.; Puri, N. Mechanism of action of G-quadruplex-forming oligonucleotide homologous to the telomere overhang in melanoma. J.Invest. Derm. 2018, 138, 903–910. [Google Scholar] [CrossRef]
- Longe, H.O.; Romesser, P.B.; Rankin, A.M.; Faller, D.V.; Eller, M.S.; Gilchrest, B.A.; Denis, G.V. Telomere homolog oligonucleotides induce apoptosis in malignant but not in normal lymphoid cells: Mechanism and therapeutic potential. Int. J. Cancer 2009, 124, 473–482. [Google Scholar] [CrossRef] [PubMed]
- Crees, Z.; Girard, J.; Rios, Z.; Botting, G.M.; Harrington, K.; Shearrow, C.; Wojdyla, L.; Stone, A.L.; Uppada, S.B.; Devito, J.T.; et al. Oligonucleotides and G-quadruplex stabilizers: Targeting telomeres and telomerase in cancer therapy. Curr. Pharm. Des. 2014, 20, 6422–6437. [Google Scholar] [CrossRef] [PubMed]
- Kolhatkar, V.; Khambati, H.; Lote, A.; Shanine, P.; Insley, T.; Sen, S.; Munirathinam, G.; Král, P.; Kolhatkar, R. Star-shaped tetraspermine enhances cellular uptake and cytotoxicity of T-oligo in prostate cancer cells. Pharm. Res. 2015, 32, 196–210. [Google Scholar] [CrossRef] [PubMed]
- Li, G.-Z.; Eller, M.S.; Hanna, K.; Gilchrest, B.A. Signaling pathway requirements for induction of senescence by telomere homolog oligonucleotides. Exp. Cell. Res. 2004, 301, 189–200. [Google Scholar] [CrossRef] [PubMed]
- Van Steensel, B.; Smogorzewska, A.; de Lange, T. TRF2 protects human telomeres from end-to-end fusions. Cell 1998, 92, 401–413. [Google Scholar] [CrossRef]
- Ivancich, M.; Schrank, Z.; Wojdyla, L.; Leviskas, B.; Kuckovic, A.; Sanjali, A.; Puri, N. Treating cancer by targeting telomeres and telomerase. Antioxidants 2017, 6, 15. [Google Scholar] [CrossRef] [PubMed]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Veverka, P.; Janovič, T.; Hofr, C. Quantitative Biology of Human Shelterin and Telomerase: Searching for the Weakest Point. Int. J. Mol. Sci. 2019, 20, 3186. https://doi.org/10.3390/ijms20133186
Veverka P, Janovič T, Hofr C. Quantitative Biology of Human Shelterin and Telomerase: Searching for the Weakest Point. International Journal of Molecular Sciences. 2019; 20(13):3186. https://doi.org/10.3390/ijms20133186
Chicago/Turabian StyleVeverka, Pavel, Tomáš Janovič, and Ctirad Hofr. 2019. "Quantitative Biology of Human Shelterin and Telomerase: Searching for the Weakest Point" International Journal of Molecular Sciences 20, no. 13: 3186. https://doi.org/10.3390/ijms20133186
APA StyleVeverka, P., Janovič, T., & Hofr, C. (2019). Quantitative Biology of Human Shelterin and Telomerase: Searching for the Weakest Point. International Journal of Molecular Sciences, 20(13), 3186. https://doi.org/10.3390/ijms20133186