The Adaptive Mechanism of Plants to Iron Deficiency via Iron Uptake, Transport, and Homeostasis
Abstract
:1. Introduction
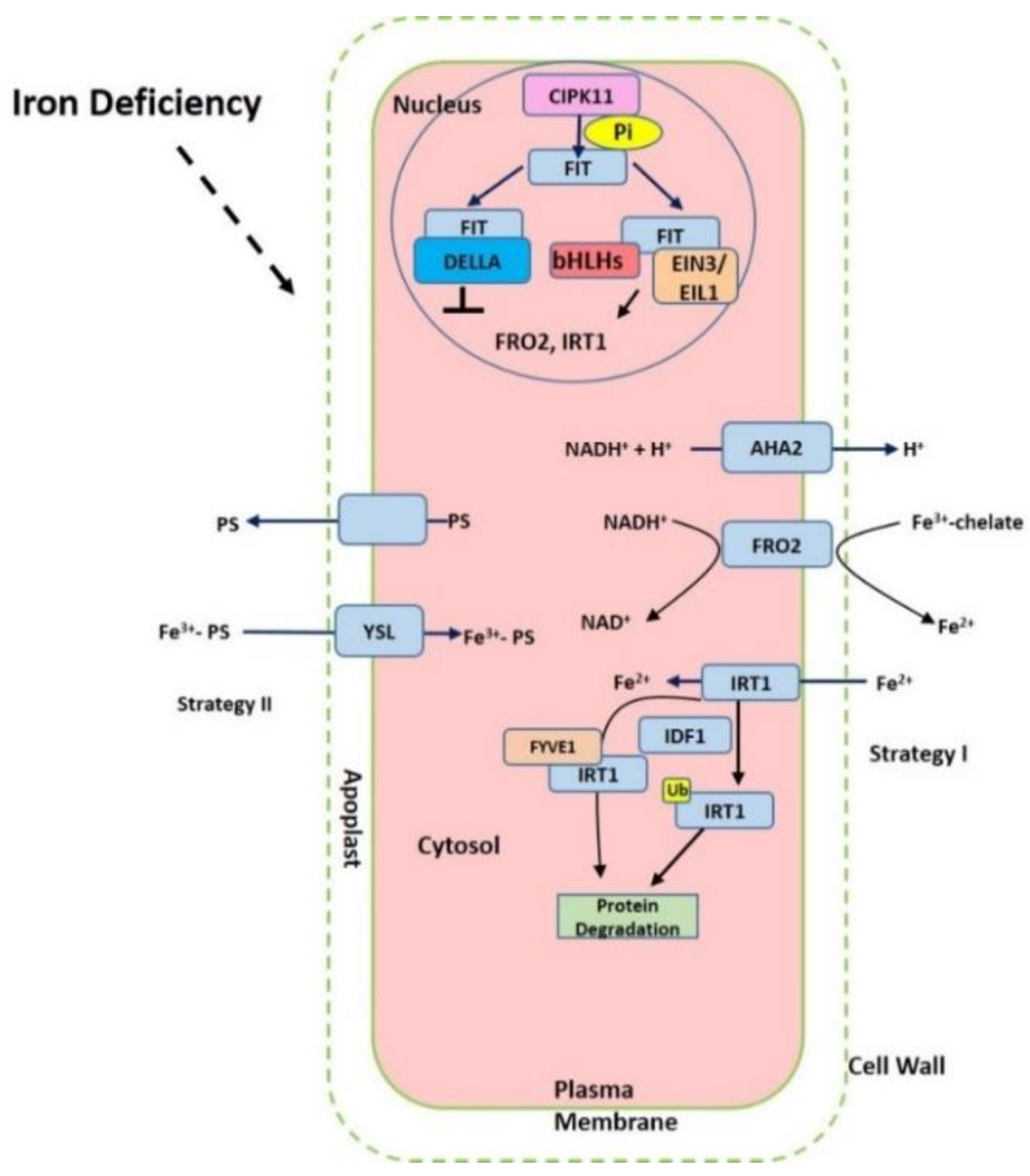
2. Iron Acquisition from Soil to Roots
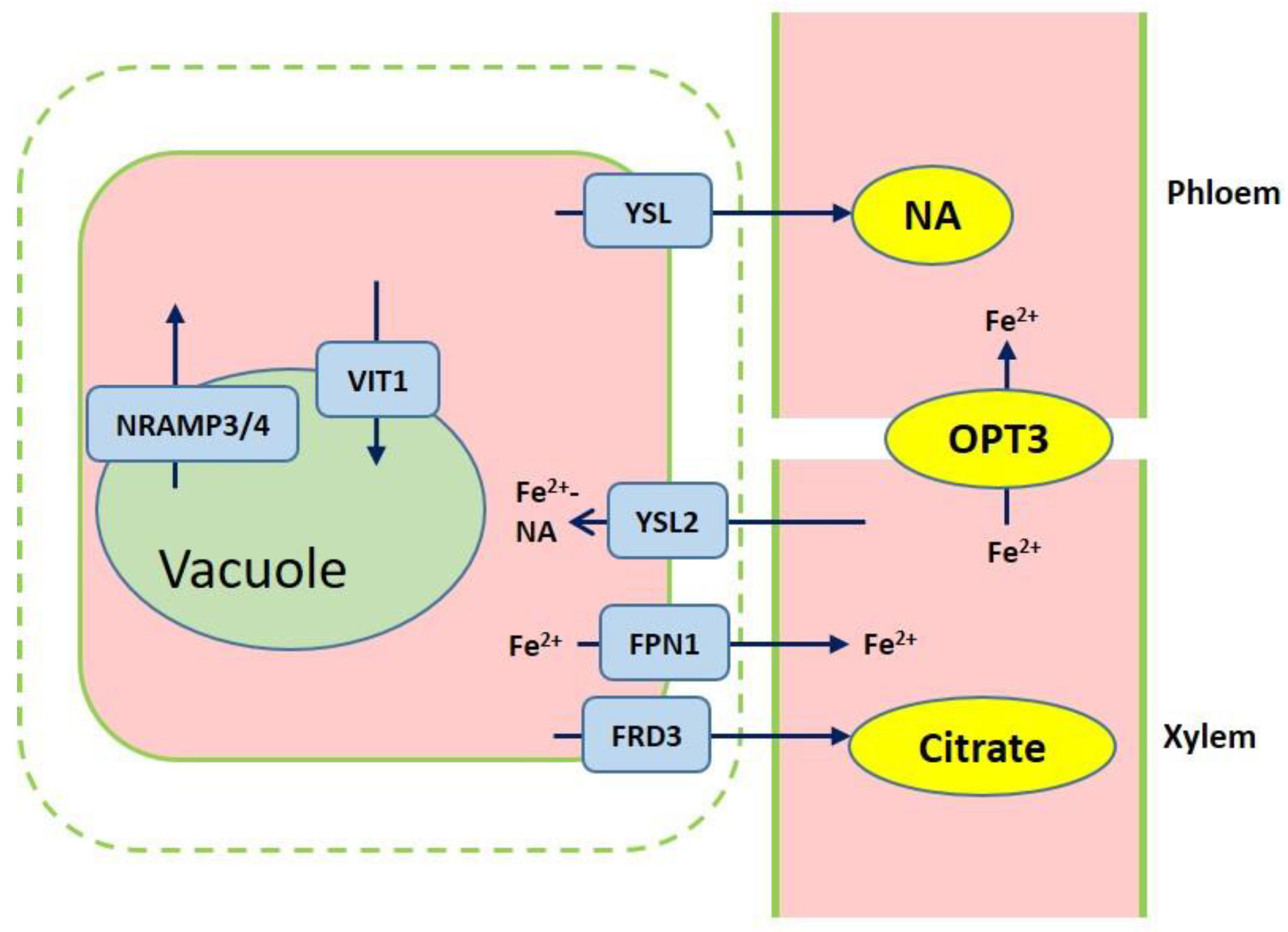
3. Iron Transport Mechanism in Plants
4. Iron Storage in Cells
5. Transcriptional and Posttranscriptional Regulation of Fe-related Genes
6. Function of Other Factors in the Iron Homeostasis
7. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Mahender, A.; Swamy, B.P.M.; Anandan, A.; Ali, J. Tolerance of iron-deficient and -toxic soil conditions in rice. Plants 2019, 8, 31. [Google Scholar] [CrossRef] [PubMed]
- Becker, M.; Asch, F. Iron toxicity in rice—Conditions and management concepts. J. Plant Nutr. Soil Sci. 2010, 168, 558–573. [Google Scholar] [CrossRef]
- Curie, C.; Briat, J.F. Iron transport and signaling in plants. Annu. Rev. Plant Biol. 2003, 54, 183–206. [Google Scholar] [CrossRef] [PubMed]
- Fageria, N.K.; Baligar, V.C.; Li, Y.C. The role of nutrient efficient plants in improving crop yields in the twenty first century. J. Plant Nutr. 2008, 31, 1121–1157. [Google Scholar] [CrossRef]
- Yi, Y.; Guerinot, M.L. Genetic evidence that induction of root Fe(III) chelate reductase activity is necessary for iron uptake under iron deficiency. Plant J. 1996, 10, 835–844. [Google Scholar] [CrossRef] [PubMed]
- Eide, D.; Broderius, M.; Fett, J.; Guerinot, M.L. A novel iron-regulated metal transporter from plants identified by functional expression in yeast. Proc. Natl. Acad. Sci. USA 1996, 93, 5624–5628. [Google Scholar] [CrossRef]
- Vert, G.; Grotz, N.; Dédaldéchamp, F.; Gaymard, F.; Guerinot, M.L.; Briat, J.F.; Curie, C. IRT1, an Arabidopsis transporter essential for iron uptake from the soil and for plant growth. Plant Cell 2002, 14, 1223–1233. [Google Scholar] [CrossRef]
- Takagi, S.-I. Naturally occurring iron-chelating compounds in oat- and rice-root washings. Soil Sci. Plant Nutr. 1976, 22, 423–433. [Google Scholar] [CrossRef]
- Takagi, S.I.; Nomoto, K.; Takemoto, T. Physiological aspect of mugineic acid, a possible phytosiderophore of graminaceous plants. J. Plant Nutr. 1984, 7, 469–477. [Google Scholar] [CrossRef]
- Curie, C.; Panaviene, Z.; Loulergue, C.; Dellaporta, S.L.; Briat, J.F.; Walker, E.L. Maize yellow stripe1 encodes a membrane protein directly involved in Fe(III) uptake. Nature 2001, 409, 346. [Google Scholar] [CrossRef] [PubMed]
- Yuan, W.; Zhang, D.; Song, T.; Xu, F.; Lin, S.; Xu, W.; Li, Q.; Zhu, Y.; Liang, J.; Zhang, J. Arabidopsis plasma membrane H+-ATPase genes AHA2 and AHA7 have distinct and overlapping roles in the modulation of root tip H+ efflux in response to low-phosphorus stress. J. Exp. Bot. 2017, 68, 1731–1741. [Google Scholar] [CrossRef]
- Oh, Y.J.; Kim, H.; Seo, S.H.; Hwang, B.G.; Chang, Y.S.; Lee, J.; Lee, D.W.; Sohn, E.J.; Lee, S.J.; Lee, Y.; et al. Cytochrome b5 reductase 1 triggers serial reactions that lead to iron uptake in plants. Mol. Plant 2016, 9, 501–513. [Google Scholar] [CrossRef] [PubMed]
- Abadía, J.; López Millán, A.F.; Rombolà, A.; Abadía, A. Organic acids and Fe deficiency: A review. Plant Soil 2002, 241, 75–86. [Google Scholar] [CrossRef]
- Jin, C.W.; You, G.Y.; He, Y.F.; Tang, C.; Wu, P.; Zheng, S.J. Iron deficiency-induced secretion of phenolics facilitates the reutilization of root apoplastic iron in red clover. Plant Physiol. 2007, 144, 278–285. [Google Scholar] [CrossRef]
- Connorton, J.M.; Balk, J.; Rodríguez Celma, J. Iron homeostasis in plants—A brief overview. Metallomics 2017, 9, 813–823. [Google Scholar] [CrossRef] [PubMed]
- Jeong, J.; Merkovich, A.; Clyne, M.; Connolly, E.L. Directing iron transport in dicots: Regulation of iron acquisition and translocation. Curr. Opin. Plant Biol. 2017, 39, 106–113. [Google Scholar] [CrossRef]
- Clemens, S.; Weber, M. The essential role of coumarin secretion for Fe acquisition from alkaline soil. Plant Signal. Behav. 2015, 11, e1114197. [Google Scholar] [CrossRef] [PubMed]
- Sisó Terraza, P.; Luis Villarroya, A.; Fourcroy, P.; Briat, J.F.; Abadía, A.; Gaymard, F.; Abadía, J.; Álvarez Fernández, A. Accumulation and secretion of coumarinolignans and other coumarins in Arabidopsis thaliana roots in response to iron deficiency at high pH. Front. Plant Sci. 2016, 7, 1711. [Google Scholar] [CrossRef] [PubMed]
- Fourcroy, P.; Tissot, N.; Gaymard, F.; Briat, J.F.; Dubos, C. Facilitated Fe nutrition by phenolic compounds excreted by the Arabidopsis ABCG37/PDR9 transporter requires the IRT1/FRO2 high affinity root Fe2+ transport system. Mol. Plant 2016, 9, 485–488. [Google Scholar] [CrossRef] [PubMed]
- Schmid, N.B.; Giehl, R.F.H.; Döll, S.; Mock, H.P.; Strehmel, N.; Scheel, D.; Kong, X.; Hider, R.C.; von Wirén, N. Feruloyl-CoA 6′-hydroxylase1-dependent coumarins mediate iron acquisition from alkaline substrates in Arabidopsis. Plant Physiol. 2014, 164, 160–172. [Google Scholar] [CrossRef]
- Robinson, N.J.; Procter, C.M.; Connolly, E.L.; Guerinot, M.L. A ferric-chelate reductase for iron uptake from soils. Nature 1999, 397, 694–697. [Google Scholar] [CrossRef] [PubMed]
- Schagerlöf, U.; Wilson, G.; Hebert, H.; Al Karadaghi, S.; Hägerhäll, C. Transmembrane topology of FRO2, a ferric chelate reductase from Arabidopsis thaliana. Plant Mol. Biol. 2006, 62, 215–221. [Google Scholar] [CrossRef] [PubMed]
- Mukherjee, I.; Campbell, N.H.; Ash, J.S.; Connolly, E.L. Expression profiling of the Arabidopsis ferric chelate reductase (FRO) gene family reveals differential regulation by iron and copper. Planta 2006, 223, 1178–1190. [Google Scholar] [CrossRef]
- Connolly, E.L.; Campbell, N.H.; Grotz, N.; Prichard, C.L.; Guerinot, M.L. Overexpression of the FRO2 ferric chelate reductase confers tolerance to growth on low iron and uncovers posttranscriptional control. Plant Physiol. 2003, 133, 1102–1110. [Google Scholar] [CrossRef] [PubMed]
- Chang, Y.L.; Li, W.Y.; Miao, H.; Yang, S.Q.; Li, R.; Wang, X.; Li, W.Q.; Chen, K.M. Comprehensive genomic analysis and expression profiling of the NOX gene families under abiotic stresses and hormones in plants. Genome Biol. Evol. 2016, 8, 791–810. [Google Scholar] [CrossRef]
- Wu, H.; Li, L.; Du, J.; Yuan, Y.; Cheng, X.; Ling, H.Q. Molecular and biochemical characterization of the Fe(III) chelate reductase gene family in Arabidopsis thaliana. Plant Cell Physiol. 2005, 46, 1505–1514. [Google Scholar] [CrossRef]
- Feng, H.; An, F.; Zhang, S.; Ji, Z.; Ling, H.Q.; Zuo, J. Light-regulated, tissue-specific, and cell differentiation-specific expression of the Arabidopsis Fe(III)-chelate reductase gene AtFRO6. Plant Physiol. 2006, 140, 1345–1354. [Google Scholar] [CrossRef]
- Li, L.-Y.; Cai, Q.-Y.; Yu, D.-S.; Guo, C.-H. Overexpression of AtFRO6 in transgenic tobacco enhances ferric chelate reductase activity in leaves and increases tolerance to iron-deficiency chlorosis. Mol. Biol. Rep. 2011, 38, 3605–3613. [Google Scholar] [CrossRef] [PubMed]
- Jeong, J.; Cohu, C.; Kerkeb, L.; Pilon, M.; Connolly, E.L.; Guerinot, M.L. Chloroplast Fe(III) chelate reductase activity is essential for seedling viability under iron limiting conditions. Proc. Natl. Acad. Sci. USA 2008, 105, 10619–10624. [Google Scholar] [CrossRef]
- Korshunova, Y.O.; Eide, D.; Clark, W.G.; Guerinot, M.L.; Pakrasi, H.B. The IRT1 protein from Arabidopsis thaliana is a metal transporter with a broad substrate range. Plant Mol. Biol. 1999, 40, 37–44. [Google Scholar] [CrossRef] [PubMed]
- Rogers, E.E.; Eide, D.J.; Guerinot, M.L. Altered selectivity in an Arabidopsis metal transporter. Proc. Natl. Acad. Sci. USA 2000, 97, 12356–12360. [Google Scholar] [CrossRef]
- Barberon, M.; Zelazny, E.; Robert, S.; Conéjéro, G.; Curie, C.; Friml, J.; Vert, G. Monoubiquitin-dependent endocytosis of the IRON-REGULATED TRANSPORTER 1 (IRT1) transporter controls iron uptake in plants. Proc. Natl. Acad. Sci. USA 2011, 108, E450–E458. [Google Scholar] [CrossRef] [PubMed]
- Shin, L.J.; Lo, J.C.; Chen, G.H.; Callis, J.; Fu, H.; Yeh, K.C. IRT1 degradation factor1, a RING E3 ubiquitin ligase, regulates the degradation of iron-regulated transporter1 in Arabidopsis. Plant Cell 2013, 25, 3039–3051. [Google Scholar] [CrossRef]
- Barberon, M.; Dubeaux, G.; Kolb, C.; Isono, E.; Zelazny, E.; Vert, G. Polarization of IRON-REGULATED TRANSPORTER 1 (IRT1) to the plant-soil interface plays crucial role in metal homeostasis. Proc. Natl. Acad. Sci. USA 2014, 111, 8293–8298. [Google Scholar] [CrossRef]
- Ivanov, R.; Brumbarova, T.; Blum, A.; Jantke, A.M.; Fink Straube, C.; Bauer, P. SORTING NEXIN1 is required for modulating the trafficking and stability of the Arabidopsis IRON-REGULATED TRANSPORTER1. Plant Cell 2014, 26, 1294–1307. [Google Scholar] [CrossRef] [PubMed]
- Agorio, A.; Giraudat, J.; Bianchi, M.W.; Marion, J.; Espagne, C.; Castaings, L.; Lelièvre, F.; Curie, C.; Thomine, S.; Merlot, S. Phosphatidylinositol 3-phosphate-binding protein AtPH1 controls the localization of the metal transporter NRAMP1 in Arabidopsis. Proc. Natl. Acad. Sci. USA 2017, 114, E3354–E3363. [Google Scholar] [CrossRef] [PubMed]
- Takemoto, T.; Nomoto, K.; Fushiya, S.; Ouchi, R.; Kusano, G.; Hikino, H.; Takagi, S.I.; Matsuura, Y.; Kakudo, M. Structure of mugineic acid, a new amino acid possessing an iron-chelating activity from roots washings of water-cultured Hordeum vulgare L. Proc. Jpn. Acad. 1978, 54, 469–473. [Google Scholar] [CrossRef]
- Mori, S. Iron acquisition by plants. Curr. Opin. Plant Biol. 1999, 2, 250–253. [Google Scholar] [CrossRef]
- Negishi, T.; Nakanishi, H.; Yazaki, J.; Kishimoto, N.; Fujii, F.; Shimbo, K.; Yamamoto, K.; Sakata, K.; Sasaki, T.; Kikuchi, S.; et al. cDNA microarray analysis of gene expression during Fe-deficiency stress in barley suggests that polar transport of vesicles is implicated in phytosiderophore secretion in Fe-deficient barley roots. Plant J. 2002, 30, 83–94. [Google Scholar] [CrossRef] [PubMed]
- Conte, S.S.; Walker, E.L. Transporters contributing to iron trafficking in plants. Mol. Plant 2011, 4, 464–476. [Google Scholar] [CrossRef] [PubMed]
- Roberts, L.A.; Pierson, A.J.; Panaviene, Z.; Walker, E.L. Yellow Stripe1 expanded roles for the maize iron-phytosiderophore transporter. Plant Physiol. 2004, 135, 112–120. [Google Scholar] [CrossRef]
- Koike, S.; Inoue, H.; Mizuno, D.; Takahashi, M.; Nakanishi, H.; Mori, S.; Nishizawa, N.K. OsYSL2 is a rice metal-nicotianamine transporter that is regulated by iron and expressed in the phloem. Plant J. 2004, 39, 415–424. [Google Scholar] [CrossRef]
- Karim, M.R. Responses of Aerobic rice (Oryza sativa L.) to iron deficiency. J. Integr. Agric. 2012, 11, 938–945. [Google Scholar]
- Morrissey, J.; Baxter, I.R.; Lee, J.; Li, L.; Lahner, B.; Grotz, N.; Kaplan, J.; Salt, D.E.; Guerinot, M.L. The ferroportin metal efflux proteins function in iron and cobalt homeostasis in Arabidopsis. Plant Cell 2009, 21, 3326–3338. [Google Scholar] [CrossRef] [PubMed]
- López-Millán, A.F.; Morales, F.; Abadía, A.; Abadía, J. Effects of iron deficiency on the composition of the leaf apoplastic fluid and xylem sap in sugar beet. Implications for iron and carbon transport. Plant Physiol. 2000, 124, 873–884. [Google Scholar] [CrossRef] [PubMed]
- Durrett, T.P.; Gassmann, W.; Rogers, E.E. The FRD3-mediated efflux of citrate into the root vasculature is necessary for efficient iron translocation. Plant Physiol. 2007, 144, 197–205. [Google Scholar] [CrossRef] [PubMed]
- Rellán Álvarez, R.; Giner Martínez Sierra, J.; Orduna, J.; Orera, I.; Rodríguez Castrillón, J.Á.; García Alonso, J.I.; Abadía, J.; Álvarez Fernández, A. Identification of a tri-iron(III), tri-citrate complex in the xylem sap of iron-deficient tomato resupplied with iron: New insights into plant iron long-distance transport. Plant Cell Physiol. 2009, 51, 91–102. [Google Scholar] [CrossRef] [PubMed]
- Rellán-Álvarez, R.; Abadía, J.; Álvarez-Fernández, A. Formation of metal-nicotianamine complexes as affected by pH, ligand exchange with citrate and metal exchange. A study by electrospray ionization time-of-flight mass spectrometry. Rapid Commun. Mass Sp. 2008, 22, 1553–1562. [Google Scholar]
- Von Wiren, N.; Klair, S.; Bansal, S.; Briat, J.F.; Khodr, H.; Shioiri, T.; Leigh, R.A.; Hider, R.C. Nicotianamine chelates both FeIII and FeII. Implications for metal transport in plants. Plant Physiol. 1999, 119, 1107–1114. [Google Scholar] [CrossRef]
- Green, L.S.; Rogers, E.E. FRD3 controls iron localization in Arabidopsis. Plant Physiol. 2004, 136, 2523–2531. [Google Scholar] [CrossRef]
- Yokosho, K.; Yamaji, N.; Ma, J.F. OsFRDL1 expressed in nodes is required for distribution of iron to grains in rice. J. Exp. Bot. 2016, 67, 5485–5494. [Google Scholar] [CrossRef] [PubMed]
- Yokosho, K.; Yamaji, N.; Ueno, D.; Mitani, N.; Ma, J.F. OsFRDL1 is a citrate transporter required for efficient translocation of iron in rice. Plant Physiol. 2009, 149, 297–305. [Google Scholar] [CrossRef] [PubMed]
- Conte, S.; Stevenson, D.; Furner, I.; Lloyd, A. Multiple antibiotic resistance in Arabidopsis is conferred by mutations in a chloroplast-localized transport protein. Plant Physiol. 2009, 151, 559–573. [Google Scholar] [CrossRef]
- Schaaf, G.; Häberle, J.; von Wirén, N.; Schikora, A.; Curie, C.; Vert, G.; Briat, J.-F.; Ludewig, U. A Putative function for the Arabidopsis Fe–phytosiderophore transporter homolog AtYSL2 in Fe and Zn homeostasis. Plant Cell Physiol. 2005, 46, 762–774. [Google Scholar] [CrossRef]
- Bonneau, J.; Baumann, U.; Beasley, J.; Li, Y.; Johnson, A.A.T. Identification and molecular characterization of the nicotianamine synthase gene family in bread wheat. Plant Biotechnol. J. 2016, 14, 2228–2239. [Google Scholar] [CrossRef]
- Inoue, H.; Higuchi, K.; Takahashi, M.; Nakanishi, H.; Mori, S.; Nishizawa, N.K. Three rice nicotianamine synthase genes, OsNAS1, OsNAS2, and OsNAS3 are expressed in cells involved in long-distance transport of iron and differentially regulated by iron. Plant J. 2003, 36, 366–381. [Google Scholar] [CrossRef]
- Kawai, S.; Kamei, S.; Matsuda, Y.; Ando, R.; Kondo, S.; Ishizawa, A.; Alam, S. Concentrations of iron and phytosiderophores in xylem sap of iron-deficient barley plants. J. Soil Sci. Plant Nutr. 2001, 47, 265–272. [Google Scholar] [CrossRef]
- Mori, S.; Nishizawa, N.; Hayashi, H.; Chino, M.; Yoshimura, E.; Ishihara, J. Why are young rice plants highly susceptible to iron deficiency? Plant Soil 1991, 130, 143–156. [Google Scholar] [CrossRef]
- Curie, C.; Cassin, G.; Couch, D.; Divol, F.; Higuchi, K.; Le Jean, M.; Misson, J.; Schikora, A.; Czernic, P.; Mari, S. Metal movement within the plant: Contribution of nicotianamine and yellow stripe 1-like transporters. Ann. Bot. 2009, 103, 1–11. [Google Scholar] [CrossRef]
- DiDonato, R.J., Jr.; Roberts, L.A.; Sanderson, T.; Eisley, R.B.; Walker, E.L. Arabidopsis Yellow Stripe-Like2 (YSL2): A metal-regulated gene encoding a plasma membrane transporter of nicotianamine–metal complexes. Plant J. 2004, 39, 403–414. [Google Scholar] [CrossRef]
- Waters, B.M.; Chu, H.H.; DiDonato, R.J.; Roberts, L.A.; Eisley, R.B.; Lahner, B.; Salt, D.E.; Walker, E.L. Mutations in Arabidopsis Yellow Stripe-Like1 and Yellow Stripe-Like3 reveal their roles in metal ion homeostasis and loading of metal ions in seeds. Plant Physiol. 2006, 141, 1446–1458. [Google Scholar] [CrossRef]
- Jean, M.L.; Schikora, A.; Mari, S.; Briat, J.F.; Curie, C. A loss-of-function mutation in AtYSL1 reveals its role in iron and nicotianamine seed loading. Plant J. 2005, 44, 769–782. [Google Scholar] [CrossRef] [PubMed]
- Chu, H.-H.; Chiecko, J.; Punshon, T.; Lanzirotti, A.; Lahner, B.; Salt, D.E.; Walker, E.L. Successful reproduction requires the function of Arabidopsis Yellow Stripe-Like1 and Yellow Stripe-Like3 metal-nicotianamine transporters in both vegetative and reproductive structures. Plant Physiol. 2010, 154, 197–210. [Google Scholar] [CrossRef]
- Ishimaru, Y.; Masuda, H.; Bashir, K.; Inoue, H.; Tsukamoto, T.; Takahashi, M.; Nakanishi, H.; Aoki, N.; Hirose, T.; Ohsugi, R.; et al. Rice metal-nicotianamine transporter, OsYSL2, is required for the long-distance transport of iron and manganese. Plant J. 2010, 62, 379–390. [Google Scholar] [CrossRef]
- Kakei, Y.; Ishimaru, Y.; Kobayashi, T.; Yamakawa, T.; Nakanishi, H.; Nishizawa, N.K. OsYSL16 plays a role in the allocation of iron. Plant Mol. Biol. 2012, 79, 583–594. [Google Scholar] [CrossRef] [PubMed]
- Aoyama, T.; Kobayashi, T.; Takahashi, M.; Nagasaka, S.; Usuda, K.; Kakei, Y.; Ishimaru, Y.; Nakanishi, H.; Mori, S.; Nishizawa, N.K. OsYSL18 is a rice iron(III)-deoxymugineic acid transporter specifically expressed in reproductive organs and phloem of lamina joints. Plant Mol. Biol. 2009, 70, 681–692. [Google Scholar] [CrossRef] [PubMed]
- Senoura, T.; Sakashita, E.; Kobayashi, T.; Takahashi, M.; Aung, M.; Masuda, H.; Nakanishi, H.; Nishizawa, N.K. The iron-chelate transporter OsYSL9 plays a role in iron distribution in developing rice grains. Plant Mol. Biol. 2017, 95, 375–387. [Google Scholar] [CrossRef] [PubMed]
- Zhai, Z.; Gayomba, S.R.; Jung, H.I.; Vimalakumari, N.K.; Piñeros, M.; Craft, E.; Rutzke, M.A.; Danku, J.; Lahner, B.; Punshon, T.; et al. OPT3 is a phloem-specific iron transporter that is essential for systemic iron signaling and redistribution of iron and cadmium in Arabidopsis. Plant Cell 2014, 26, 2249–2264. [Google Scholar] [CrossRef]
- Mendoza-Cózatl, D.G.; Xie, Q.; Akmakjian, G.Z.; Jobe, T.O.; Patel, A.; Stacey, M.G.; Song, L.; Demoin, D.W.; Jurisson, S.S.; Stacey, G.; et al. OPT3 is a component of the iron-signaling network between leaves and roots and misregulation of OPT3 leads to an over-accumulation of cadmium in seeds. Mol. Plant 2014, 7, 1455–1469. [Google Scholar] [CrossRef] [PubMed]
- Wintz, H.; Fox, T.; Wu, Y.-Y.; Feng, V.; Chen, W.; Chang, H.-S.; Zhu, T.; Vulpe, C. Expression profiles of Arabidopsis thaliana in mineral deficiencies reveal novel transporters involved in metal homeostasis. J. Biol. Chem. 2003, 278, 47644–47653. [Google Scholar] [CrossRef] [PubMed]
- Leaden, L.; Pagani, M.A.; Balparda, M.; Busi, M.V.; Gomez-Casati, D.F. Altered levels of AtHSCB disrupts iron translocation from roots to shoots. Plant Mol. Biol. 2016, 92, 613–628. [Google Scholar] [CrossRef] [PubMed]
- Jain, A.; Wilson, G.T.; Connolly, E.L. The diverse roles of FRO family metalloreductases in iron and copper homeostasis. Front. Plant Sci. 2014, 5, 100. [Google Scholar] [CrossRef] [PubMed]
- Ravet, K.; Touraine, B.; Boucherez, J.; Briat, J.-F.; Gaymard, F.; Cellier, F. Ferritins control interaction between iron homeostasis and oxidative stress in Arabidopsis. Plant J. 2009, 57, 400–412. [Google Scholar] [CrossRef] [PubMed]
- Briat, J.-F.; Duc, C.; Ravet, K.; Gaymard, F. Ferritins and iron storage in plants. BBA-Gen. Subj. 2010, 1800, 806–814. [Google Scholar] [CrossRef]
- Marentes, E.; Grusak, M.A. Iron transport and storage within the seed coat and embryo of developing seeds of pea (Pisum sativum L.). Seed Sci. Res. 1998, 8, 367–375. [Google Scholar] [CrossRef]
- López-Millán, A.F.; Duy, D.; Philippar, K. Chloroplast iron transport proteins—Function and impact on plant physiology. Front. Plant Sci. 2016, 7, 178. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.A.; Punshon, T.; Lanzirotti, A.; Li, L.; Alonso, J.M.; Ecker, J.R.; Kaplan, J.; Guerinot, M.L. Localization of iron in Arabidopsis seed requires the vacuolar membrane transporter VIT1. Science 2006, 314, 1295–1298. [Google Scholar] [CrossRef] [PubMed]
- Lanquar, V.; Lelièvre, F.; Bolte, S.; Hamès, C.; Alcon, C.; Neumann, D.; Vansuyt, G.; Curie, C.; Schröder, A.; Krämer, U.; et al. Mobilization of vacuolar iron by AtNRAMP3 and AtNRAMP4 is essential for seed germination on low iron. EMBO J. 2005, 24, 4041–4051. [Google Scholar] [CrossRef] [PubMed]
- Thomine, S.; Lelièvre, F.; Debarbieux, E.; Schroeder, J.I.; Barbier-Brygoo, H. AtNRAMP3, a multispecific vacuolar metal transporter involved in plant responses to iron deficiency. Plant J. 2003, 34, 685–695. [Google Scholar] [CrossRef] [PubMed]
- Bashir, K.; Takahashi, R.; Akhtar, S.; Ishimaru, Y.; Nakanishi, H.; Nishizawa, N.K. The knockdown of OsVIT2 and MIT affects iron localization in rice seed. Rice 2013, 6, 31. [Google Scholar] [CrossRef] [PubMed]
- Bashir, K.; Hanada, K.; Shimizu, M.; Seki, M.; Nakanishi, H.; Nishizawa, N.K. Transcriptomic analysis of rice in response to iron deficiency and excess. Rice 2014, 7, 18. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Xu, Y.H.; Yi, H.Y.; Gong, J.M. Vacuolar membrane transporters OsVIT1 and OsVIT2 modulate iron translocation between flag leaves and seeds in rice. Plant J. 2012, 72, 400–410. [Google Scholar] [CrossRef]
- Nevo, Y.; Nelson, N. The NRAMP family of metal-ion transporters. Biochim. Biophys. Acta 2006, 1763, 609–620. [Google Scholar] [CrossRef] [PubMed]
- Harrison, P.M.; Arosio, P. The ferritins: Molecular properties, iron storage function and cellular regulation. Biochim. Biophys. Acta 1996, 1275, 161–203. [Google Scholar] [CrossRef]
- Reyt, G.; Boudouf, S.; Boucherez, J.; Gaymard, F.; Briat, J.F. Iron-and ferritin-dependent reactive oxygen species distribution: Impact on Arabidopsis root system architecture. Mol. Plant 2015, 8, 439–453. [Google Scholar] [CrossRef] [PubMed]
- Jain, A.; Connolly, E.L. Mitochondrial iron transport and homeostasis in plants. Front. Plant Sci. 2013, 4, 348. [Google Scholar] [CrossRef] [PubMed]
- Bashir, K.; Ishimaru, Y.; Shimo, H.; Nagasaka, S.; Fujimoto, M.; Takanashi, H.; Tsutsumi, N.; An, G.; Nakanishi, H.; Nishizawa, N.K. The rice mitochondrial iron transporter is essential for plant growth. Nat. Commun. 2011, 2, 322. [Google Scholar] [CrossRef]
- Conte, S.S.; Chu, H.H.; Rodriguez, D.C.; Punshon, T.; Vasques, K.A.; Salt, D.E.; Walker, E.L. Arabidopsis thaliana Yellow Stripe1-Like4 and Yellow Stripe1-Like6 localize to internal cellular membranes and are involved in metal ion homeostasis. Front. Plant Sci. 2013, 4, 283. [Google Scholar] [CrossRef] [PubMed]
- Duy, D.; Wanner, G.; Meda, A.R.; von Wirén, N.; Soll, J.; Philippar, K. PIC1, an ancient permease in Arabidopsis chloroplasts, mediates iron transport. Plant Cell 2007, 19, 986–1006. [Google Scholar] [CrossRef] [PubMed]
- Gong, X.; Guo, C.; Terachi, T.; Cai, H.; Yu, D. Tobacco PIC1 mediates iron transport and regulates chloroplast development. Plant Mol. Biol. Rep. 2015, 33, 401–413. [Google Scholar] [CrossRef]
- Ling, H.Q.; Pich, A.; Scholz, G.; Ganal, M.W. Genetic analysis of two tomato mutants affected in the regulation of iron metabolism. Mol. Gen. Genet. 1996, 252, 87–92. [Google Scholar] [CrossRef] [PubMed]
- Bauer, P.; Ling, H.Q.; Guerinot, M.L. FIT, the FER-LIKE IRON DEFICIENCY INDUCED TRANSCRIPTION FACTOR in Arabidopsis. Plant Physiol. Biochem. 2007, 45, 260–261. [Google Scholar] [CrossRef]
- Lingam, S.; Mohrbacher, J.; Brumbarova, T.; Potuschak, T.; Fink-Straube, C.; Blondet, E.; Genschik, P.; Bauer, P. Interaction between the bHLH transcription factor FIT and ETHYLENE INSENSITIVE3/ETHYLENE INSENSITIVE3-LIKE1 reveals molecular linkage between the regulation of iron acquisition and ethylene signaling in Arabidopsis. Plant Cell 2011, 23, 1815–1829. [Google Scholar] [CrossRef]
- Colangelo, E.P.; Guerinot, M.L. The essential basic Helix-Loop-Helix protein FIT1 is required for the iron deficiency response. Plant Cell 2004, 16, 3400–3412. [Google Scholar] [CrossRef]
- Jakoby, M.; Wang, H.Y.; Reidt, W.; Weisshaar, B.; Bauer, P. FRU (BHLH029) is required for induction of iron mobilization genes in Arabidopsis thaliana. FEBS Lett. 2004, 577, 528–534. [Google Scholar] [CrossRef] [PubMed]
- Yuan, Y.X.; Zhang, J.; Wang, D.W.; Ling, H.Q. AtbHLH29 of Arabidopsis thaliana is a functional ortholog of tomato FER involved in controlling iron acquisition in strategy I plants. Cell Res. 2005, 15, 613. [Google Scholar] [CrossRef]
- Wang, H.-Y.; Klatte, M.; Jakoby, M.; Bäumlein, H.; Weisshaar, B.; Bauer, P. Iron deficiency-mediated stress regulation of four subgroup Ib BHLH genes in Arabidopsis thaliana. Planta 2007, 226, 897–908. [Google Scholar] [CrossRef] [PubMed]
- Wang, N.; Cui, Y.; Liu, Y.; Fan, H.; Du, J.; Huang, Z.; Yuan, Y.; Wu, H.; Ling, H.Q. Requirement and functional redundancy of Ib subgroup bHLH proteins for iron deficiency responses and uptake in Arabidopsis thaliana. Mol. Plant 2013, 6, 503–513. [Google Scholar] [CrossRef] [PubMed]
- Yuan, Y.; Wu, H.; Wang, N.; Li, J.; Zhao, W.; Du, J.; Wang, D.; Ling, H.Q. FIT interacts with AtbHLH38 and AtbHLH39 in regulating iron uptake gene expression for iron homeostasis in Arabidopsis. Cell Res. 2008, 18, 385. [Google Scholar] [CrossRef] [PubMed]
- Wild, M.; Davière, J.M.; Regnault, T.; Sakvarelidze Achard, L.; Carrera, E.; Lopez Diaz, I.; Cayrel, A.; Dubeaux, G.; Vert, G.; Achard, P. Tissue-specific regulation of gibberellin signaling fine-tunes Arabidopsis iron-deficiency responses. Dev. Cell 2016, 37, 190–200. [Google Scholar] [CrossRef] [PubMed]
- Long, T.A.; Tsukagoshi, H.; Busch, W.; Lahner, B.; Salt, D.E.; Benfey, P.N. The bHLH transcription factor POPEYE regulates response to iron deficiency in Arabidopsis roots. Plant Cell 2010, 22, 2219–2236. [Google Scholar] [CrossRef] [PubMed]
- Palmer, C.M.; Hindt, M.N.; Schmidt, H.; Clemens, S.; Guerinot, M.L. MYB10 and MYB72 are required for growth under iron-limiting conditions. PLoS Genet. 2013, 9, e1003953. [Google Scholar] [CrossRef] [PubMed]
- Zamioudis, C.; Hanson, J.; Pieterse, C.M.J. β-Glucosidase BGLU42 is a MYB72-dependent key regulator of rhizobacteria-induced systemic resistance and modulates iron deficiency responses in Arabidopsis roots. New Phytol. 2014, 204, 368–379. [Google Scholar] [CrossRef] [PubMed]
- Yan, J.Y.; Li, C.X.; Sun, L.; Ren, J.Y.; Li, G.X.; Ding, Z.J.; Zheng, S.J. A WRKY transcription factor regulates fe translocation under Fe deficiency. Plant Physiol. 2016, 171, 2017–2027. [Google Scholar] [CrossRef]
- Kobayashi, T.; Ogo, Y.; Aung, M.S.; Nozoye, T.; Itai, R.N.; Nakanishi, H.; Yamakawa, T.; Nishizawa, N.K. The spatial expression and regulation of transcription factors IDEF1 and IDEF2. Ann. Bot. 2010, 105, 1109–1117. [Google Scholar] [CrossRef] [PubMed]
- Ogo, Y.; Nakanishi Itai, R.; Nakanishi, H.; Kobayashi, T.; Takahashi, M.; Mori, S.; Nishizawa, N.K. The rice bHLH protein OsIRO2 is an essential regulator of the genes involved in Fe uptake under Fe-deficient conditions. Plant J. 2007, 51, 366–377. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Zhu, X.; Zhang, F.S. Role of shoot in regulation of iron deficiency responses in cucumber and bean plants. J. Plant Nutr. 2000, 23, 1809–1818. [Google Scholar] [CrossRef]
- Li, X.; Li, C. Is ethylene involved in regulation of root ferric reductase activity of dicotyledonous species under iron deficiency? Plant Soil 2004, 261, 147–153. [Google Scholar] [CrossRef]
- Shao, J.Z.; Tang, C.; Arakawa, Y.; Masaoka, Y. The responses of red clover (Trifolium pratense L.) to iron deficiency: A root Fe(III) chelate reductase. Plant Sci. 2003, 164, 679–687. [Google Scholar]
- Lei, G.J.; Zhu, X.F.; Wang, Z.W.; Dong, F.; Dong, N.Y.; Zheng, S.J. Abscisic acid alleviates iron deficiency by promoting root iron reutilization and transport from root to shoot in Arabidopsis. Plant Cell Environ. 2014, 37, 852–863. [Google Scholar] [CrossRef]
- Matsuoka, K.; Bidadi, H.; Furukawa, J.; Satoh, S.; Asahina, M.; Yamaguchi, S. Gibberellin-induced expression of Fe uptake-related genes in Arabidopsis. Plant Cell Physiol. 2013, 55, 87–98. [Google Scholar] [CrossRef]
- Séguéla, M.; Briat, J.-F.; Vert, G.; Curie, C. Cytokinins negatively regulate the root iron uptake machinery in Arabidopsis through a growth-dependent pathway. Plant J. 2008, 55, 289–300. [Google Scholar] [CrossRef]
- Maurer, F.; Müller, S.; Bauer, P. Suppression of Fe deficiency gene expression by jasmonate. Plant Physiol. Biochem. 2011, 49, 530–536. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.W.; Yang, J.L.; Qin, C.; Jin, C.W.; Mo, J.H.; Ye, T.; Zheng, S.J. Nitric oxide acts downstream of auxin to trigger root ferric-chelate reductase activity in response to iron deficiency in Arabidopsis. Plant Physiol. 2010, 154, 810–819. [Google Scholar] [CrossRef]
- Ye, Y.Q.; Jin, C.W.; Fan, S.K.; Mao, Q.Q.; Sun, C.L.; Yu, Y.; Lin, X.Y. Elevation of NO production increases Fe immobilization in the Fe-deficiency roots apoplast by decreasing pectin methylation of cell wall. Sci. Rep. 2015, 5, 10746. [Google Scholar] [CrossRef]
- Kudla, J.; Xu, Q.; Harter, K.; Gruissem, W.; Luan, S. Genes for calcineurin B-like proteins in Arabidopsis are differentially regulated by stress signals. Proc. Natl. Acad. Sci. USA 1999, 96, 4718–4723. [Google Scholar] [CrossRef] [PubMed]
- Weinl, S.; Kudla, J. The CBL–CIPK Ca2+-decoding signaling network: Function and perspectives. New Phytol. 2009, 184, 517–528. [Google Scholar] [CrossRef] [PubMed]
- Tian, Q.; Zhang, X.; Yang, A.; Wang, T.; Zhang, W.-H. CIPK23 is involved in iron acquisition of Arabidopsis by affecting ferric chelate reductase activity. Plant Sci. 2016, 246, 70–79. [Google Scholar] [CrossRef]
- Fuglsang, A.T.; Guo, Y.; Cuin, T.A.; Qiu, Q.; Song, C.; Kristiansen, K.A.; Bych, K.; Schulz, A.; Shabala, S.; Schumaker, K.S. Arabidopsis protein kinase PKS5 inhibits the plasma membrane H+-ATPase by preventing interaction with 14-3-3 protein. Plant Cell 2007, 19, 1617–1634. [Google Scholar] [CrossRef] [PubMed]
- Gratz, R.; Manishankar, P.; Ivanov, R.; Köster, P.; Mohr, I.; Trofimov, K.; Steinhorst, L.; Meiser, J.; Mai, H.-J.; Drerup, M.; et al. CIPK11-dependent phosphorylation modulates FIT activity to promote Arabidopsis iron acquisition in response to calcium signaling. Dev. Cell 2019, 48, 726–740. [Google Scholar] [CrossRef] [PubMed]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, X.; Zhang, D.; Sun, W.; Wang, T. The Adaptive Mechanism of Plants to Iron Deficiency via Iron Uptake, Transport, and Homeostasis. Int. J. Mol. Sci. 2019, 20, 2424. https://doi.org/10.3390/ijms20102424
Zhang X, Zhang D, Sun W, Wang T. The Adaptive Mechanism of Plants to Iron Deficiency via Iron Uptake, Transport, and Homeostasis. International Journal of Molecular Sciences. 2019; 20(10):2424. https://doi.org/10.3390/ijms20102424
Chicago/Turabian StyleZhang, Xinxin, Di Zhang, Wei Sun, and Tianzuo Wang. 2019. "The Adaptive Mechanism of Plants to Iron Deficiency via Iron Uptake, Transport, and Homeostasis" International Journal of Molecular Sciences 20, no. 10: 2424. https://doi.org/10.3390/ijms20102424
APA StyleZhang, X., Zhang, D., Sun, W., & Wang, T. (2019). The Adaptive Mechanism of Plants to Iron Deficiency via Iron Uptake, Transport, and Homeostasis. International Journal of Molecular Sciences, 20(10), 2424. https://doi.org/10.3390/ijms20102424




