Src and ROCK Kinases Differentially Regulate Mineralization of Human Osteosarcoma Saos-2 Cells
Abstract
1. Introduction
2. Results
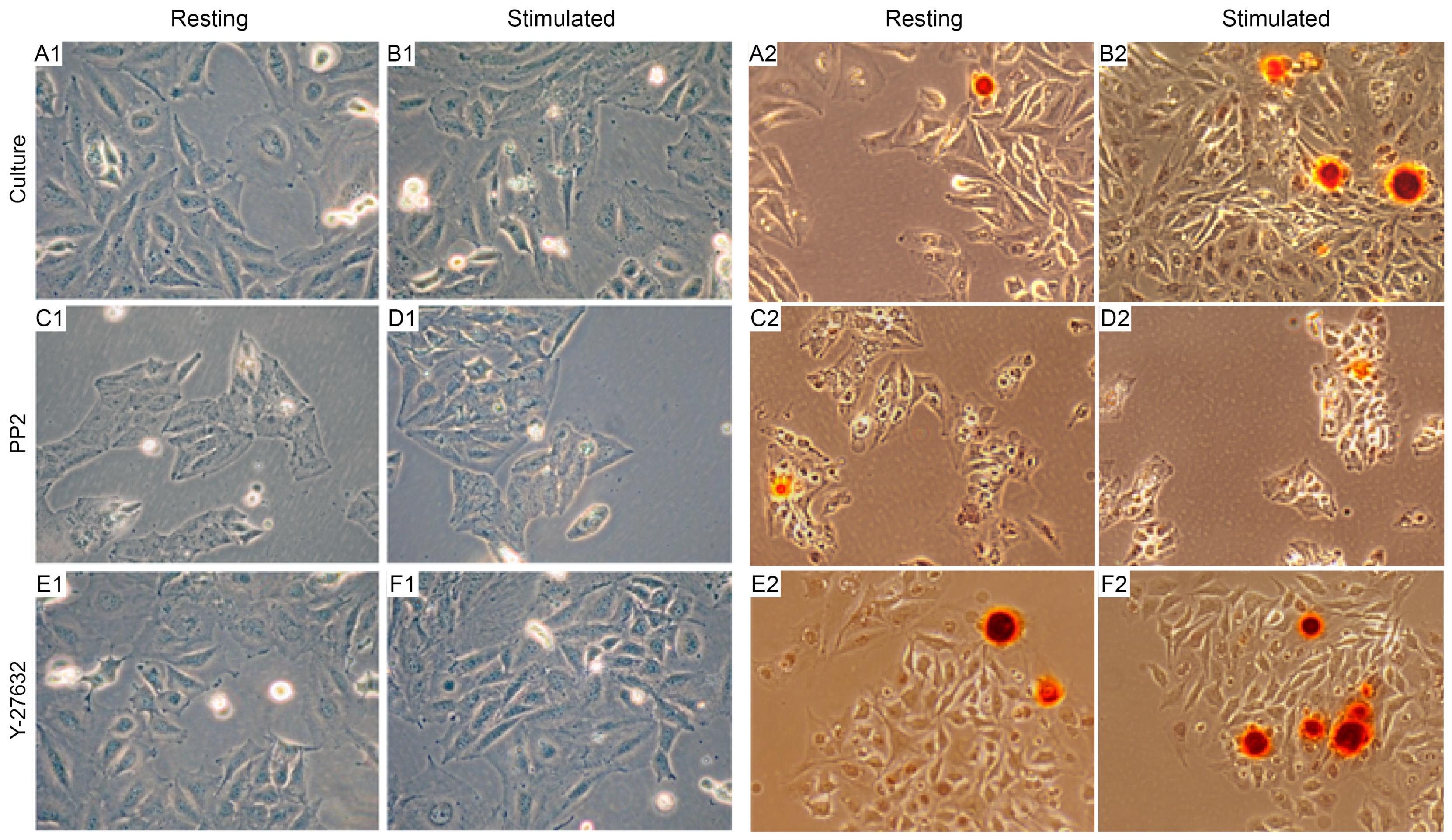
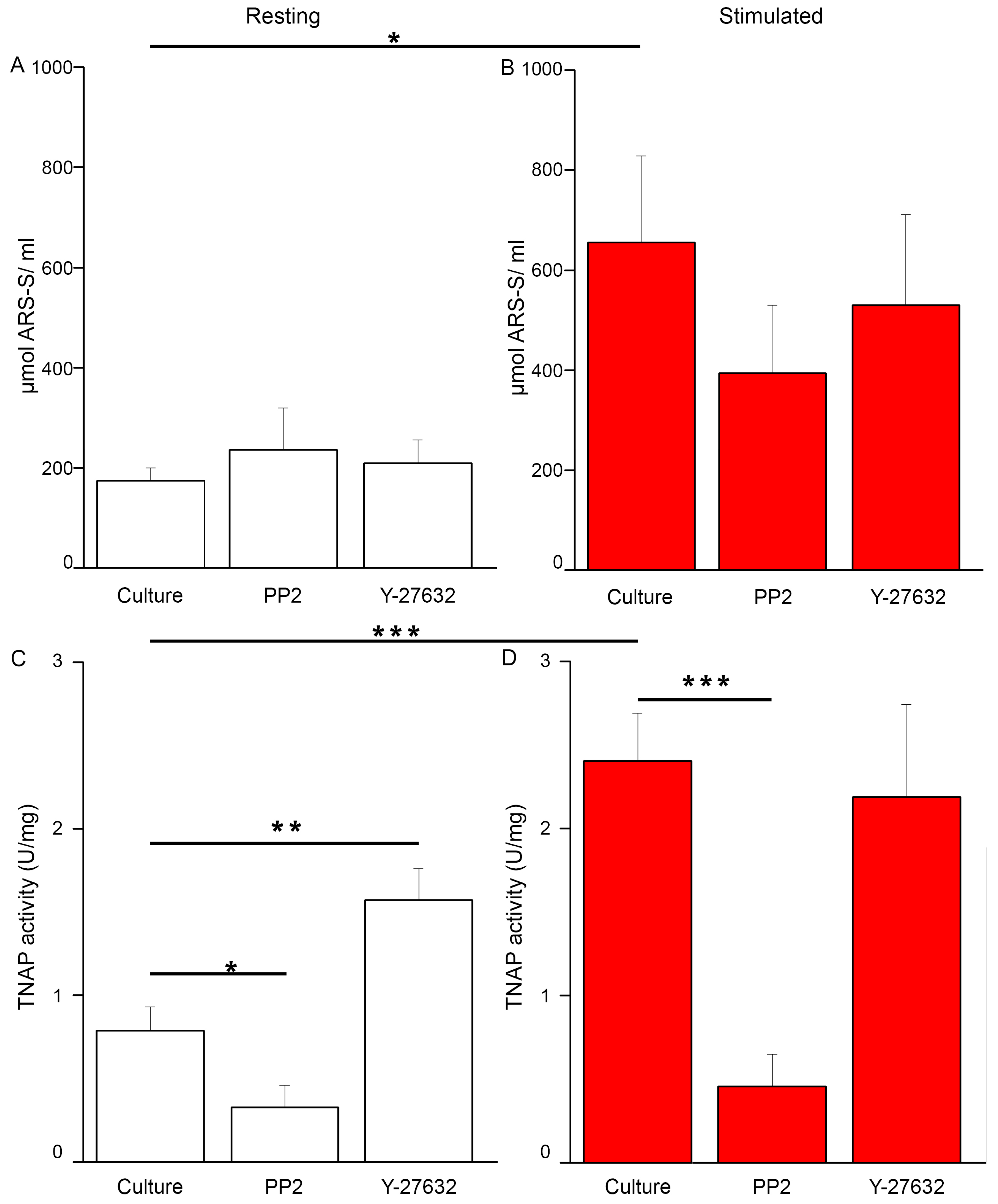
2.1. Characterization of the Mineralization Process in Human Osteosarcoma Saos-2 Cells
2.2. Saos-2 Cells Viability and Proliferation during Inhibition of the Mineralization Process
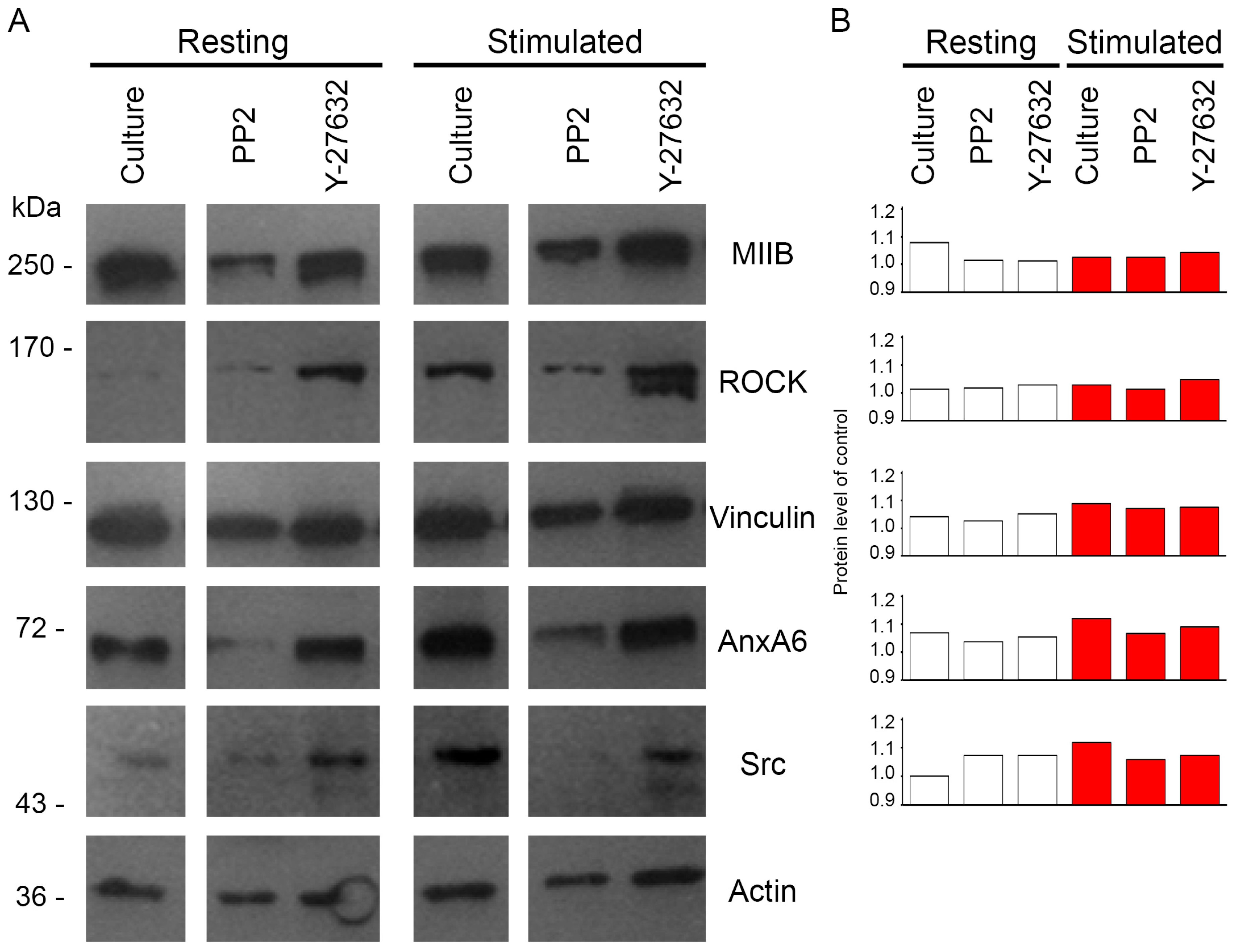
2.3. Protein Profile of Mineralizing Saos-2 Cells
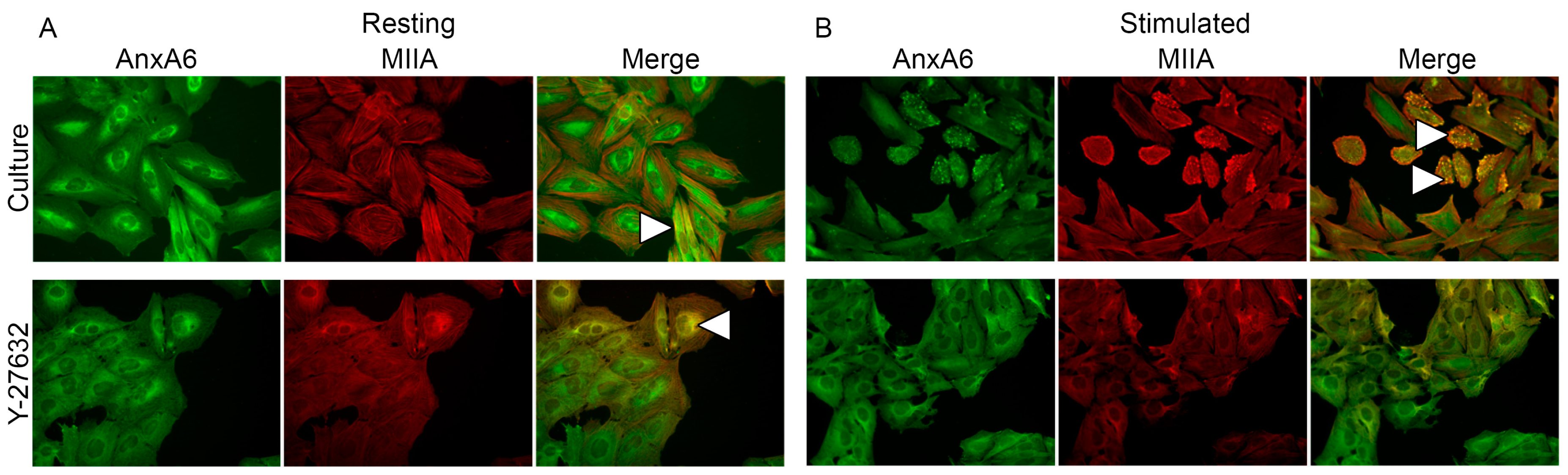
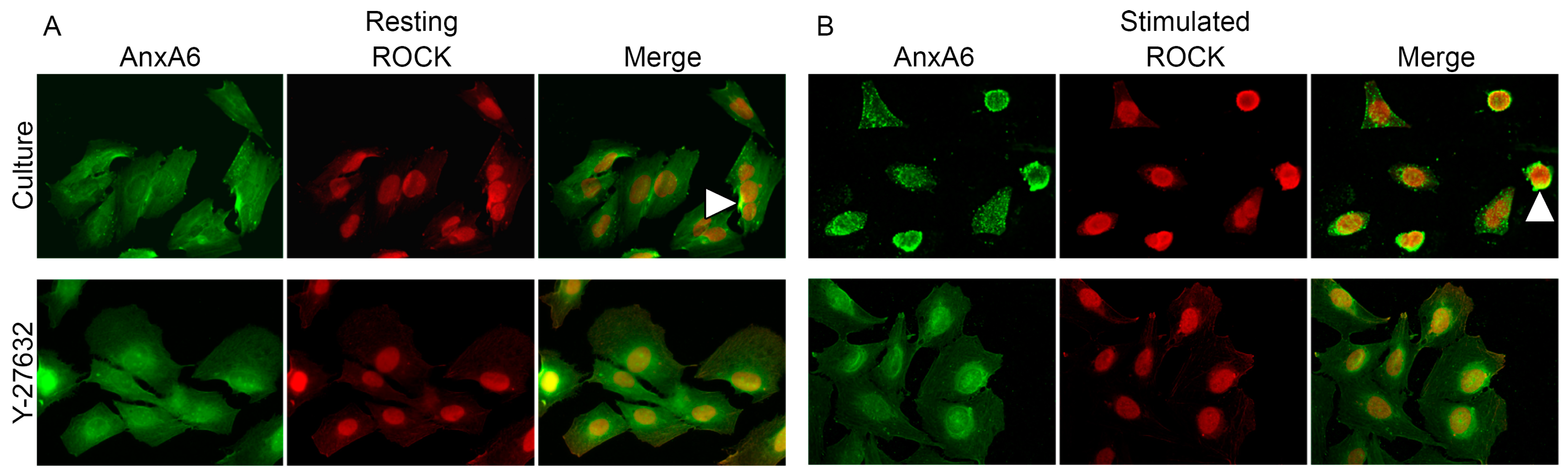
2.4. Annexins Distribution during Stimulation of Saos-2 Cells to Mineralize
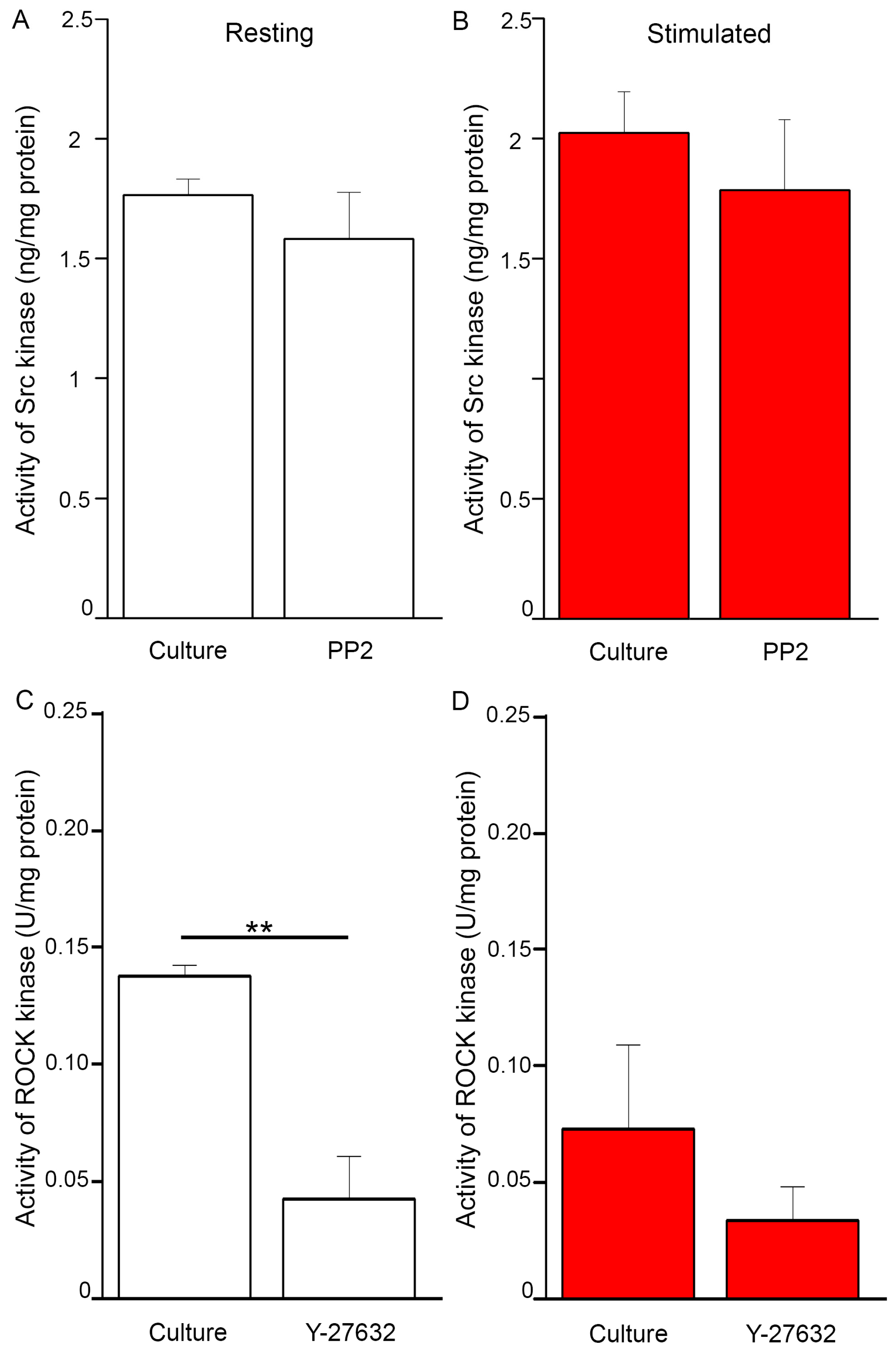
2.5. Changes of Src and ROCK Kinase Activities during Mineral Formation by Saos-2 Cells
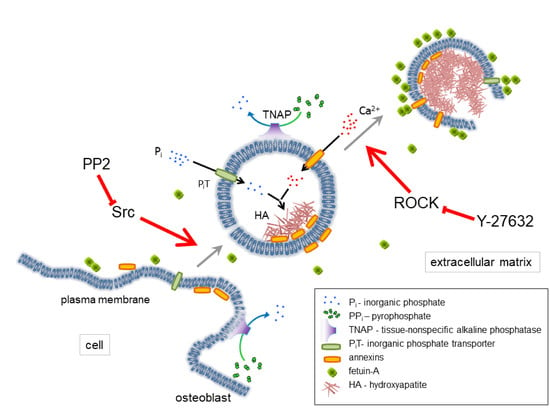
3. Discussion
4. Materials and Methods
4.1. Cell Culture and Treatment
4.2. Calcium Mineral Detection
4.3. TNAP Activity Assay
4.4. Cell Lysis, SDS-PAGE, and Immunoblot Analysis
4.5. Immunochemistry and Fluorescent Microscopy
4.6. Src Activity Assay
4.7. ROCK Activity Assay
4.8. Cell Viability and Cell Cycle Analysis by Flow Cytometry
4.9. Statistical Analysis
Supplementary Materials
Author Contributions
Acknowledgments
Conflicts of Interest
Abbreviations
| AA | ascorbic acid |
| ALP | alkaline phosphatase |
| AnxA | vertebrate annexin |
| AR-S | Alizarin Red-S |
| BSP | bone sialoprotein |
| ECM | extracellular matrix |
| β-GP | β-glycerophosphate |
| CPC | cetylpyridinium chloride |
| FAK | focal adhesion kinase |
| FN | fibronectin |
| MII | myosin II |
| MVs | matrix vesicles |
| Pi | inorganic phosphate |
| pNPP | paranitrophenyl phosphate |
| PP2 | 4-amino-5-(4-chlorophenyl)-7-(t-butyl)pyrazolo [3,4-d] pyrimidine |
| PPi | inorganic pyrophosphate |
| ROCK | Rho-associated coiled-coil kinase |
| Src | sarcoma proto-oncogene tyrosine-protein kinase |
| TNAP | tissue non-specific alkaline phosphatase |
| Y-27632 | (R)-(+)-trans-4-(1-aminoethyl)-N-(4-pyridyl) cyclohexane carboxamide dihydrochloride |
References
- Tatosyan, A.G.; Mizenina, O.A. Kinases of the Src family: structure and functions. Biochemistry (Mosc) 2000, 65, 49–58. [Google Scholar] [PubMed]
- Schlessinger, J. New roles for Src kinases in control of cell survival and angiogenesis. Cell 2000, 100, 293–296. [Google Scholar] [CrossRef]
- Riento, K.; Ridley, A.J. Rocks: multifunctional kinases in cell behaviours. Nat. Rev. Mol. Cell. Biol. 2003, 4, 446–456. [Google Scholar] [CrossRef] [PubMed]
- Gill, K.S.; Beier, F.; Goldberg, H.A. Rho-ROCK signaling differentially regulates chondrocyte spreading on fibronectin and bone sialoprotein. Am. J. Physiol. Cell Physiol. 2008, 295, C38–C49. [Google Scholar] [CrossRef][Green Version]
- Strzelecka-Kiliszek, A.; Bozycki, L. Cross-talk between Src kinases and Rho small GTPases regulates biomineralization and simplify imaging of the mineralization process. Postepy Biochem. 2017, 63, 93–109. [Google Scholar]
- Strzelecka-Kiliszek, A.; Mebarek, S.; Roszkowska, M.; Buchet, R.; Magne, D.; Pikula, S. Functions of Rho family of small GTPases and Rho-associated coiled-coil kinases in bone cells during differentiation and mineralization. Biochem. Biophys. Acta Gen. Sub. 2017, 1861, 1009–1023. [Google Scholar] [CrossRef]
- Bottini, M.; Mebarek, S.; Anderson, K.L.; Strzelecka-Kiliszek, A.; Bozycki, L.; Sper Simao, A.M.; Bolean, M.; Ciancaglini, P.; Bandorowicz-Pikula, J.; Pikula, S.; et al. Matrix vesicles from chondrocytes and osteoblasts: their biogenesis, properties, functions and biomimetic models. Biochem. Biophys. Acta Gen. Sub. 2018, 1862, 532–546. [Google Scholar] [CrossRef]
- Thouverey, C.; Strzelecka-Kiliszek, A.; Balcerzak, M.; Buchet, R.; Pikula, S. Matrix vesicles originate from apical membrane microvilli of mineralizing osteoblast-like Saos-2 cells. J. Cell. Biochem. 2009, 106, 127–138. [Google Scholar] [CrossRef]
- Thouverey, C.; Malinowska, A.; Balcerzak, M.; Strzelecka-Kiliszek, A.; Buchet, R.; Dadlez, M.; Pikula, S. Proteomic characterization of biogenesis and functions of matrix vesicles released from mineralizing human osteoblast-like cells. J. Proteomics 2011, 74, 1123–1134. [Google Scholar] [CrossRef]
- Strzelecka-Kiliszek, A.; Bozycki, L.; Mebarek, S.; Buchet, R.; Pikula, S. Characteristics of minerals in matrix vesicles produced by human osteoblasts hFOB 1.19 and osteosarcoma Saos-2 cells stimulated for mineralization. J. Inorg. Biochem. 2017, 171, 100–107. [Google Scholar] [CrossRef]
- Wang, W.; Xu, J.; Kirsch, T. Annexin-mediated Ca2+ influx regulates growth plate chondrocyte maturation and apoptosis. J. Biol. Chem. 2003, 278, 3762–3769. [Google Scholar] [CrossRef] [PubMed]
- Genetos, D.C.; Wong, A.; Weber, T.J.; Karin, N.J.; Yellowley, C.E. Impaired osteoblast differentiation in annexin A2- and -A5-deficient cells. PLoS ONE 2014, 15, e107482. [Google Scholar] [CrossRef] [PubMed]
- Sekrecka, A.; Balcerzak, M.; Thouverey, C.; Buchet, R.; Pikula, S. Annexins in mineralization process. Postepy Biochem. 2007, 53, 159–163. [Google Scholar] [PubMed]
- Kundrada, M.N.; Ray, S.; Saria, M.; Friedman, D.; Matrisian, L.M.; Lukyanov, P.; Ochieng, J. Annexins expressed on the cell surface serve as receptors for adhesion to immobilized fetuin-A. Biochim. Biophys. Acta 2004, 1693, 111–123. [Google Scholar] [CrossRef] [PubMed]
- Genge, B.R.; Wu, L.N.; Wuthier, R.E. Mineralization of annexin-5-containing lipid-calcium-phosphate complexes: modulation by varying lipid composition and incubation with cartilage collagens. J. Biol. Chem. 2008, 283, 9737–9748. [Google Scholar] [CrossRef]
- Kirsh, T.; Harrison, G.; Golub, E.E.; Nah, H.D. The roles of annexins and type II and X collagen in matrix vesicle-mediated mineralization of growth plate cartilage. J. Biol. Chem. 2000, 275, 35577–35583. [Google Scholar] [CrossRef]
- Wuthier, R.E.; Lipscomb, G.F. Matrix vesicles: structure, composition, formation and function in calcification. Front. Biosci. 2011, 16, 2812–2902. [Google Scholar] [CrossRef]
- Wang, W.; Xu, J.; Kirsch, T. Annexin V and terminal differentiation of growth plate chondrocytes. Exp. Cell Res. 2005, 305, 156–165. [Google Scholar] [CrossRef]
- Golczak, M.; Kicinska, A.; Bandorowicz-Pikula, J.; Buchet, R.; Szewczyk, A.; Pikula, S. Acidic pH-induced folding of annexin VI is a prerequisite for its insertion into lipid bilayers and formation of ion channels by the protein molecules. FASEB J. 2001, 15, 1083–1085. [Google Scholar] [CrossRef]
- Cmoch, A.; Strzelecka-Kiliszek, A.; Palczewska, M.; Groves, P.; Pikula, S. Matrix vesicles isolated from mineralization-competent Saos-2 cells are selectively enriched with annexins and S100 proteins. Biochem. Biophys. Res. Commun. 2011, 412, 683–687. [Google Scholar] [CrossRef]
- Wang, G.; Woods, A.; Sabari, S.; Pagnotta, L.; Stanton, L.-A.; Beier, F. RhoA/ROCK signaling suppresses hypertrophic chondrocyte differentiation. J. Biol. Chem. 2004, 279, 13205–13214. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Lemay, S.; Aoudjit, L.; Kawachi, H.; Takano, T. Src-family kinase Fyn phosphorylates the cytoplasmic domain of nephrin and modulates its interaction with podocin. J. Am. Soc. Nephrol. 2004, 15, 3006–3015. [Google Scholar] [CrossRef] [PubMed]
- Minashima, T.; Small, W.; Moss, S.E.; Kirsch, T. Intracellular modulation of signaling pathways by annexin A6 regulates terminal differentiation of chondrocytes. J. Biol. Chem. 2012, 287, 14803–14815. [Google Scholar] [CrossRef] [PubMed]
- Lv, Z.; Hu, M.; Ren, X.; Fan, M.; Zhen, J.; Chen, L.; Lin, J.; Ding, N.; Wang, Q.; Wang, R. Fyn mediates high glucose-induced actin cytoskeleton reorganization of podocytes via promoting ROCK activation in vitro. J. Diabetes Res. 2016, 2016, 5671803. [Google Scholar] [CrossRef] [PubMed]
- Xiong, N.; Li, S.; Tang, K.; Bai, H.; Peng, Y.; Yang, H.; Wu, C.; Liu, Y. Involvement of caveolin-1 in low shear stress-induced breast cancer cell motility and adhesion: Roles of FAK/Src and ROCK/p-MLC pathways. Biochim. Biophys. Acta 2017, 1864, 12–22. [Google Scholar] [CrossRef] [PubMed]
- Vaingankar, S.M.; Fitzpatrick, T.A.; Johnson, K.; Goding, J.W.; Maurice, M.; Terkeltaub, R. Subcellular targeting and function of osteoblast nucleotide pyrophosphatase phosphodiesterase 1. Am. J. Phys. Cell Physiol. 2004, 286, C1177–C1187. [Google Scholar] [CrossRef]
- Gillette, J.M.; Nielsen-Preiss, S.M. The role of annexin 2 in osteoblastic mineralization. J. Cell Sci. 2004, 117, 441–449. [Google Scholar] [CrossRef][Green Version]
- Stanford, C.M.; Jacobson, P.A.; Eanes, E.D.; Lembke, L.A.; Midura, R.J. Rapidly forming apatitic mineral in an osteoblastic cell line (UMR 106-01 BSP). J. Biol. Chem. 1995, 270, 9420–9428. [Google Scholar] [CrossRef]
- Gregory, C.A.; Gunn, W.G.; Peister, A.; Prockop, D.J. An Alizarin red-based assay of mineralization by adherent cells in culture: comparison with cetylpyridinium chloride extraction. Anal. Biochem. 2004, 329, 77–84. [Google Scholar] [CrossRef]
- Chen, N.X.; O’Neill, K.D.; Chen, X.; Moe, S.M. Annexin-mediated matrix vesicle calcification in vascular smooth muscle cells. J. Bone Miner. Res. 2008, 23, 1798–1805. [Google Scholar] [CrossRef]
- Laemmli, U.K. Cleavage of structural proteins during assembly of the head of bacteriophage T4. Nature 1970, 277, 680–685. [Google Scholar] [CrossRef]
- Towbin, H.; Staehelin, T.; Gordon, J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: Procedure and some applications. Proc. Natl. Acad. Sci. USA 1979, 76, 4350–4354. [Google Scholar] [CrossRef] [PubMed]
- Krishan, A. Rapid flow cytometric analysis of mammalian cell cycle by propidium iodide staining. J. Cell Biol. 1975, 66, 188–193. [Google Scholar] [CrossRef] [PubMed]
- Dressler, L.G. DNA flow cytometry and prognostic factors in 1331 frozen breast cancer speciments. Cancer 1988, 61, 420–427. [Google Scholar] [CrossRef]
- Strzelecka-Kiliszek, A.; Buszewska, M.E.; Podszywalow-Bartnicka, P.; Pikula, S.; Otulak, K.; Bandorowicz-Pikula, J. Calcium- and pH-dependent localization of annexin A6 isoforms in Balb/3T3 fibroblasts reflecting their potential participation in vesicular transport. J. Cell Biochem. 2008, 104, 418–434. [Google Scholar] [CrossRef] [PubMed]
- Kraus, D. Consolidated data analysis and presentation using an open-source add-in for the Microsoft Excel® spreadsheet software. Medical Writing 2014, 23, 25–28. [Google Scholar] [CrossRef]


© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Strzelecka-Kiliszek, A.; Romiszewska, M.; Bozycki, L.; Mebarek, S.; Bandorowicz-Pikula, J.; Buchet, R.; Pikula, S. Src and ROCK Kinases Differentially Regulate Mineralization of Human Osteosarcoma Saos-2 Cells. Int. J. Mol. Sci. 2019, 20, 2872. https://doi.org/10.3390/ijms20122872
Strzelecka-Kiliszek A, Romiszewska M, Bozycki L, Mebarek S, Bandorowicz-Pikula J, Buchet R, Pikula S. Src and ROCK Kinases Differentially Regulate Mineralization of Human Osteosarcoma Saos-2 Cells. International Journal of Molecular Sciences. 2019; 20(12):2872. https://doi.org/10.3390/ijms20122872
Chicago/Turabian StyleStrzelecka-Kiliszek, Agnieszka, Marta Romiszewska, Lukasz Bozycki, Saida Mebarek, Joanna Bandorowicz-Pikula, Rene Buchet, and Slawomir Pikula. 2019. "Src and ROCK Kinases Differentially Regulate Mineralization of Human Osteosarcoma Saos-2 Cells" International Journal of Molecular Sciences 20, no. 12: 2872. https://doi.org/10.3390/ijms20122872
APA StyleStrzelecka-Kiliszek, A., Romiszewska, M., Bozycki, L., Mebarek, S., Bandorowicz-Pikula, J., Buchet, R., & Pikula, S. (2019). Src and ROCK Kinases Differentially Regulate Mineralization of Human Osteosarcoma Saos-2 Cells. International Journal of Molecular Sciences, 20(12), 2872. https://doi.org/10.3390/ijms20122872