Abstract
Most commercial cultivars of lily are sensitive to abiotic stresses. However, tiger lily (Lilium lancifolium L.), one of the most widely distributed wild lilies in Asia, has strong abiotic stresses resistance. Thus, it is indispensable to identify stress-responsive candidate genes in tiger lily for the stress resistance improvement of plants. In this study, a MYB related homolog (LlMYB3) from tiger lily was functionally characterized as a positive regulator in plant stress tolerance. LlMYB3 is a nuclear protein with transcriptional activation activity at C-terminus. The expression of LlMYB3 gene was induced by multiple stress treatments. Several stress-related cis-acting regulatory elements (MYBRS, MYCRS, LTRE and DRE/CRT) were located within the promoter of LlMYB3; however, the promoter activity was not induced sufficiently by various stresses treatments. Overexpressing LlMYB3 in Arabidopsis thaliana L. transgenic plants showed ABA hypersensitivity and enhanced tolerance to cold, drought, and salt stresses. Furthermore, we found LlMYB3 highly co-expressed with LlCHS2 gene under cold treatment; yeast one-hybrid (Y1H) assays demonstrated LlMYB3 was able to bind to the promoter of LlCHS2. These findings suggest that the stress-responsive LlMYB3 may be involved in anthocyanin biosynthesis pathway to regulate stress tolerance of tiger lily.
1. Introduction
Environmental stresses, such as drought, high salinity, and extreme temperatures, adversely affect the growth, development, and productivity of plants. To adapt to these environmental stressors, plants have evolved complex signaling cascades to regulate the expression of stress-related genes that can improve stress tolerance either directly or indirectly []. Many proteins and genes in the complex signaling networks are regulated by multiple transcription factor (TF) families. As one of the largest TF groups in plants, the MYB family has been proven to be essential for responding to abiotic stresses [,].
MYB proteins are characterized by their highly conserved DNA-binding domains [,]. According to the number of imperfect repeats of the SANT (for SWI3, ADA2, N-CoR, and TFIIIB) domain (50–53 amino acids) in MYB DNA-binding domains, plant MYB proteins can be mainly sub divided into three subfamilies: MYB-related (one single SANT domain), the R2R3-MYB (two SANT domains) and R1R2R3-MYB (three SANT domains) []. Accordingly, research on MYB genes has mainly focused on the R2R3-MYB gene family, which has been shown to play important roles in many plant-specific processes including the response to abiotic stress in past decades []. In Arabidopsis thaliana L., AtMYB14 and AtMYB15 enhance freezing tolerance by regulating CBF and its downstream target genes [,]; AtMYB20 and AtMYB44 confer salt and drought resistance respectively by downregulating the expression of PP2Cs [,]; AtMYB2, AtMYB15 and AtMYB96 function in the ABA-mediated drought and salt stress response [,,]. In rice (Oryza sativa L.), OsMYB4 was reported to be a positive regulator in transgenic Arabidopsis, tomato (Solanum lycopersicum L. cv. Tondino), and apple (Malus pumila Mill.) to cold and drought tolerance [,,]; Ectopic expression of OsMYB2 facilitated salt, cold, dehydration tolerance in rice []. In wheat (Triticum aestivum L.), overexpression of TaMYBsm1, TaMYB33, TaMYB2A and TaMYB30-B have been shown to improve the drought tolerance in Arabidopsis [,,,]; TaMYB73 can improve salinity stress tolerance in Arabidopsis []; TaMYB19 has been found to participate in responses to abiotic stress in transgenic Arabidopsis []. However, compared to 2R-MYB genes, there are few reports of functional studies of other MYB subfamilies in abiotic stress response in plants [].
MYB TFs have also been identified to be the major determinant regulators in anthocyanin biosynthesis []. Interestingly, a cross-talk exists between abiotic stresses responses and anthocyanin biosynthesis. For instance, low temperature induced and high temperature suppressed anthocyanin biosynthesis in Arabidopsis, which involved the altered regulation of AtMYB3, AtMYB6 and AtMYBL2 [,]. Overexpression of AtPAP1 or AtMYB12, two flavonol synthesis regulators, enhances oxidative and drought tolerance in Arabidopsis []. MaMYB10 in apple, PcMYB10 in pear (Pyrus communis L.) and BrMYB2-2 in Brassica rapa are responsible for the temperature affected anthocyanin accumulation [,,]. Thus, it is pertinent to propose that some MYB proteins may be involved in the correlation between anthocyanin accumulation and abiotic stress tolerance.
As a wild stress-resistant plant, tiger lily (Lilium lancifolium L.) has been shown to have capacity for resisting multiple abiotic stresses [,], which could be an ideal model to study stress tolerance mechanisms and signaling regulation of a stress-resistant plant. Based on our RNA-seq data published previously [], we isolated a MYB-related type gene, LlMYB3, from L. lancifolium, to further study the role of LlMYB3 in plant stress response. Our work showed that LlMYB3 was induced by cold, drought, salt and exogenous ABA treatments. Ectopic expression of LlMYB3 could improve cold, drought and salt tolerance by upregulating the transcription of several stress-related genes in Arabidopsis. Moreover, LlMYB3 TF might be involved in anthocyanin biosynthesis, which can bind to the promoter of LlCHS2. These results provide valuable insights into the role of the LlMYB3 in regulating plant stress response.
2. Results
2.1. Isolation of LlMYB3 and Sequence Analysis
LlMYB3 gene comprises 1205 bp nucleotides with 462 bp open reading frame, 341 bp 5′ UTR, 402 bp 3′ UTR. It encodes a putative protein of 153 amino acids with a calculated molecular mass of 17.71kD and a pI of 9.54. Amino acid analysis revealed that the LlMYB3 is a MYB-related type protein with one single conserved SANT domain at the N-terminus between 39 and 86 amino acids (Figure 1a). A phylogenetic tree based on the amino acid sequences of some well-studied MYB proteins was constructed [], which revealed that LlMYB was clustered closely to OsMYB4, OsMYB2 and AtMYB41 (Figure 1b). Furthermore, we BLAST the DNA sequence of LlMYB3 to the whole-genome sequence of Arabidopsis thaliana using The Arabidopsis Information Resource (TAIR) to locate the chromosomal location. The BLAST result showed that the highest score (bits) significant alignment of LlMYB3 was AT5G62320.1 which was located in No.5 chromosome (Figure S1).
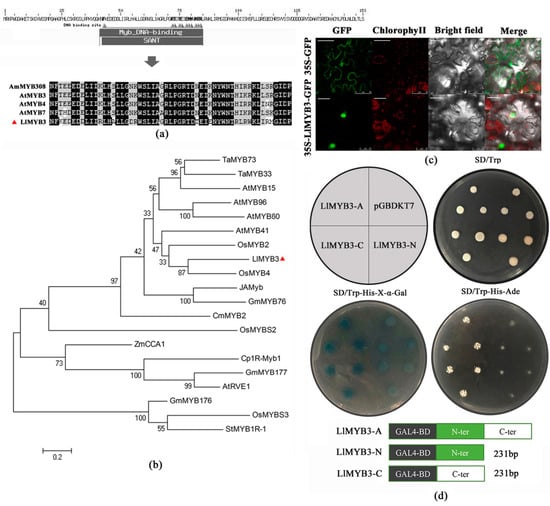
Figure 1.
Characterization of tiger lily LlMYB3 protein. (a) Alignment of LlMYB3 with Antirrhinum majus AmMYB308, Arabidopsis AtMYB3, AtMYB4 and AtMYB5. The conserved MYB domain is marked and identical amino acids are shaded in black. (b) Phylogenetic tree analysis of LlMYB3 with other known stress-responsive MYB proteins. The LlMYB3 is marked with red triangle. (c) GFP and GFP-LlMYB3 fusion proteins were transiently expressed in tobacco leaves under control of the CaMV35S promoter and observed under a laser scanning confocal microscope. Scale bars for 35S-GFP, 50 μm; for 35S-LlMYB3-GFP, 25 μm. (d) Full-length protein (LlMYB3-A), N-terminal fragment (LlMYB3-N) and C-terminal fragment (LlMYB3-C) were fused with GAL4 DNA binding domain and expressed in yeast strain Y2HGold. The pGBDKT7 vector was used as a negative control. Transformed yeasts were dripped on the SD/-Trp, SD/-Trp-His-x-gal and SD/-Trp-His-Ade, after being cultured for 3 days in the growth chamber.
2.2. Subcellular Localization and Transactivation Assay of LlMYB3
The GFP-LlMYB3 fusion construct and the GFP control in pBI121-GFP vector driven by CaMV35S promoter were transiently expressed in tobacco epidermal cells and visualized under a laser scanning confocal microscope to determine the subcellular localization of LlMYB3. Results showed that the fluorescence signals from GFP alone were widely distributed throughout the cells, whereas, the GFP-LlMYB3 fusion protein fluorescence signal was mainly detected in the nucleus (Figure 1c). Thus, these results demonstrated that LlMYB3 is a nuclear protein.
To investigate the transcriptional activity of LlMYB3 protein, the entire coding region, N-terminal and C-terminal domain coding sequence were inserted into the pGBDKT7 vector, which contains the GAL4 DNA-binding domain. The transactivation results showed that all transformed yeast cells grew well on SD/-Trp medium (Figure 1d). The yeast strain containing the full-length LlMYB3 (LlMYB3-A) and the C-terminus of LlMYB3 (LlMYB3-C) could grow well on the selection medium SD/-Trp/-His/-Ade, while the cells with the N-terminus of LlMYB3 (LlMYB3-N) and pGBDKT7 empty vector could not grow normally (Figure 1d). Furthermore, the yeast cells that grew well on the SD/-Trp/-His-x-α-gal medium appeared blue in the presence of α-galactosidase, indicating the activation of the reporter gene Mel1 (Figure 1d). These results indicated that LlMYB3 is a transcriptional activator, and its transactivation domain locates in the C-terminal region.
2.3. Expression Patterns of LlMYB3 under Multiple Stresses and ABA
To explore the possible involvement of LlMYB3 in abiotic stress response, we analyzed the expression patterns of LlMYB3 in tiger lily plants after treatment with abiotic stresses and ABA. The qRT-PCR analyses revealed that the LlMYB3 gene has relatively high expression levels in bulb and flower petal, while its expression was low in the leaf and stem (Figure 2a). The expression of LlMYB3 was significantly and rapidly induced within 1 h after cold, salt and drought treatment, leading to sevenfold to ninefold increase, but only in the salt treatment, the transcript level of LlMYB3 increased again during 6–24 h (Figure 2c–e). Treatment of tiger lily plants with ABA induced the expression of LlMYB3, showing fivefold to sixfold increases at 24 h after treatment (Figure 2b). These data indicate that LlMYB3 is a stress-responsive MYB-related gene in tiger lily, and its expression is sensitive to cold, drought and salt signaling molecules.
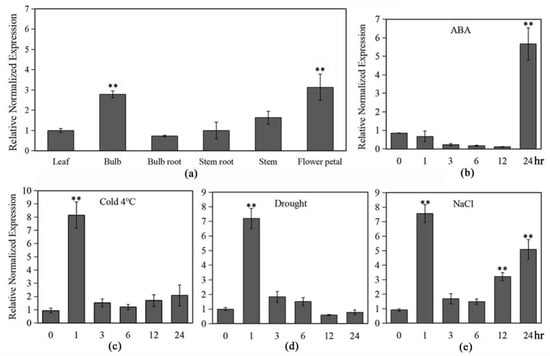
Figure 2.
Expression patterns of LlMYB3 in tiger lily seedlings under different stress treatments. Expression patterns of LlMYB3 in leaves, bulbs, roots, stems, and flower petal (a), and after ABA (b), cold (c), drought (d) and NaCl (e) treatments in leaves by qRT-PCR analysis. Transcript levels were normalized to LlTIP1. Values are means ± SD of three replicates. Three independent experiments were performed. Asterisks indicate a significant difference (** p < 0.01) compared with the corresponding controls.
2.4. Promoter Analysis of LlMYB3 under Multiple Stresses and ABA
To clarify the mechanism underlying the stress-inducible expressions of LlMYB3, the 1374 bp upstream of ATG start codon LlMYB3 promoter sequence was cloned and used to drive the GUS expression in Arabidopsis. Sequences of various putative stress-related cis-acting regulatory elements were identified, including MYB, MYC recognition sites, LTRE, DRE/CRT, TGA and ERE elements (Table 1).

Table 1.
Stress-related cis-acting regulatory elements identified in the promoter region of LlMYB3.
By histochemical GUS staining, we only observed prominent GUS staining in leaf veins of over 2-week old transgenic LlMYB3 promoter plants (Figure 3a). There was no obvious difference in GUS staining between stress-treated and non-treated transgenic plants. Thus, the stress inducible activity of the LlMYB3 promoter was revealed by measuring GUS gene expression levels in the transgenic Arabidopsis through qRT-PCR analyses. The result showed that GUS gene transcript level could be induced by cold, ABA, salt and drought treatments with a maximal level at 2 and 12 h, respectively (Figure 3b). By GUS enzyme activity assay, however, only an extremely weak fluorescence signal was detected. These results indicated that the activity of LlMYB3 promoter can be induced by multiple stress treatments, while it is not strong enough to mediate the GUS enzyme activity.
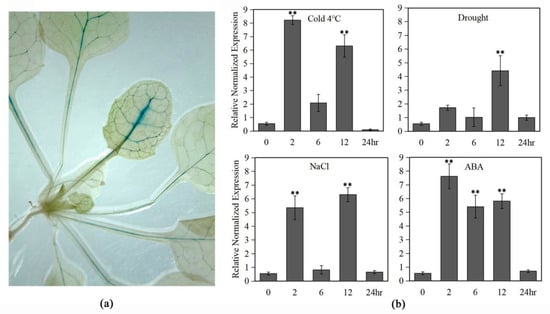
Figure 3.
GUS activity of the LlMYB3 promoter in transgenic Arabidopsis plants. (a) Beta-glucuronidase (GUS) expression in untreated transgenic Arabidopsis plants. (b) The GUS transcript levels in the leaves of the transgenic Arabidopsis under cold (4 °C), drought, salt and ABA treatments. The untreated transformants served as controls. There were 12 transgenic lines acquired. Values are means ±SD of three replicates. Three independent experiments were performed. Asterisks indicate a significant difference (** p < 0.01) compared with the corresponding controls.
2.5. Overexpression of LlMYB3 in Arabidopsis Improves Tolerance to Cold and Drought Stresses
To explore the function of LlMYB3 in providing tolerance to abiotic stress in plants, transgenic Arabidopsis plants overexpressing LlMYB3 driven by the CaMV35S promoter were generated. Two independent homozygous lines LlMYB3-5 (L5) and LlMYB3-8 (L8) with relatively high expression levels (Figure S2) were selected for the analysis.
To study the effect of LlMYB3 overexpression on cold stress, LlMYB3 transgenic lines and wild-type (WT) plants were grown in equal amounts of potting soil for 4 weeks under normal conditions, and cold stress was applied by being exposed to −4, −6, −8 °C for 12 h. The results showed that all plants grew well under −4 °C treatment as the same as under normal temperature 22 °C (Figure 4a). When the temperature decreased to −6 °C, most of WT plants were dead with a survival rate at approximately 20%, but over half of transgenic plants survived (Figure 4a,b). Furthermore, all WT plants were dead, whereas the survival rate for transgenic plants was observed at 30–35% under −8 °C treatment (Figure 4a,b). In a further experiment, 4-week-old plants were treated at 4 °C for 3 h, and the relative electrolyte leakage and soluble sugars were measured after treatment. As a result, the electrolyte leakage was lower in transgenic plants relative to WT plants (Figure 4c); and transgenic plants produced remarkably higher levels of soluble sugars under a chilling condition compared to WT plants (Figure 4d).
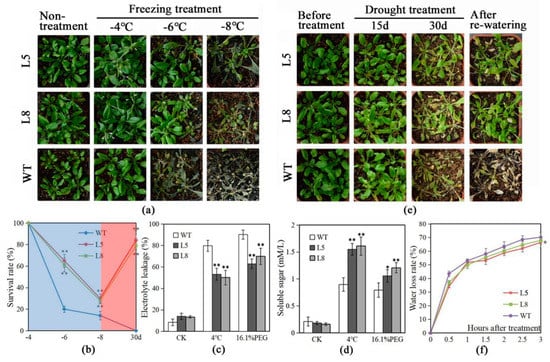
Figure 4.
Overexpression of LlMYB3 in Arabidopsis improves the freezing and drought tolerance. Performance of wild-type (WT) and LlMYB3 transgenic lines after freezing (a) and drought (e) treatments. (b) Survival rate of plants in (a) under freezing temperatures (blue region) and in (e) after drought treatment for 30 days (red region). Each data point is the mean of four experiments, and each experiment comprises 30 plants. Relative electrolyte leakage (c) and soluble sugar content (d) in WT and LlMYB3 transgenic lines after 4 °C and 16.1% PEG6000 treatments for 3 h. (f) Water loss rate of leaves from WT and transgenic Arabidopsis. The data represent the means from three experiments. The bars show the SD. Significant differences between the transgenic and WT lines are indicated as * 0.01 < p < 0.05 and ** p < 0.01.
Similarly, to study drought stress tolerance, after withholding water for 30 days, WT plants showed visible symptoms of drought-induced damage, such as drying, wilting, and even death while some transgenic plants remained green with expanded leaves (Figure 4e). Further analyses showed that after re-watering, few WT plants survived, whereas about 78–82% of transgenic plants continued to grow (Figure 4e,f). Additionally, after being treated with 16.1% PEG6000 (−0.5 MPa) for 3 h, transgenic plants showed lower electrolyte leakage and higher levels of soluble sugars compared to WT plants (Figure 4c,d). The water-loss rates were also slightly lower in transgenic plants (L5) than in WT plants after 3 h treatment (Figure 4f).
2.6. Overexpression of LlMYB3 in Arabidopsis Increases Seed Sensitivity to ABA and Tolerance to NaCl
The salt tolerance and ABA sensitivity of LlMYB3 transgenic plants was assessed. NaCl significantly inhibited Arabidopsis germination when the seeds were cultivated on MS medium supplemented with 50 mM NaCl (Figure 5a). Only about 30% of the WT seeds germinated in MS medium containing 50 mM NaCl while about 60–65% of transgenic plants seeds were able to germinate (Figure 5a,b). In contrast, except that WT seeds germinated slower in MS medium containing 2 µM ABA, no obvious difference was observed between the germination of WT seeds cultivated on MS medium supplemented with 0 and 2 µM ABA. However, the germination ratio of transgenic plants seeds was remarkably lower than that of WT seeds in MS medium containing 2 µM ABA (Figure 5a,b). Therefore, we suggested that LlMYB3 transgenic plants are more tolerant to salt stresses and more hypersensitive to ABA than WT plants.
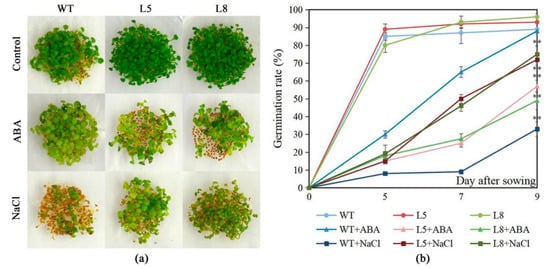
Figure 5.
Hypersensitivity and enhancing tolerance of LlMYB3 transgenic lines to ABA and NaCl. Germination of WT seeds of Col-0 and 35S:LlMYB3 on MS supplemented with 50 mM NaCl and 2 µM ABA after 9 days of incubation at 22 °C (a). Germination rate of seeds counted after 5, 7 and 9 days after sowing. The data represent the means from three experiments. The bars show the SD. Significant differences between the transgenic and WT lines are indicated as ** p < 0.01.
2.7. Altered Expression of Stress-Responsive Genes in LlMYB3 Transgenic Plants
The LlMYB3 transgenic plants exhibited an improved tolerance to freezing, drought and salt stresses. We then measured the expression levels of genes involving stress response in the transgenic plants under normal conditions. Except for AtCOR47, transcripts of AtRD29A, AtRD29B, AtRD20, AtABI5, AtGolS1, AtLEA14 and AtAPX2 genes (NCBI accession numbers are shown in Table S1) accumulated in LlMYB3 transgenic plants compared to WT plants (Figure 6). The enhanced expression of these genes in transgenic plants might contribute to the stronger stress tolerance, which also implied that LlMYB3 TF may confer stress tolerance through regulating various stress-responsive genes.
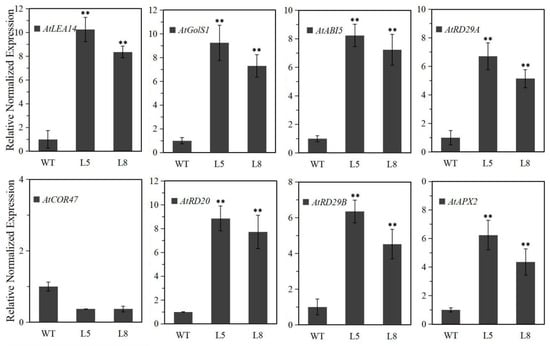
Figure 6.
Expression levels of the stress-associated genes under normal condition in WT and LlMYB3 transgenic plants. Gene-specific primers were used to detect the relative transcript levels of the stress-related genes. Values are means ± SD of three replicates. Three independent experiments were performed. Asterisks indicate a significant difference (** p < 0.01) compared with the corresponding controls.
2.8. LlMYB3 Can Bind to the Promoter of LlCHS2
In our previous study, through the analysis of gene co-expression networks involved in cold resistance of tiger lily, we found that the LlMYB3 was highly co-expressed with genes involved in anthocyanin biosynthesis pathway, including phenylalanine ammonia-lyase (PAL), cinnamic acid 4-hydroxylase (C4H), 4-hydroxycinnamoyl-CoA ligase (4CL), chalcone synthases (CHS) and flavonol synthase (FLS) []. In this study, the results of qRT-PCR and Pearson’s correlation coefficient (r) confirmed that the expression pattern of LlMYB3 was significantly similar to LlCHS2’s (chalcone synthase2) (r > 0.8) under continuous cold treatment (Figure 7a and Table S4). The LlCHS2 gene information is shown in Figure S3. Thus, we performed the Y1H assay to explore whether there is an interaction between LlMYB3 protein and LlCHS2 promoter. The 932 bp upstream of ATG start codon LlCHS2 promoter sequence was cloned, and the fragment (−820 to −553) of the LlCHS2 promoter containing four MYB binding sites was isolated (Figure S3 and Table S3). The minimal inhibitory concentration of Aureobasidin A (AbA) for bait yeast strains was found to be 200 ng·mL−1 (Figure S4). Yeast cells transformed with pGADT7-LlMYB3 and pAbAi-proLlCHS2 grew well on SD/Leu plates with 200 and 250 ng·mL−1 AbA (Figure 7b). This showed that LlMYB3 could bind to the promoter of LlCHS2; suggesting LlMYB3 is involved in the regulatory pathway of LlCHS2.
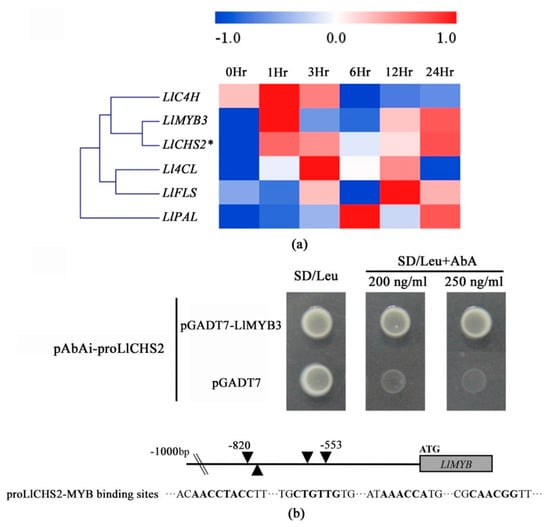
Figure 7.
Yeast one-hybrid analysis of LlMYB3 binding to LlCHS2 promoter. (a) Correlation analysis of expression patterns of LlMYB3 and some structural genes involved in anthocyanin biosynthesis pathway under continuous cold treatment. (b) Yeast one-hybrid analysis and schematic representation of MYB binding sites in the LlCHS2 promoter. Yeast strain Y1HGold was co-transformed with bait (pAbAi-proLlCHS2) and a prey (pGADT7 or pGADT7-LlMYB3) construct. Interaction between bait and prey was determined by cell growth on SD medium lacking Leu in the presence of 200 and 250 ng·mL−1 AbA.
3. Discussion
Considerable studies indicate that plant MYB family members play critical roles in response to abiotic stresses. However, the evidence of improved stress resistance for most MYB genes is mainly from model species as Arabidopsis and rice. In this study, a cold stress-responsive gene LlMYB3 was cloned and characterized from tiger lily (Lilium lancifolium L.), to investigate the role of this MYB gene response to various abiotic stresses. Sequence analysis shows that LlMYB3 is a MYB-related type protein with one single conserved SANT domain and displays high identity with reported stress responsive MYB member OsMYB4, OsMYB2 and AtMYB41. As predicted that LlMYB3 is a TF, LlMYB3 protein is localized in the nucleus and both the C-terminal and full-length LlMYB3 have high transactivation ability in yeast. Our previous transcriptome data analysis identified a unigene contig 10,499 coding for LlMYB3, which showed significant changes in expression in tiger lily under cold stress [,]. Here, we further confirmed that LlMYB3 was also up-regulated by drought, salt and ABA treatments.
Several stress-related cis-elements are present in the promoter of LlMYB3. The LlMYB3 promoter activity is shown to be induced by multiple stress treatments; however, it is not strong enough to mediate the GUS activity significantly. It indicates that the LlMYB3 promoter is not an effective stress-inducible promoter, and the expression of LlMYB3 responding to abiotic stresses might mainly be regulated by the upstream regulatory factors. Furthermore, the promoter of LlMYB3 shows vascular vein specific expression in transgenic Arabidopsis leaves. Usually, an expression pattern of a gene by promoter analysis could reflect its function []. The gene expression in leaf veins is usually regulated by the alteration of environments and physiological metabolic signals of this tissue during leaf development and growth []. Thus, it is assumed that LlMYB3 gene might function in vascular tissues in leaves during stress response.
Compared to R2R3-MYB genes, few studies of the MYB-related genes in abiotic stress response have been reported in plants []. For instance, AtMYBC1, a 1R-MYB protein, was reported to be a repressor of freezing tolerance in a CBF independent pathway in Arabidopsis []. In rice, MYBS3 was shown to be essential for cold stress tolerance []; and overexpression of OsMYB48-1 enhanced drought and salinity tolerance in rice []. In potato, the single MYB domain TF StMYB1R-1 has been shown to involve in drought tolerance by activation of drought-related genes []. Overexpression of a single-repeat MYB TF AmMYB1 from grey mangrove conferred salt tolerance in transgenic tobacco []. In the present study, we generated transgenic Arabidopsis plants overexpressing LlMYB3 gene under the control of the constitutive CaMV35S promoter. Both morphological and physiological evidence strongly demonstrated that transgenic lines had more pronounced tolerances to cold, drought and salt than WT. At gene transcription level, qRT-PCR analysis showed that the expression level of 7 of picked 8 stress-responsive genes, including AtRD29A, AtRD29B, AtRD20, AtGSTF6, AtGolS1, AtLEA14 and AtAPX2 genes were higher in the transgenic plants compared with those of WT. It suggests that these genes might be transcriptionally regulated directly or indirectly by LlMYB3, and overexpressed LlMYB3 gene may enhance stress tolerance by regulating downstream stress-responsive genes in Arabidopsis.
Moreover, we showed overexpression of LlMYB3 resulted in enhanced ABA sensitivity, which was also observed on many MYB TFs from Arabidopsis, such as AtMYB2, AtMYB15, AtMYB96 and AtDIV2 [,,,]. Given that the expression level and promoter activity of LlMYB3 can be induced by ABA treatment, LlMYB3 might be involved in ABA signaling pathway to response to stresses. However, promoter analysis showed that there are no known ABA responsive related cis-acting elements located in the promoter of LlMYB3. We thus assume that there are two possible reasons. The first is that novel ABA responsive related cis-elements might exist in the promoter of LlMYB3; the second is that the transport of ABA signaling molecules to LlMYB3 is through a complex signaling network rather than by directly recognizing ABA responsive related cis-elements in LlMYB3 promoter.
On the other hand, MYB proteins play essential roles by regulating the expression of a large number of anthocyanin biosynthesis genes. For example, in Arabidopsis, the anthocyanin regulators MYB75/PAP1, MYB90/PAP2, MYB113, and MYB114 control the expression of the late biosynthetic genes DFR and LDOX/ANS [,,]; the flavonol regulators MYB12/PFG1, MYB11/PFG2, and MYB111/PFG3 regulate expression of the four early biosynthetic genes CHS, CHI, F3H, F3’H and FLS [,]. More importantly, some anthocyanin biosynthetic genes are even the direct targets of MYB proteins in response to abiotic stresses []. MYB1 in carrot can bind to the box-L-like sequences of phenylalanine ammonia-lyase 1 (PAL1) promoter specifically and activates PAL1 under UV-B irradiation []. MYB134 in poplar, which is essential for wound and UV-B tolerance, regulates stress-responsive proanthocyanidin biosynthesis by binding to the promoter of proanthocyanidin biosynthetic genes, such as ANR2 []. In rice, OsC1-MYB protein is shown to bind to the MYB responsive elements in the promoters of stress-induced flavonoid pathway genes OsDFR and OsANS []; overexpression of OsMYB4 in transgenic Arabidopsis increases chilling and freezing tolerance by transactivating PAL2 and other cold inducible genes []. Here, we found LlMYB3 can bind to the MYB binding sites in the promoter of cold-responsive LlCHS2 gene, suggesting LlMYB3 protein may also function in the correlation between anthocyanin accumulation and cold stress tolerance.
In conclusion, LlMYB3 is a nucleus-localized transcriptional activator which is regulated by cold, drought and salt stresses and sensitive to ABA. Overexpressing LlMYB3 in Arabidopsis showed ABA hypersensitivity and enhancing tolerance of transgenic plants to freezing, dehydration and salt conditions by up-regulate many stress-responsive genes. Furthermore, cold-responsive LlCHS2 is a direct target of LlMYB3 TF in response to abiotic stresses. Therefore, our findings provide a novel MYB-related gene, which plays a positive role in plant stress resistance and might be involved in anthocyanin biosynthesis pathway in response to cold stress. Our future efforts will be focused on investigating the role of upstream regulatory factors in regulating expression and modulating the function of LlMYB3 under various stress conditions.
4. Materials and Methods
4.1. Plant Materials
The tiger lily seedlings preparation method is described in our previous study []. The bulbs of tiger lily were cleaned, disinfected, and then stored at 4 °C; in March, the bulbs were box-cultivated in a greenhouse (116.3° E, 40.0° N) under controlled conditions. The model plant Arabidopsis thaliana L. Columbia-0 (Col-0) was selected for the transgenic study of LlMYB3. Arabidopsis plants were grown in 8 cm × 8 cm plastic pots containing a 1:1 mixture of sterile peat soil and vermiculite under controlled conditions (22/16 °C, 16 h light/8 h dark, 65% relative humidity, and 1000lx light intensity). Seeds of Nicotiana benthamiana L. were planted and cultured under the same conditions.
4.2. Cloning and Sequence Analysis of LlMYB3
The complete sequence cDNA of LlMYB3 gene was obtained from the transcriptome data of cold-treated tiger lily leave in our laboratory. Primer pairs (Table S1) were designed to amplify the coding sequence (CDS). The PCR products were cloned into pEASYT1-Blunt vector (TransGen Biotech, Beijing, China). After confirmation by sequencing, plasmid pEASYT1-LlMYB3 was used as a template for all experiments. The homolog genes of LlMYB3 were searched through BLAST (http://www.ncbi.nlm.nih.gov/BLAST/) database []. Multiple sequence alignments were performed using DNAMAN (version 7). Phylogenetic tree analysis was performed using neighbor-joining method in MEGA5 software with 1000 replications []. The NCBI accession numbers of genes used in multiple sequence alignments and phylogenetic tree analysis are shown in Table S2. The theoretical molecular weight and isoelectric point were calculated using ExPASy (http://expasy.org/tools/protparam.html) [].
Genomic DNA was extracted from tiger lily leaves using the DNeasy Plant Mini Kit (Qiagen, Valencia, CA, USA). The promoter of LlMYB3 gene was isolated using a Genome Walker Kit (Clontech, Mountain View, CA, USA) with nest PCR according to the manufacturer’s instructions. Conserved cis-element motifs of LlMYB3 promoter were predicted using PLACE (http://www.dna.affrc.go.jp/PLACE/signalscan.html) databases [].
4.3. Abiotic Stresses Treatment and Quantitative Real-Time PCR Analysis
For expression analysis of LlMYB3 in response to abiotic stress and ABA treatment, 8-week-old tiger lily seedlings were treated with 4 °C, 16.1% PEG6000 (−0.5 MPa), 100 mM NaCl and 100 µM exogenous ABA for 0, 1, 3, 6, 12 and 24 h, respectively. Leaf samples were collected and immediately frozen with liquid nitrogen and stored at −80 °C for RNA isolation.
Total RNA was isolated using an RNAisomate RNA Easyspin isolation system (Aidlab Biotech, Beijing, China). First-strand cDNA synthesis was performed using Prime Script II 1st strand cDNA Synthesis Kit (Takara, Shiga, Japan). The qRT-PCR was performed using a Bio-Rad/CFX Connect™ Real-Time PCR Detection System (Bio-Rad, San Diego, CA, USA) with SYBR® qPCR mix (Takara, Shiga, Japan). Relative mRNA content was calculated using the 2−ΔΔCt method [] against the internal reference gene encoding tiger lily tonoplast intrinsic protein 1 (LlTIP1) [] and Arabidopsis Atactin gene (NCBI accession No. NM_112764). The primers used in this study were designed with Primer Premier 5 and are listed in Table S1. All reactions were performed in three biological replicates. Student’s t-test was performed for all statistical analysis in this study. The heat-map was generated using MeV4.9 and clustered by hierarchical clustering (HCL) with default parameters []. Pearson’s correlation coefficient (r) to define similarity of expression levels between LlMYB3 and structural genes involved in anthocyanin biosynthesis pathway.
4.4. Subcellular Localization and Transactivation Assay
To determine its subcellular localization, the whole LlMYB3 coding region without stop codon was amplified and cloned into pBI121-GFP at XhoI and SalI by using ClonExpressII One Step Cloning Kits (Vazyme, Piscataway, NJ, USA) to produce LlMYB3-GFP fusion construct driven by CaMV35S promoter. The recombinant constructs and empty GFP vector were transformed into Agrobacterium tumefaciens GV3101 and infiltrated separately into tobacco (N. benthamiana) epidermal cells. After agro-infiltration for 32–48 h, GFP fluorescence signals were excited at 488nm and detected under Leica TCS SP8 Confocal Laser Scanning Platform (Leica SP8, Leica, Wetzlar, Germany) using a 500–530 nm emission filter.
The transactivation experiment was carried out according to the manual of Yeast Protocols Handbook (Clontech). The full-length coding region and truncated fragments N-terminus (1–231 bp) and C-terminus (232–462) of LlMYB3 generated by PCR amplification were fused in frame to the GAL4 DNA binding domain in the vector of pGBKT7 (Invitrogen, Carlsbad, CA, USA). These constructs and negative control pGBKT7 were transformed into yeast strain Y2HGold by using Quick Easy Yeast Transformation Mix (Clontech). The transformed yeast strains were screened on the selective medium plates SD/-Trp, SD/-Trp/-His-Ade and SD/-Trp-His-x-α-gal plates. The transactivation activity was detected according to their growth status and α-galactosidase activity.
4.5. Yeast One-Hybrid (Y1H) Assays
Y1H assay was carried out using the Matchmaker™ Gold Yeast One-Hybrid System (Clontech). The LlCHS2 promoter was amplified by genome walking nested PCR method described previously for LIMYB3 promoter, and the fragment (−820 to −553) of LlCHS2 promoter containing four MYB binding sites was isolated and cloned into the pAbAi (bait) vector (shown in Figure 7b and Figure S3). Full-length LlMYB3 was inserted into pGADT7 (prey) vector yielding plasmid pGADT7-LlMYB3. The bait plasmids were linearized and transformed into the yeast strain Y1HGold. Positive yeast cells were then transformed with pGADT7-LlMYB3 plasmid. The DNA-protein interaction was determined based on the growth ability of the co-transformants on SD/-Leu medium with Aureobasidin A (AbA) according to the manual.
4.6. Generation of Transgenic Arabidopsis
The LlMYB3 open read frame (ORF) was cloned into pBI121 vector under control of a CaMV35S promoter; the LlMYB3 promoter region was inserted into CaMV35S-GUS vector by replacing the CaMV35S promoter. The recombinant vectors and empty GUS vector were transformed into 5-week-old Arabidopsis ecotype Col-0 plants by using Agrobacterium tumefaciens GV3101 and the floral-dip method []. Transformed seeds were selected on MS medium containing 50 mg/L kanamycin. T3-generation homozygous lines were selected for gene functional analysis.
4.7. Histochemical Staining and Fluorometric GUS Assay
Histochemical staining and fluorometric GUS assay analysis for GUS activity was carried out as described before []. To understand the effects of different stresses on GUS expression mediated by the LlMYB3 promoter, transgenic LlMYB3 Arabidopsis plants were treated with 4 °C, 16.1% PEG6000 (−0.5 MPa), 100 mM NaCl and 100 µM exogenous ABA for different durations before sampling. The leaves of stress-treated transgenic LlMYB3 Arabidopsis were incubated in GUS reaction buffer (3 mg/mL X-gluc, 40 mM sodium phosphate pH 7, 10 mM EDTA, 0.1% Triton X-100, 0.5 mM ferricyanatum kalium, 0.5 mM ferrocyanatum kalium and 20% methanol). After overnight incubation at 37 °C, the stained samples were bleached with 70% (v/v) ethanol to remove chlorophyll. Photos of those stained samples were obtained by a Leica TL3000 Ergo microscope under white light. Leaves of stress-treated transgenic Arabidopsis were also used to exam GUS gene expression level by qRT-PCR, and determine GUS enzyme activity and measuring the fluorescence of 4-methylumbelliferone produced by GUS cleavage of 4-methylumbelliferyl-β-d-glucuronide (4-MUG). Protein amount was determined using a Protein Assay kit (Bio-Rad, Hercules, CA, USA) using bovine serum albumin as a standard.
4.8. Evaluation of Transgenic Plants Abiotic Stress Tolerance and ABA Sensitivity
The seeds of LlMYB3 T3-generation homozygous lines and the wild type (WT) were sown on vermiculite soil in pots for freezing and drought treatment. There were 3-week-old seedlings at 4 °C for 3 h, then at −4, −6 or −8 °C, respectively, for 12 h. After that, the plants were kept at 4 °C for 3 h before transferring to a normal condition at 22 °C. For the drought treatment, the water intake of 3-week-old potted Arabidopsis plants in water-saturated substrate was withheld for 30 days, followed by rehydrating the seedlings for 7 days. The survival rates of transgenic and WT seedlings were statistical analyzed.
For determining the salt tolerance and ABA sensitivity in transgenic plants, Arabidopsis seeds were cultivated on MS medium supplemented with 0 and 2 µM ABA or 50 mM NaCl, respectively, under continuous light at 22 °C in a growth chamber. The germination rate was scored on the 9th day after planting on the plates.
4.9. Measurements of Relative Electrolyte Leakage, Soluble Sugar, and Water Loss Rate
The relative electrolyte leakage, soluble sugar content and water loss rate were evaluated following the method described previously [,]. The relative electrolyte leakage was evaluated by determining the relative conductivity of fresh leaves (100 mg) in solution using a conductivity detector. The anthrone-sulfuric acid colorimetry was used for determining the soluble sugar. The water loss rate was calculated related to the initial fresh weight of the leaf samples; the samples were placed on the lab bench (20−22 °C, humidity 45−60%) and weighed at designated time points. All the measurements were performed with ten plants in triplicate.
Supplementary Materials
Supplementary materials can be found at https://www.mdpi.com/1422-0067/20/13/3195/s1. Table S1 Primers used in this study. Table S2 NCBI accession numbers of genes used in multiple sequence alignments and phylogenetic tree analysis. Table S3 MYB binding sites identified in the promoter region of LlCHS2. Table S4 Expression pattern correlation between LlMYB3 and anthocyanin biosynthesis structural genes under continuous cold stress. Asterisks indicate a significant difference (0.01 < * p < 0.05). Figure S1 Chromosomal location of AT5G62320.1. The highest score (bits) significant alignment of LlMYB3 in the whole-genome sequence of Arabidopsis thaliana was AT5G62320.1 which located in No.5 chromosome. Figure S2 Overexpression of LlMYB3 confirmed by qRT-PCR. 12 independent homozygous were selected for the analysis. Wild type Arabidopsis is served as negative control. Values are means ± SD of three replicates. Three independent experiments were performed. Asterisks indicate a significant difference (** p < 0.01) compared with the corresponding controls. The lines 5 and 8, which showed relative high expression levels of LlMYB3 transcripts, were selected for further study. Figure S3 Sequence information about chalcone synthase 2 gene LlCHS2 from tiger lily. The fragment (−820 to −553) of the LlCHS2 promoter containing four MYB binding sites used in Y1H assay is underlined. The MYB binding sites described in Table S3 are highlighted in yellow. The partial ORF sequence of LlCHS2 is shaded in grey. Figure S4 Minimal inhibitory concentration of Aureobasidin A (AbA) selected for bait yeast strains. 200 ng·mL−1 of Aureobasidin A (AbA) was shown to be the minimal inhibitory concentration for bait (pAbAi-proLlCHS2) yeast strains.
Author Contributions
Y.Y. designed of the study and performed the statistical analysis and molecular experiments. Y.Z. conceived of the study, and helped to draft the manuscript. All authors read and approved the final manuscript.
Funding
This research was funded by China National Key Research & Development Project, grant number 2018YFD1000402 and China National Natural Science Foundation (grant no. 31672190, 31872138, 31071815 and No. 31272204).
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Abbreviations
| ABA | abscisic acid |
| HCL | hierarchical clustering HCL |
| GFP | Green fluorescent protein |
| AbA | Aureobasidin A |
| GUS | β-glucuronidase |
| CHS | chalcone synthase |
| CHI | chalcone isomerase |
| F3H | flavanone 3-hydroxylase |
| F3’H | flavonoid 3′-hydroxylase |
| FLS | flavonol synthase |
| DFR | late biosynthetic genes dihydroflavonol reductase |
| LDOX/ANS | leucoanthocyanidin dioxygenase/anthocyanidin synthase |
| PAL1 | phenylalanine ammonia-lyase 1 |
| ANR2 | anthocyanidin reductase2 |
References
- Huang, G.T.; Ma, S.L.; Bai, L.P.; Zhang, L.; Ma, H.; Jia, P.; Liu, J.; Zhong, M.; Guo, Z.F. Signal transduction during cold, salt, and drought stresses in plants. Mol. Biol. Rep. 2012, 39, 969–987. [Google Scholar] [CrossRef] [PubMed]
- Dubos, C.; Stracke, R.; Grotewold, E.; Weisshaar, B.; Martin, C.; Lepiniec, L. MYB transcription factors in Arabidopsis. Trends Plant Sci. 2010, 15, 573–581. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Ng, C.K.Y.; Fan, L.M. MYB transcription factors, active players in abiotic stress signaling. Environ. Exp. Bot. 2015, 114, 80–91. [Google Scholar] [CrossRef]
- Yanhui, C.; Xiaoyuan, Y.; Kun, H.; Meihua, L.; Jigang, L.; Zhaofeng, G.; Zhiqiang, L.; Yunfei, Z.; Xiaoxiao, W.; Xiaoming, Q.; et al. The MYB transcription factor superfamily of Arabidopsis: Expression analysis and phylogenetic comparison with the rice MYB family. Plant Mol. Biol. 2006, 60, 107–124. [Google Scholar] [CrossRef] [PubMed]
- Xiong, H.; Li, J.; Liu, P.; Duan, J.; Zhao, Y.; Guo, X.; Li, Y.; Zhang, H.; Ali, J.; Li, Z. Overexpression of OsMYB48-1, a novel MYB-related transcription factor, enhances drought and salinity tolerance in rice. PLoS ONE 2014, 9, e92913. [Google Scholar] [CrossRef] [PubMed]
- Du, H.; Wang, Y.B.; Xie, Y.; Liang, Z.; Jiang, S.J.; Zhang, S.S.; Huang, Y.B.; Tang, Y.X. Genome-wide identification and evolutionary and expression analyses of MYB-related genes in land plants. DNA Res. 2013, 20, 437–448. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Zhangliang, C.; Kang, J.; Kang, D.; Gu, H.; Qin, G. AtMYB14 regulates cold tolerance in Arabidopsis. Plant Mol. Biol. Rep. 2013, 31, 87–97. [Google Scholar] [CrossRef]
- Agarwal, M.; Hao, Y.; Kapoor, A.; Dong, C.-H.; Fujii, H.; Zheng, X.; Zhu, J.K. A R2R3 type MYB transcription factor is involved in the cold regulation of CBF genes and in acquired freezing tolerance. J. Biol. Chem. 2007, 281, 37636–37645. [Google Scholar] [CrossRef]
- Cui, M.H.; Yoo, K.S.; Hyoung, S.; Nguyen, H.T.K.; Kim, Y.Y.; Kim, H.J.; Ok, S.H.; Yoo, S.D.; Shin, J.S. An Arabidopsis R2R3-MYB transcription factor, AtMYB20, negatively regulates type 2C serine/threonine protein phosphatases to enhance salt tolerance. FEBS Lett. 2013, 587, 1773–1778. [Google Scholar] [CrossRef]
- Jung, C.; Seo, J.S.; Won Han, S.; Koo, Y.; Ho Kim, C.; Ik Song, S.; Hie Nahm, B.; Do Choi, Y.; Cheong, J.J. Overexpression of AtMYB44 Enhances stomatal closure to confer abiotic stress tolerance in transgenic Arabidopsis. Plant Physiol. 2008, 146, 623–635. [Google Scholar] [CrossRef]
- Abe, H. Arabidopsis AtMYC2 (bHLH) and AtMYB2 (MYB) function as transcriptional activators in abscisic acid signaling. Plant Cell 2003, 15, 63–78. [Google Scholar] [CrossRef] [PubMed]
- Seo, P.J.; Xiang, F.; Qiao, M.; Park, J.Y.; Na Lee, Y.; Kim, S.G.; Lee, Y.H.; Park, W.J.; Park, C.M. The MYB96 transcription factor mediates abscisic acid signaling during drought stress response in Arabidopsis. Plant Physiol. 2009, 151, 275–289. [Google Scholar] [CrossRef] [PubMed]
- Ding, Z.; Li, S.; An, X.; Liu, X.; Huanju, Q.; Wang, D. Transgenic expression of MYB15 confers enhanced sensitivity to abscisic acid and improved drought tolerance in Arabidopsis thaliana. J. Genet. Genom. 2009, 36, 17–29. [Google Scholar] [CrossRef]
- Pasquali, G.; Biricolti, S.; Locatelli, F.; Baldoni, E.; Mattana, M. OsMYB4 expression improves adaptive responses to drought and cold stress in transgenic apples. Plant Cell Rep. 2008, 27, 1677–1686. [Google Scholar] [CrossRef] [PubMed]
- Vannini, C.; Campa, M.; Iriti, M.; Genga, A.; Faoro, F.; Carravieri, S.; Rotino, G.; Rossoni, M.; Spinardi, A.; Bracale, M. Evaluation of transgenic tomato plants ectopically expressing the rice Osmyb4 gene. Plant Sci. 2007, 173, 231–239. [Google Scholar] [CrossRef]
- Vannini, C.; Locatelli, F.; Bracale, M.; Magnani, E.; Marsoni, M.; Osnato, M.; Mattana, M.; Baldoni, E.; Coraggio, I. Overexpression of the rice OsMYB4 gene increases chilling and freezing tolerance of Arabidopsis thaliana plants. Plant J. 2004, 37, 115–127. [Google Scholar] [CrossRef]
- Yang, A.; Dai, X.; Zhang, W.H. A R2R3-type MYB gene, OsMYB2, is involved in salt, cold, and dehydration tolerance in rice. J. Exp. Bot. 2012, 63, 2541–2556. [Google Scholar] [CrossRef]
- Mao, X.; Jia, D.; Li, A.; Zhang, H.; Tian, S.; Zhang, X.; Jia, J.; Jing, R. Transgenic expression of TaMYB2A confers enhanced tolerance to multiple abiotic stresses in Arabidopsis. Funct. Integr. Genom. 2011, 11, 445. [Google Scholar] [CrossRef]
- Qin, Y.; Wang, M.; Tian, Y.; He, W.; Han, L.; Xia, G. Over-expression of TaMYB33 encoding a novel wheat MYB transcription factor increases salt and drought tolerance in Arabidopsis. Mol. Biol. Rep. 2012, 39, 7183–7192. [Google Scholar] [CrossRef]
- Li, M.J.; Qiao, Y.; Li, Y.Q.; Shi, Z.L.; Zhang, N.; Bi, C.; Guo, J.K. A R2R3-MYB transcription factor gene in common wheat (namely TaMYBsm1) involved in enhancement of drought tolerance in transgenic Arabidopsis. J. Plant Res. 2016, 129. [Google Scholar] [CrossRef]
- Zhang, L.; Zhao, G.; Xia, C.; Jia, J.; Liu, X.; Kong, X. A wheat R2R3-MYB gene, TaMYB30-B, improves drought stress tolerance in transgenic Arabidopsis. J. Exp. Bot. 2012, 63, 5873–5885. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Li, W.; Lv, J.; Jia, Y.; Wang, M.; Xia, G. Ectopic expression of a wheat MYB transcription factor gene, TaMYB73, improves salinity stress tolerance in Arabidopsis thaliana. J. Exp. Bot. 2011, 63, 1511–1522. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Liu, G.; Zhao, G.; Xia, C.; Jia, J.; Liu, X.; Kong, X. Characterization of a wheat R2R3-MYB transcription factor gene, TaMYB19, involved in enhanced abiotic stresses in Arabidopsis. Plant Cell Physiol. 2014, 55. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Hu, B.; Qin, Y.; Hu, G.; Zhao, J. Advance of the negative regulation of anthocyanin biosynthesis by MYB transcription factors. Plant Physiol. Biochem. 2019, 136, 178–187. [Google Scholar] [CrossRef] [PubMed]
- Leyva, A.; Jarillo, J.A.; Salinas, J.; Martinez-Zapater, J.M. Low Temperature induces the accumulation of phenylalanine ammonia-lyase and chalcone synthase mRNAs of Arabidopsis thaliana in a light-dependent manner. Plant Physiol. 1995, 108, 39–46. [Google Scholar] [CrossRef] [PubMed]
- Rowan, D.; Cao, M.; Lin-Wang, K.; Cooney, J.; Jensen, D.; Austin, P.; B Hunt, M.; Norling, C.; Hellens, R.; J Schaffer, R.; et al. Environmental regulation of leaf colour in red 35S:PAP1 Arabidopsis thaliana. New Phytol. 2009, 182, 102–115. [Google Scholar] [CrossRef] [PubMed]
- Nakabayashi, R.; Yonekura-Sakakibara, K.; Urano, K.; Suzuki, M.; Yamada, Y.; Nishizawa, T.; Matsuda, F.; Kojima, M.; Sakakibara, H.; Shinozaki, K.; et al. Enhancement of oxidative and drought tolerance in Arabidopsis by over accumulation of antioxidant flavonoids. Plant J. 2013, 77. [Google Scholar] [CrossRef]
- Lin-Wang, K.; Micheletti, D.; Palmer, J.; Volz, R.; Lozano, L.; Espley, R.; Hellens, R.; Chagne, D.; Rowan, D.; Troggio, M.; et al. High temperature reduces apple fruit colour via modulation of the anthocyanin regulatory complex. Plant Cell Environ. 2011, 34, 1176–1190. [Google Scholar] [CrossRef]
- Li, L.; Ban, Z.; Li, X.H.; Wu, M.Y.; Wang, A.L.; Jiang, Y.; Jiang, Y.H. Differential expression of anthocyanin biosynthetic genes and transcription factor PcMYB10 in pears (Pyrus communis L.). PLoS ONE 2012, 7, e46070. [Google Scholar] [CrossRef]
- Ahmed, N.U.; Park, J.I.; Jung, H.J.; Hur, Y.; Nou, I.S. Anthocyanin biosynthesis for cold and freezing stress tolerance and desirable color in Brassica rapa. Funct. Integr. Genom. 2015, 15, 383–394. [Google Scholar] [CrossRef]
- Wang, J.; Wang, Q.; Yang, Y.; Liu, X.; Gu, J.; Li, W.; Ma, S.; Lu, Y. De novo assembly and characterization of stress transcriptome and regulatory networks under temperature, salt and hormone stresses in Lilium lancifolium. Mol. Biol. Rep. 2014, 41, 8231–8245. [Google Scholar] [CrossRef] [PubMed]
- Yong, Y.B.; Li, W.Q.; Wang, J.M.; Zhang, Y.; Lu, Y.M. Identification of gene co-expression networks involved in cold resistance of Lilium lancifolium. Biol. Plant 2018, 62, 287–298. [Google Scholar] [CrossRef]
- Huang, P.; Chen, H.; Mu, R.; Yuan, X.; Zhang, H.S.; Huang, J. OsMYB511 encodes a MYB domain transcription activator early regulated by abiotic stress in rice. Genet. Mol. Res. 2015, 14, 9506–9517. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Huang, R.; Liu, Z.; Zhang, G. Functional characterization of cis-elements conferring vascular vein expression of At4g34880 amidase family protein gene in Arabidopsis. PLoS ONE 2013, 8, e67562. [Google Scholar] [CrossRef] [PubMed]
- Zhai, H.; Bai, X.; Zhu, Y.; Li, Y.; Cai, H.; Ji, W.; Ji, Z.; Liu, X.; Liu, X.; Li, J. A single-repeat R3-MYB transcription factor MYBC1 negatively regulates freezing tolerance in Arabidopsis. Biochem. Biophys. Res. Commun. 2010, 394, 1018–1023. [Google Scholar] [CrossRef] [PubMed]
- Su, C.F.; Wang, Y.C.; Hsieh, T.H.; Lu, C.A.; Tseng, T.H.; Yu, S.M. A novel MYBS3-dependent pathway confers cold tolerance in rice. Plant Physiol. 2010, 153, 145–158. [Google Scholar] [CrossRef]
- Shin, D.; Moon, S.J.; Han, S.; Kim, B.G.; Park, S.R.; Lee, S.K.; Yoon, H.J.; Lee, H.E.; Kwon, H.B.; Baek, D.; et al. Expression of StMYB1R-1, a novel potato single MYB-like domain transcription factor, increases drought tolerance. Plant Physiol. 2011, 155, 421–432. [Google Scholar] [CrossRef]
- Ganesan, G.; Sankararamasubramanian, H.M.; Harikrishnan, M.; Parida, A.; Ashwin, G. A MYB transcription factor from the grey mangrove is induced by stress and confers NaCl tolerance in tobacco. J. Exp. Bot. 2012, 63, 4549–4561. [Google Scholar] [CrossRef]
- Fang, Q.; Wang, Q.; Mao, H.; Xu, J.; Wang, Y.; Hu, H.; He, S.; Tu, J.; Cheng, C.; Tian, G.; et al. AtDIV2, an R-R-type MYB transcription factor of Arabidopsis, negatively regulates salt stress by modulating ABA signaling. Plant Cell Rep. 2018, 37, 1499–1511. [Google Scholar] [CrossRef]
- Borevitz, J.O.; Xia, Y.; Blount, J.; Dixon, R.; Lamb, C. Activation Tagging identifies a conserved MYB regulator of phenylpropanoid biosynthesis. Plant Cell 2001, 12, 2383–2394. [Google Scholar] [CrossRef]
- Gonzalez, A.; Zhao, M.; M Leavitt, J.; Lloyd, A. Regulation of the anthocyanin biosynthetic pathway by the TTG1/BHLH/MYB transcriptional complex in Arabidopsis seedlings. Plant J. 2008, 53, 814–827. [Google Scholar] [CrossRef]
- Stracke, R.; Ishihara, H.; Huep, G.; Barsch, A.; Mehrtens, F.; Niehaus, K.; Weisshaar, B. Differential regulation of closely related R2R3-MYB transcription factors controls flavonol accumulation in different parts of the Arabidopsis thaliana seedling. Plant J. 2007, 50, 660–677. [Google Scholar] [CrossRef] [PubMed]
- Stracke, R.; Favory, J.J.; Gruber, H.; Bartelniewoehner, L.; Bartels, S.; Binkert, M.; Funk, M.; Weisshaar, B.; Ulm, R. The Arabidopsis bZIP transcription factor HY5 regulates expression of the PFG1/MYB12 gene in response to light and ultraviolet-B radiation. Plant Cell Environ. 2009, 33, 88–103. [Google Scholar] [CrossRef] [PubMed]
- Maeda, K.; Kimura, S.; Demura, T.; Takeda, J.; Ozeki, Y. DcMYB1 acts as a transcriptional activator of the carrot phenylalanine ammonia-lyase Gene (DcPAL1) in response to elicitor treatment, UV-B irradiation and the dilution effect. Plant Mol. Biol. 2005, 59, 739–752. [Google Scholar] [CrossRef] [PubMed]
- Mellway, R.D.; T Tran, L.; Prouse, M.B.; Campbell, M.M.; Peter Constabel, C. The wound-, pathogen-, and ultraviolet B-responsive MYB134 gene encodes an R2R3 MYB transcription factor that regulates proanthocyanidin synthesis in poplar. Plant Physiol. 2009, 150, 924–941. [Google Scholar] [CrossRef] [PubMed]
- Ithal, N.; Reddy, A.R. Rice flavonoid pathway genes, OsDfr and OsAns, are induced by dehydration, high salt and ABA, and contain stress responsive promoter elements that interact with the transcription activator, OsC1-MYB. Plant Sci. 2004, 166, 1505–1513. [Google Scholar] [CrossRef]
- Altschul, S.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic Local Aligment Search Tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Hall, B.G. Building Phylogenetic Trees from Molecular Data with MEGA. Mol. Biol. Evol. 2013, 30, 1229–1235. [Google Scholar] [CrossRef]
- Artimo, P.; Jonnalagedda, M.; Arnold, K.; Baratin, D.; Csardi, G.; de Castro, E.; Duvaud, S.; Flegel, V.; Fortier, A.; Gasteiger, E.; et al. ExPASy: SIB bioinformatics resource portal. Nucleic Acids Res. 2012, 40, W597–W603. [Google Scholar] [CrossRef]
- Higo, K.; Ugawa, Y.; Iwamoto, M.; Korenaga, T. Plant cis-acting regulatory DNA elements (PLACE) database: 1999. Nucleic Acids Res. 1999, 27, 297–300. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Saeed, A.I.; Sharov, V.; White, J.; Li, J.; Liang, W.; Bhagabati, N.; Braisted, J.; Klapa, M.; Currier, T.; Thiagarajan, M.; et al. TM4: A Free, Open-Source System for Microarray Data Management and Analysis. Biotechniques 2003, 34, 374–378. [Google Scholar] [CrossRef] [PubMed]
- Bent, A. Arabidopsis thaliana floral dip transformation method. Methods Mol. Biol. 2006, 343, 87–103. [Google Scholar] [CrossRef] [PubMed]
- Jefferson, R.A.; Kavanagh, T.A.; Bevan, M.W. GUS fusions: Beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 1987, 6, 3901–3907. [Google Scholar] [CrossRef] [PubMed]
- Cao, W.H.; Liu, J.; He, X.J.; Mu, R.-L.; Zhou, H.L.; Chen, S.Y.; Zhang, J.S. Modulation of ethylene responses affects plant salt-stress responses. Plant Physiol. 2007, 143, 707–719. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Zhang, L.; Xia, C.; Zhao, G.; Jia, J.; Kong, X. The novel wheat transcription factor TaNAC47 enhances multiple abiotic stress tolerances in transgenic plants. Front. Plant Sci. 2015, 6, 1174. [Google Scholar] [CrossRef] [PubMed]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).