Role of Green Macroalgae Enteromorpha Prolifera Polyphenols in the Modulation of Gene Expression and Intestinal Microflora Profiles in Type 2 Diabetic Mice
Abstract
1. Introduction
2. Results
2.1. Characterization of Potent Major Compounds
2.2. Effects of EPE3k on Fasting Blood Glucose (FBG) Levels and Oral Glucose Tolerance Test (OGTT)
2.3. Effect of EPE3k on the Histopathology of Liver in Diabetic Mice
2.4. Effect of EPE3k on Gene Expression of Signal Pathway in Liver
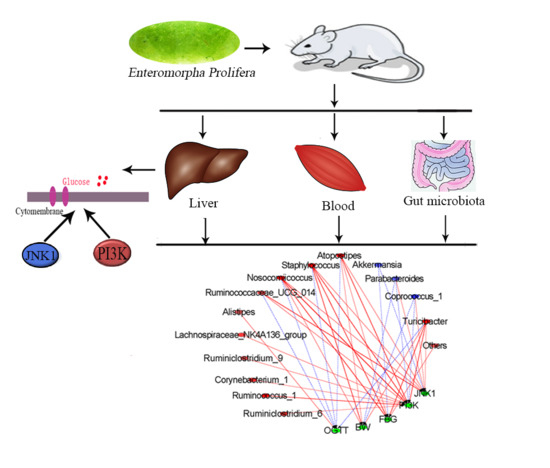
2.5. Effect of EPE3k on the Intestinal Microflora
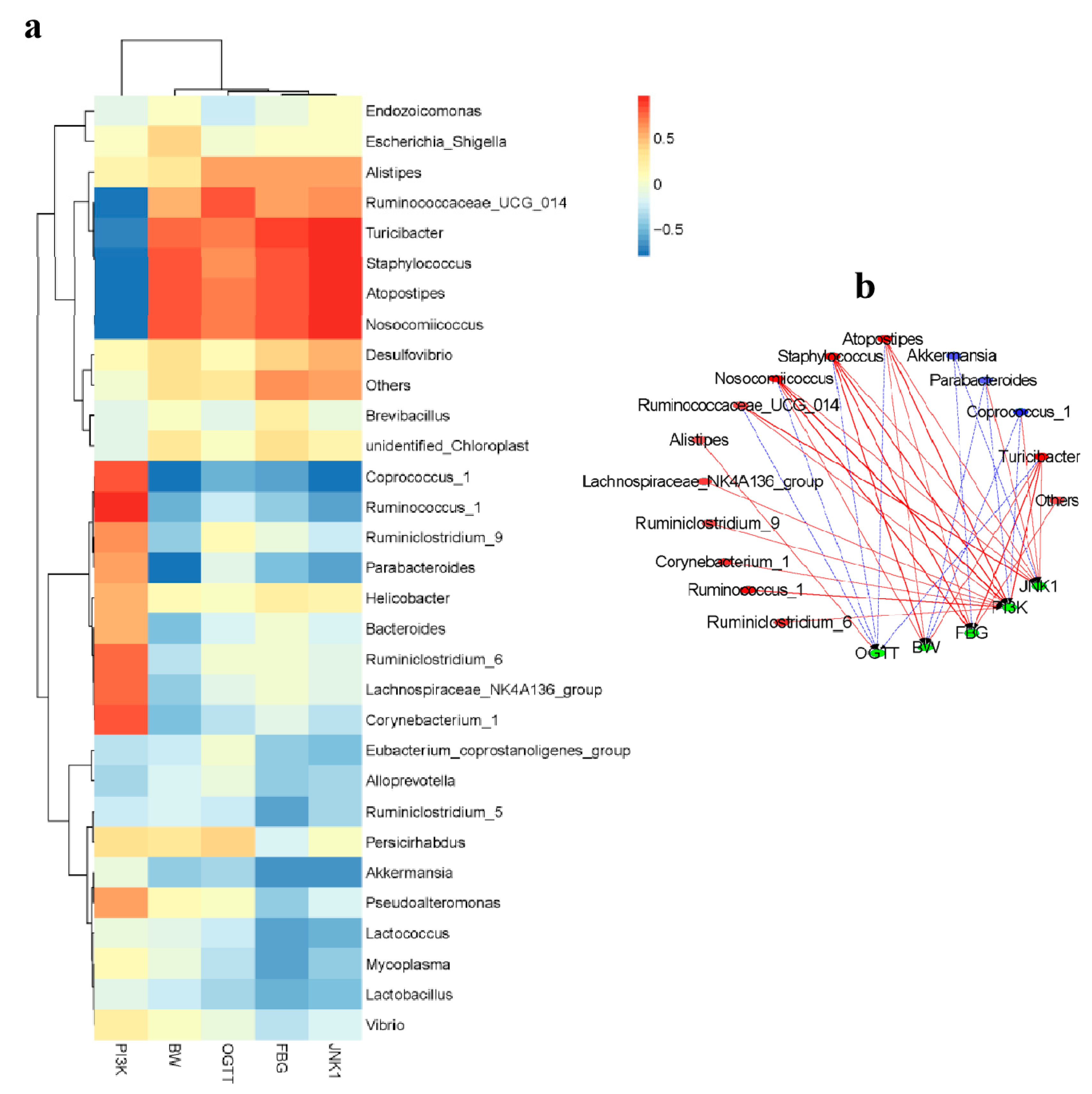
2.6. Correlation between Biological Indexes, Gene Expression Levels and Cecal Microbiota
3. Discussion
4. Materials and Methods
4.1. Preparation of Ethanolic Extract from E. prolifera
4.2. Ultra-Performance Liquid Chromatography-Mass Spectrometric (UPLC-MS) Analysis
4.3. Animals Experiments
4.4. Blood Sample Collection and Oral Glucose Tolerance Test
4.5. Histopathological Analysis
4.6. Dynamic Profile of Intestinal Microflora in Response to EPE3k
4.7. Real-Time PCR
4.8. Protein Extraction and Western Blotting
4.9. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Acknowledgments
Conflicts of Interest
References
- Zheng, Y.; Ley, S.H.; Hu, F.B. Global aetiology and epidemiology of type 2 diabetes mellitus and its complications. Nat. Rev. Endocrinol. 2018, 14, 88–98. [Google Scholar] [CrossRef] [PubMed]
- Zhao, C.; Wu, Y.; Yang, C.; Liu, B.; Huang, Y. Hypotensive, hypoglycaemic and hypolipidaemic effects of bioactive compounds from microalgae and marine micro-organisms. Int. J. Food Sci. Technol. 2015, 50, 1705–1717. [Google Scholar] [CrossRef]
- Brunkwall, L.; Orho-Melander, M. The gut microbiome as a target for prevention and treatment of hyperglycaemia in type 2 diabetes: From current human evidence to future possibilities. Diabetologia 2017, 60, 943–951. [Google Scholar] [CrossRef] [PubMed]
- Zhao, C.; Yang, C.F.; Liu, B.; Lin, L.; Sarker, S.D.; Nahar, L.; Yu, H.; Cao, H.; Xiao, J. Bioactive compounds from marine macroalgae and their hypoglycemic benefits. Trends Food Sci. Technol. 2018, 72, 1–12. [Google Scholar] [CrossRef]
- Fernando, I.P.; Kim, M.; Son, K.T.; Jeong, Y.; Jeon, Y.J. Antioxidant activity of marine algal polyphenolic compounds: A mechanistic approach. J. Med. Food 2016, 19, 615–628. [Google Scholar] [CrossRef]
- Murray, M.; Dordevic, A.L.; Bonham, M.P.; Ryan, L. Do marine algal polyphenols have antidiabetic, anti-hyperlipidaemic or anti-inflammatory effects in humans? A systematic review. Crit. Rev. Food Sci. Nutr. 2018, 58, 2039–2054. [Google Scholar] [CrossRef]
- Moussavou, G.; Kwak, D.H.; Obiang-Obonou, B.W.; Maranguy, C.A.; Dinzouna-Boutamba, S.D.; Lee, D.H.; Pissibanganga, O.G.; Ko, K.; Seo, J.I.; Choo, Y.K. Anticancer effects of different seaweeds on human colon and breast cancers. Mar. Drugs 2014, 12, 4898–4911. [Google Scholar] [CrossRef]
- Athukorala, Y.; Lee, K.W.; Kim, S.K.; Jeon, Y.J. Anticoagulant activity of marine green and brown algae collected from Jeju Island in Korea. Bioresour. Technol. 2007, 98, 1711–1716. [Google Scholar] [CrossRef]
- Charoensiddhi, S.; Conlon, M.A.; Franco, C.M.M.; Zhang, W. The development of seaweed-derived bioactive compounds for use as prebiotics and nutraceuticals using enzyme technologies. Trends Food Sci. Technol. 2017, 70, 20–33. [Google Scholar] [CrossRef]
- Hardouin, K.; Bedoux, G.; Burlot, A.S.; Donnay-Moreno, C.; Bergé, J.P.; Nyvall-Collén, P.; Bourgougnon, N. Enzyme-assisted extraction (EAE) for the production of antiviral and antioxidant extracts from the green seaweed Ulva armoricana (Ulvales, Ulvophyceae). Algal Res. 2016, 16, 233–239. [Google Scholar] [CrossRef]
- Abu-Farha, M.; Cherian, P.; Al-Khairi, I.; Tiss, A.; Khadir, A.; Kavalakatt, S.; Warsame, S.; Dehbi, M.; Behbehani, K.; Abubaker, J. DNAJB3/HSP-40 cochaperone improves insulin signaling and enhances glucose uptake in vitro through JNK repression. Sci. Rep. 2015, 5, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Gaster, M.; Staehr, P.; Beck-Nielsen, H.; Schroder, H.D.; Handberg, A. GLUT4 is reduced in slow muscle fibers of type 2 diabetic patients: Is insulin resistance in type 2 diabetes a slow, type 1 fiber disease? Diabetes 2001, 50, 1324–1329. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Nishina, P.M.; Naggert, J.K. Degradation of IRS1 leads to impaired glucose uptake in adipose tissue of the type 2 diabetes mouse model TALLYHO/Jng. J. Endocrinol. 2009, 203, 65–74. [Google Scholar] [CrossRef] [PubMed]
- Cordero-Herrera, I.; Martín, M.A.; Bravo, L.; Goya, L.; Ramos, S. Cocoa flavonoids improve insulin signalling and modulate glucose production via Akt and AMPK in HepG2 cells. Mol. Nutr. Food Res. 2013, 57, 974–985. [Google Scholar] [CrossRef] [PubMed]
- Cui, J.; Zhang, M.; Zhang, Y.Q.; Xu, Z.H. JNK pathway: Diseases and therapeutic potential. Acta Pharmacol. Sin. 2007, 28, 601–608. [Google Scholar] [CrossRef] [PubMed]
- Yan, X.; Yang, C.F.; Lin, G.P.; Chen, Y.Q.; Miao, S.; Liu, B.; Zhao, C. Antidiabetic potential of green seaweed Enteromorpha prolifera flavonoids regulating insulin signaling pathway and gut microbiota in type 2 diabetic mice. J. Food Sci. 2018. [Google Scholar] [CrossRef]
- Sharma, B.R.; Karki, R.; Lee, E.; Zhu, Q.; Gurung, P.; Kanneganti, T.D. Innate immune adaptor MyD88 deficiency prevents skin inflammation in SHARPIN-deficient mice. Cell Death Differ. 2018. [Google Scholar] [CrossRef] [PubMed]
- Zhao, C.; Yang, C.F.; Chen, M.J.; Lv, X.C.; Liu, B.; Yi, L.Z.; Cornara, L.; Wei, M.C.; Yang, Y.C.; Tundis, R.; et al. Regulatory efficacy of brown seaweed Lessonia nigrescens extract on the gene expression profile and intestinal microflora in type 2 diabetic mice. Mol. Nutr. Food Res. 2018, 62, 1700730. [Google Scholar] [CrossRef]
- Huo, J.H.; Du, X.W.; Sun, G.D.; Dong, W.T.; Wang, W.M. Identification and characterization of major constituents in Juglans mandshurica using ultra performance liquid chromatography coupled with time-of-flight mass spectrometry (UPLC-ESI-Q-TOF/MS). Chin. J. Nat. Med. 2018, 16, 525–545. [Google Scholar] [CrossRef]
- Zeng, X.; Su, W.; Zheng, Y.; Liu, H.; Li, P.; Zhang, W.; Liang, Y.; Bai, Y.; Peng, W.; Yao, H. UFLC-Q-TOF-MS/MS-Based screening and identification of flavonoids and derived metabolites in human urine after oral administration of exocarpium Citri Grandis extract. Molecules 2018, 23, 895. [Google Scholar] [CrossRef]
- Sharma, B.R.; Kim, H.J.; Rhyu, D.Y. Caulerpa lentillifera extract ameliorates insulin resistance and regulates glucose metabolism in C57BL/KsJ-db/db mice via PI3K/Akt signaling pathway in myocytes. J. Transl. Med. 2015, 13, 62. [Google Scholar] [CrossRef]
- Shang, Q.; Shi, J.; Song, G.; Zhang, M.; Cai, C.; Hao, J.; Li, G.; Yu, G. Structural modulation of gut microbiota by chondroitin sulfate and its oligosaccharide. Int. J. Biol. Macromol. 2016, 89, 489–498. [Google Scholar] [CrossRef] [PubMed]
- Dias, M.F.; Reis, M.P.; Acurcio, L.B.; Carmo, A.O.; Diamantino, C.F.; Motta, A.M.; Kalapothakis, E.; Nicoli, J.R.; Nascimento, A.M.A. Changes in mouse gut bacterial community in response to different types of drinking water. Water Res. 2018, 132, 79–89. [Google Scholar] [CrossRef] [PubMed]
- Mardinoglu, A.; Boren, J.; Smith, U. Confounding effects of metformin on the human gut microbiome in type 2 diabetes. Cell Metab. 2016, 23, 10–12. [Google Scholar] [CrossRef] [PubMed]
- Larsen, N.; Vogensen, F.K.; Van Den Berg, F.W.J.; Nielsen, D.S.; Andreasen, A.S.; Pedersen, B.K.; Al-Soud, W.A.; Sørensen, S.J.; Hansen, L.H.; Jakobsen, M. Gut microbiota in human adults with type 2 diabetes differs from non-diabetic adults. PLoS ONE 2010, 5, e9085. [Google Scholar] [CrossRef]




| Peak No. | RT (min) | [M + H]+ | Fragment Ions | Formula | Identified Compounds | Ref. |
|---|---|---|---|---|---|---|
| 1 | 0.65 | 277 | 277[M + H]+, 235[M + H-C3H6]+, 151[M + H-C5H14O]+, 133[M + H-C8H16O2 | C18H28O2 | Estr-5(10)-ene-3,17-diol | [19] |
| 2 | 4.99 | 179 | 179[M + H]+,161[M + H-H2O]+, 133[M + H-H2O-CO]+, 105[M + H-H2O-2CO]+ | C10H10O3 | Regiolone | [19] |
| 3 | 5.64 | 449 | 449[M + H]+, 299[M + H-Glc]+ | C21H20O11 | Luteolin-6-C-glucoside | [20] |
| 4 | 6.59 | 597 | 399[M + H-Orha-2H2O]+, 135[M-H-Rha-Glc-C7H4O4]−, 107[M-H-Rha-Glc-C8H8O2-CO2]− | C27H32O15 | Neoeriocitrin | [20] |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lin, G.; Liu, X.; Yan, X.; Liu, D.; Yang, C.; Liu, B.; Huang, Y.; Zhao, C. Role of Green Macroalgae Enteromorpha Prolifera Polyphenols in the Modulation of Gene Expression and Intestinal Microflora Profiles in Type 2 Diabetic Mice. Int. J. Mol. Sci. 2019, 20, 25. https://doi.org/10.3390/ijms20010025
Lin G, Liu X, Yan X, Liu D, Yang C, Liu B, Huang Y, Zhao C. Role of Green Macroalgae Enteromorpha Prolifera Polyphenols in the Modulation of Gene Expression and Intestinal Microflora Profiles in Type 2 Diabetic Mice. International Journal of Molecular Sciences. 2019; 20(1):25. https://doi.org/10.3390/ijms20010025
Chicago/Turabian StyleLin, Guopeng, Xiaoyan Liu, Xin Yan, Dan Liu, Chengfeng Yang, Bin Liu, Yifan Huang, and Chao Zhao. 2019. "Role of Green Macroalgae Enteromorpha Prolifera Polyphenols in the Modulation of Gene Expression and Intestinal Microflora Profiles in Type 2 Diabetic Mice" International Journal of Molecular Sciences 20, no. 1: 25. https://doi.org/10.3390/ijms20010025
APA StyleLin, G., Liu, X., Yan, X., Liu, D., Yang, C., Liu, B., Huang, Y., & Zhao, C. (2019). Role of Green Macroalgae Enteromorpha Prolifera Polyphenols in the Modulation of Gene Expression and Intestinal Microflora Profiles in Type 2 Diabetic Mice. International Journal of Molecular Sciences, 20(1), 25. https://doi.org/10.3390/ijms20010025