Exploring Novel Functions of the Small GTPase Ypt1p under Heat-Shock by Characterizing a Temperature-Sensitive Mutant Yeast Strain, ypt1-G80D
Abstract
1. Introduction
2. Results
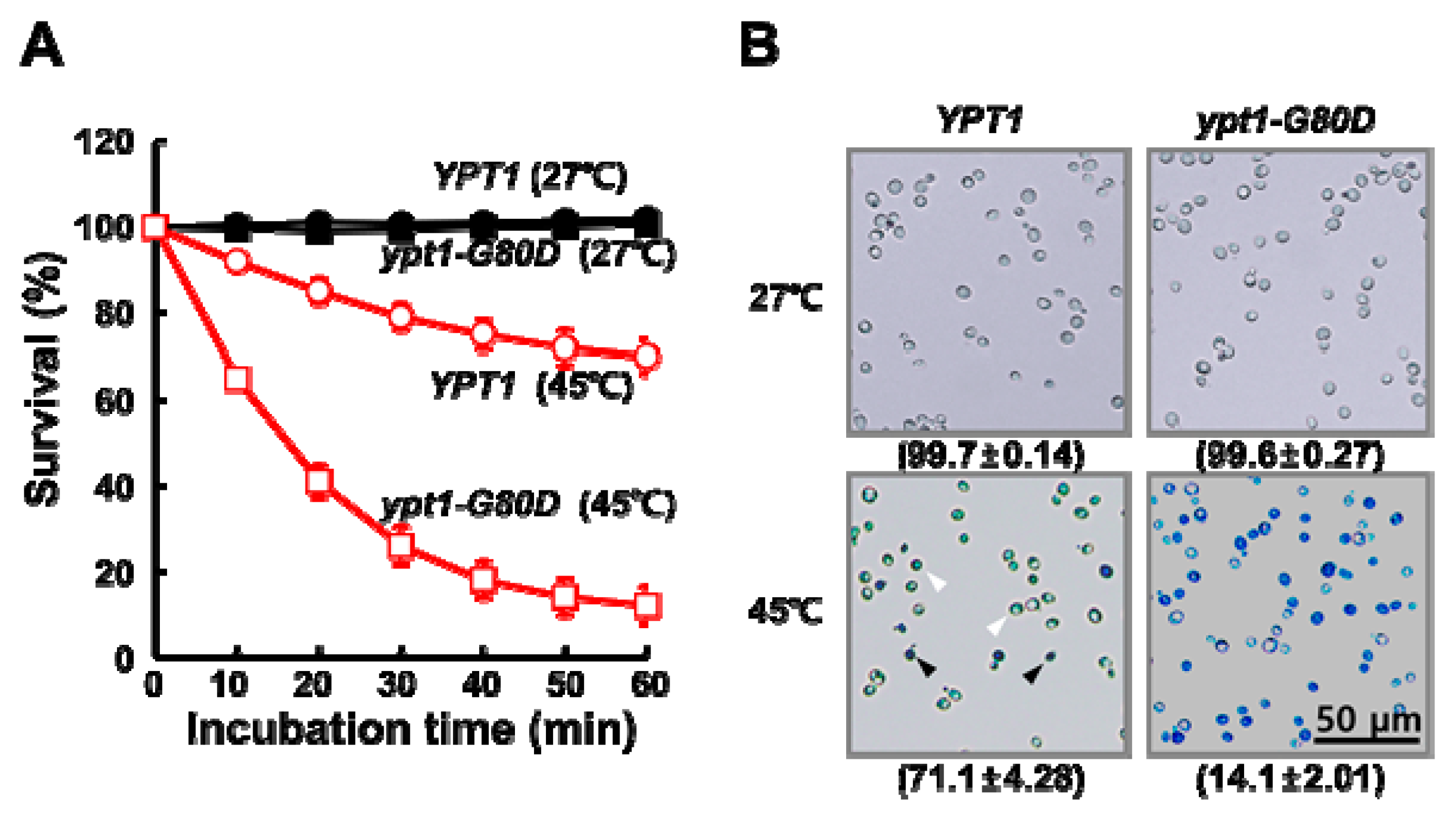
2.1. Mutant ypt1-G80D Yeast Cells Are Sensitive to Heat-Shock
2.2. Heat-Shock Induces Cytosolic Protein Aggregation in ypt1-G80D Yeast Cells
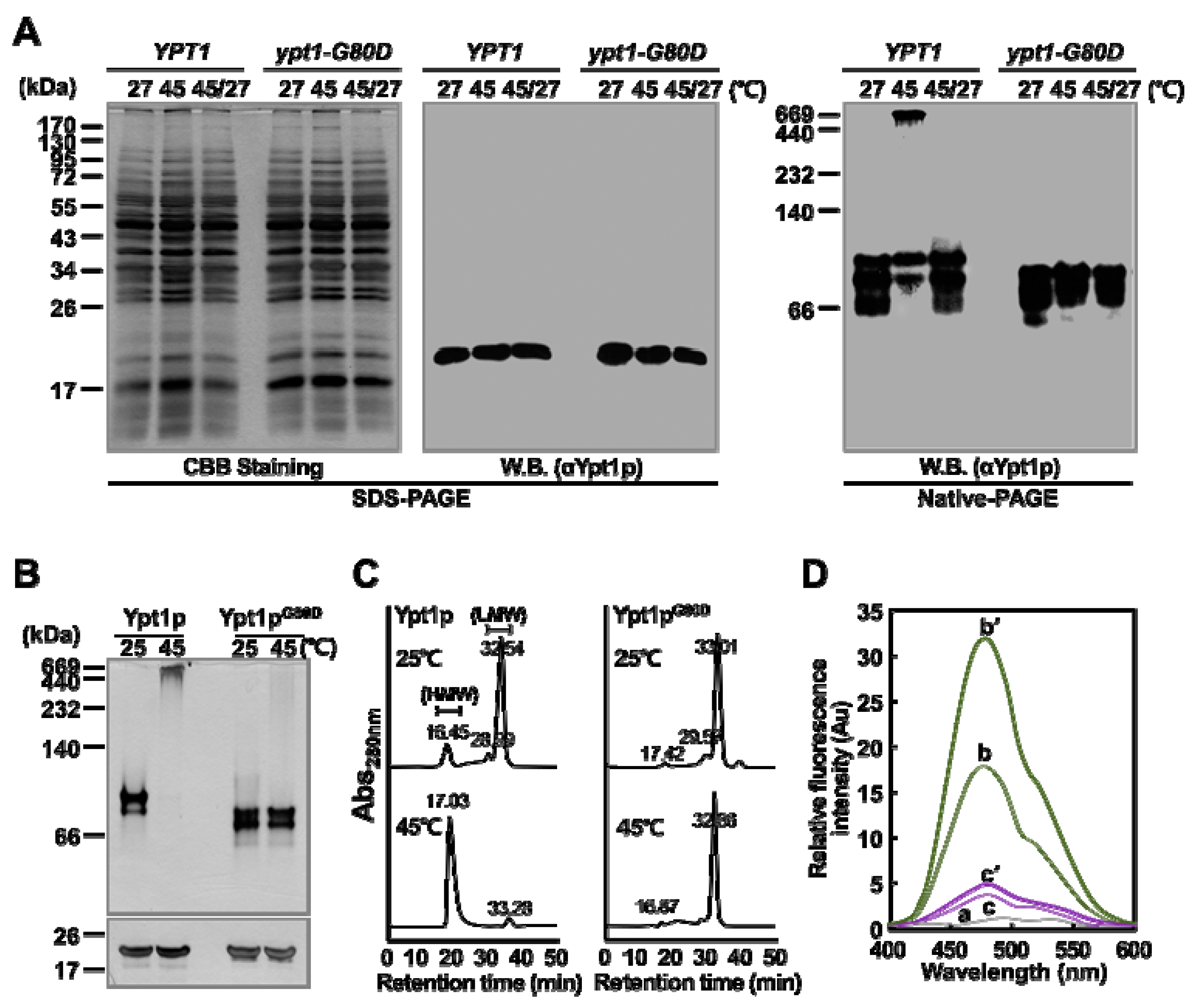
2.3. Ypt1pG80D does not Undergo a Heat-Shock-Induced Structural Change In Vivo
2.4. Ypt1pG80D does not Undergo a Heat-Shock-Induced Structural Change In Vitro
2.5. Ypt1pG80D Retains GTPase Activity but Loses Molecular Chaperone Activity
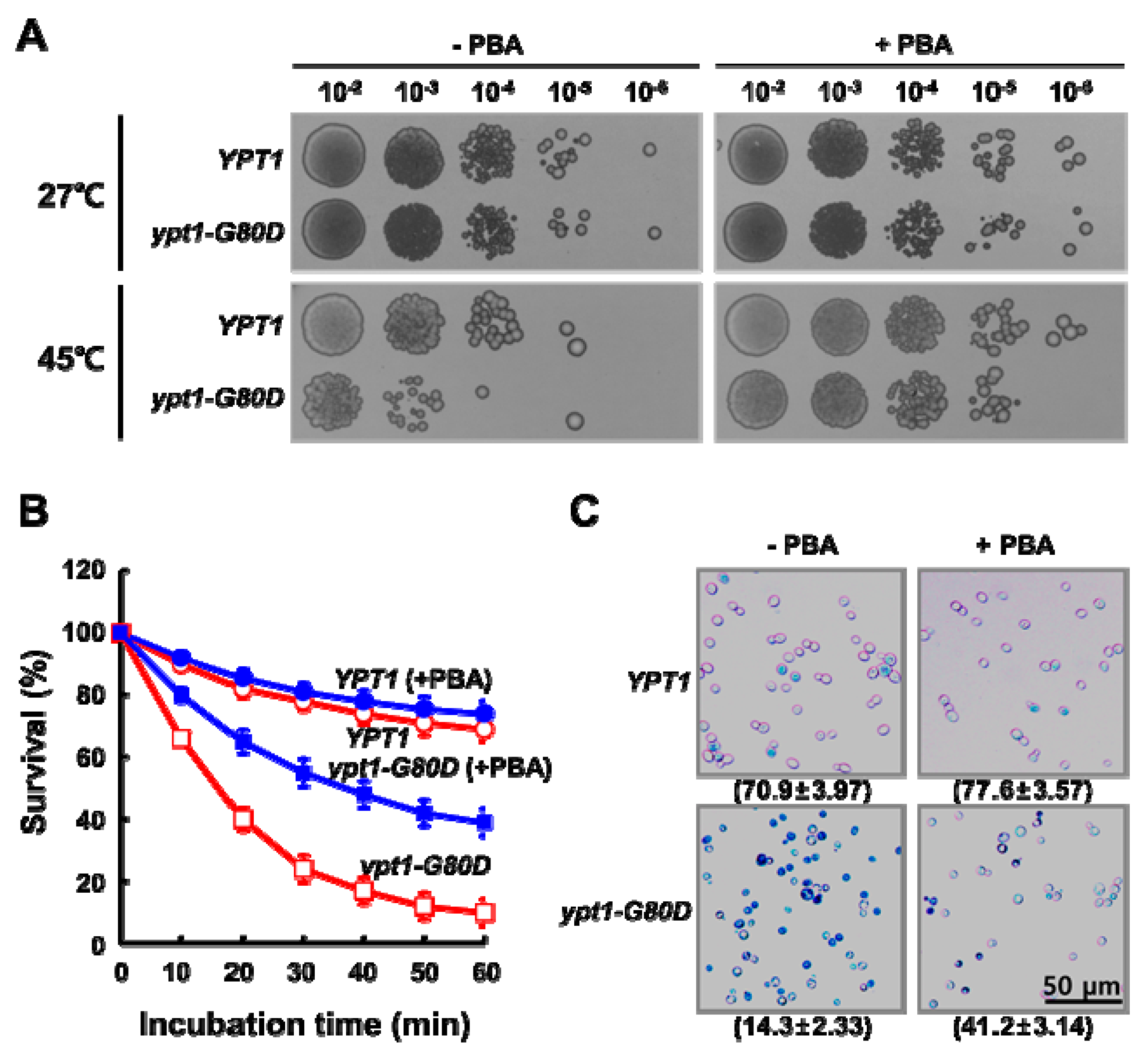
2.6. PBA Increases the Thermo Tolerance of ypt1-G80D Cells
2.7. Identification of Putative Ypt1p-Regulatory Proteins under Heat-Shock
3. Discussion
4. Materials and Methods
4.1. Yeast Strains, Survivability Assays, and TB Exclusion Assay
4.2. Analysis of Heat-Shock-Induced Cytosolic Protein Aggregation in Yeast Cells
4.3. Construction of Expression Plasmids
4.4. Purification of Recombinant Proteins and Production of the Polyclonal Antibody
4.5. Size Exclusion Chromatography
4.6. Assay of GTPase Activity
4.7. Measurement of bis-ANS Fluorescence
4.8. LC/MS Analysis
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| bis-ANS | 4,4′-bis (1-anilinonaphthalene 8-sulfonate) |
| CBB | Coomassie Brilliant Blue |
| CS | citrate synthase |
| GST | glutathione-S-transferase |
| HMW | high molecular weight |
| LC-MS | Liquid chromatography-mass spectrometry |
| LMW | low molecular weight |
| MDH | malate dehydrogenase |
| PBA | sodium 4-phenylbutyric acid |
| Prx | peroxiredoxin |
| SEC | size exclusion chromatography |
| TB | trypan blue |
| TLC | thin layer chromatography |
References
- Jedd, G.; Richardson, C.; Litt, R.; Segev, N. The Ypt1 GTPase is essential for the first two steps of the yeast secretory pathway. J. Cell Biol. 1995, 131, 583–590. [Google Scholar] [CrossRef] [PubMed]
- Segev, N.; Botstein, D. The ras-like yeast YPT1 gene is itself essential for growth, sporulation, and starvation response. Mol. Cell Biol. 1987, 7, 2367–2377. [Google Scholar] [CrossRef] [PubMed]
- Nuoffer, C.; Davidson, H.W.; Matteson, J.; Meinkoth, J.; Balch, W.E. A GDP-bound of rab1 inhibits protein export from the endoplasmic reticulum and transport between Golgi compartments. J. Cell Biol. 1994, 125, 225–237. [Google Scholar] [CrossRef] [PubMed]
- Richardson, C.J.; Jones, S.; Litt, R.J.; Segev, N. GTP hydrolysis is not important for Ypt1 GTPase function in vesicular transport. Mol. Cell Biol. 1998, 18, 827–838. [Google Scholar] [CrossRef] [PubMed]
- Du, L.L.; Novick, P. Yeast rab GTPase-activating protein Gyp1p localizes to the Golgi apparatus and is a negative regulator of Ypt1p. Mol. Biol. Cell 2001, 12, 1215–1226. [Google Scholar] [CrossRef] [PubMed]
- Ballew, N.; Liu, Y.; Barlowe, C. A Rab requirement is not bypassed in SLY1-20 suppression. Mol. Biol. Cell 2005, 16, 1839–1849. [Google Scholar] [CrossRef] [PubMed]
- Frigieri, M.C.; Joao Luiz, M.V.; Apponi, L.H.; Zanelli, C.F.; Valentini, S.R. Synthetic lethality between eIF5A and Ypt1 reveals a connection between translation and the secretory pathway in yeast. Mol. Genet. Genom. 2008, 280, 211–221. [Google Scholar] [CrossRef] [PubMed]
- Lynch-Day, M.A.; Bhandari, D.; Menon, S.; Huang, J.; Cai, H.; Bartholomew, C.R.; Brumell, J.H.; Ferro-Novick, S.; Klionsky, D.J. Trs85 directs a Ypt1 GEF, TRAPPIII, to the phagophore to promote autophagy. Proc. Natl. Acad. Sci. USA 2010, 107, 7811–7816. [Google Scholar] [CrossRef]
- Kang, C.H.; Lee, S.Y.; Park, J.H.; Lee, Y.; Jung, H.S.; Chi, Y.H.; Jung, Y.J.; Chae, H.B.; Shin, M.R.; Kim, W.Y.; et al. Stress-driven structural and functional switching of Ypt1p from a GTPase to a molecular chaperone mediates thermo tolerance in Saccharomyces cerevisiae. FASEB J. 2015, 29, 4424–4434. [Google Scholar] [CrossRef]
- Jang, H.H.; Lee, K.O.; Chi, Y.H.; Jung, B.G.; Park, S.K.; Park, J.H.; Lee, J.R.; Lee, S.S.; Moon, J.C.; Yun, J.W.; et al. Two enzymes in one; two yeast peroxiredoxins display oxidative stress-dependent switching from a peroxidase to a molecular chaperone function. Cell 2004, 117, 625–635. [Google Scholar] [CrossRef]
- Strober, W. Trypan blue exclusion test of cell viability. Curr. Protoc. Immunol. 2001, Appendix 3, Appendix 3B. [Google Scholar] [CrossRef]
- Tomoyasu, T.; Mogk, A.; Langen, H.; Goloubinoff, P.; Bukau, B. Genetic dissection of the roles of chaperones and proteases in protein folding and degradation in the Escherichia coli cytosol. Mol. Microbiol. 2001, 40, 397–413. [Google Scholar] [CrossRef] [PubMed]
- Faust, A.M.; Wong, C.C.; Yates, J.R., 3rd; Drubin, D.G.; Barnes, G. The FEAR protein Slk19 restricts Cdc14 phosphatase to the nucleus until the end of anaphase, regulating its participation in mitotic exit in Saccharomyces cerevisiae. PLoS ONE 2013, 8, e73194. [Google Scholar] [CrossRef] [PubMed]
- Paul, K.R.; Molliex, A.; Cascarina, S.; Boncella, A.E.; Taylor, J.P.; Ross, E.D. Effects of Mutations on the Aggregation Propensity of the Human Prion-Like Protein hnRNPA2B1. Mol. Cell Biol. 2017, 37. [Google Scholar] [CrossRef]
- Sharma, K.K.; Kaur, H.; Kumar, G.S.; Kester, K. Interaction of 1,1′-bi(4-anilino)naphthalene-5,5′-disulfonic acid with alpha-crystallin. J. Biol. Chem. 1998, 273, 8965–8970. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.R.; Lee, S.S.; Jang, H.H.; Lee, Y.M.; Park, J.H.; Park, S.C.; Moon, J.C.; Park, S.K.; Kim, S.Y.; Lee, S.Y.; et al. Heat-shock dependent oligomeric status alters the function of a plant-specific thioredoxin-like protein, AtTDX. Proc. Natl. Acad. Sci. USA 2009, 106, 5978–5983. [Google Scholar] [CrossRef]
- Basseri, S.; Lhotak, S.; Sharma, A.M.; Austin, R.C. The chemical chaperone 4-phenylbutyrate inhibits adipogenesis by modulating the unfolded protein response. J. Lipid Res. 2009, 50, 2486–2501. [Google Scholar] [CrossRef]
- Kim, D.Y.; Scalf, M.; Smith, L.M.; Vierstra, R.D. Advanced proteomic analyses yield a deep catalog of ubiquitylation targets in Arabidopsis. Plant Cell 2013, 25, 1523–1540. [Google Scholar] [CrossRef]
- Butterfield, D.A.; Yatin, S.M.; Varadarajan, S.; Koppal, T. Amyloid beta-peptide-associated free radical oxidative stress, neurotoxicity, and Alzheimer’s disease. Methods Enzymol. 1999, 309, 746–768. [Google Scholar]
- Kim, K.S.; Choi, S.Y.; Kwon, H.Y.; Won, M.H.; Kang, T.C.; Kang, J.H. Aggregation of alpha-synuclein induced by the Cu,Zn-superoxide dismutase and hydrogen peroxide system. Free Radic. Biol. Med. 2002, 32, 544–550. [Google Scholar] [CrossRef]
- Hendrick, J.P.; Hartl, F.U. Molecular chaperone functions of heat-shock proteins. Annu. Rev. Biochem. 1993, 62, 349–384. [Google Scholar] [CrossRef] [PubMed]
- Mayer, M.P. The unfolding story of a redox chaperone. Cell 2012, 148, 843–844. [Google Scholar] [CrossRef] [PubMed]
- Chae, H.B.; Moon, J.C.; Shin, M.R.; Chi, Y.H.; Jung, Y.J.; Lee, S.Y.; Nawkar, G.M.; Jung, H.S.; Hyun, J.K.; Kim, W.Y.; et al. Thioredoxin reductase type C (NTRC) orchestrates enhanced thermotolerance to Arabidopsis by its redox-dependent holdase chaperone function. Mol. Plant 2013, 6, 323–336. [Google Scholar] [CrossRef] [PubMed]
- Park, J.H.; Lee, S.Y.; Kim, W.Y.; Jung, Y.J.; Chae, H.B.; Jung, H.S.; Kang, C.H.; Shin, M.R.; Kim, S.Y.; Su’udi, M.; et al. Heat-induced chaperone activity of serine/threonine protein phosphatase 5 enhances thermotolerance in Arabidopsis thaliana. New Phytol. 2011, 191, 692–705. [Google Scholar] [CrossRef] [PubMed]
- Park, S.K.; Jung, Y.J.; Lee, J.R.; Lee, Y.M.; Jang, H.H.; Lee, S.S.; Park, J.H.; Kim, S.Y.; Moon, J.C.; Lee, S.Y.; et al. Heat-shock and redox-dependent functional switching of an h-type Arabidopsis thioredoxin from a disulfide reductase to a molecular chaperone. Plant Physiol. 2009, 150, 552–561. [Google Scholar] [CrossRef] [PubMed]
- Bell, S.L.; Chiang, A.N.; Brodsky, J.L. Expression of a malarial Hsp70 improves defects in chaperone-dependent activities in ssa1 mutant yeast. PLoS ONE 2011, 6, e20047. [Google Scholar] [CrossRef] [PubMed]
- Ruiz-Roig, C.; Vieitez, C.; Posas, F.; de Nadal, E. The Rpd3L HDAC complex is essential for the heat stress response in yeast. Mol. Microbiol. 2010, 76, 1049–1062. [Google Scholar] [CrossRef] [PubMed]
- Sinha, H.; David, L.; Pascon, R.C.; Clauder-Munster, S.; Krishnakumar, S.; Nguyen, M.; Shi, G.; Dean, J.; Davis, R.W.; Oefner, P.J.; et al. Sequential elimination of major-effect contributors identifies additional quantitative trait loci conditioning high-temperature growth in yeast. Genetics 2008, 180, 1661–1670. [Google Scholar] [CrossRef] [PubMed]
- Feldman, D.E.; Frydman, J. Protein folding in vivo: The importance of molecular chaperones. Curr. Opin. Struct. Biol. 2000, 10, 26–33. [Google Scholar] [CrossRef]
- Slavotinek, A.M.; Biesecker, L.G. Unfolding the role of chaperones and chaperonins in human disease. Trends Genet. 2001, 17, 528–535. [Google Scholar] [CrossRef]
- Aoki, Y.; Niihori, T.; Inoue, S.; Matsubara, Y. Recent advances in RASopathies. J. Hum. Genet. 2016, 61, 33–39. [Google Scholar] [CrossRef]
- Hor, C.H.H.; Tang, B.L.; Goh, E.L.K. Rab23 and developmental disorders. Rev. Neurosci. 2018. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Huang, T.Y.; Yancey, J.; Luo, H.; Zhang, Y.W. Role of Rab GTPases in Alzheimer’s Disease. ACS Chem. Neurosci. 2018. [Google Scholar] [CrossRef] [PubMed]
- Bobori, C.; Theocharopoulou, G.; Vlamos, P. Molecular Chaperones in Neurodegenerative Diseases: A Short Review. Adv. Exp. Med. Biol. 2017, 987, 219–231. [Google Scholar] [CrossRef] [PubMed]
- Ebrahimi-Fakhari, D.; Saidi, L.J.; Wahlster, L. Molecular chaperones and protein folding as therapeutic targets in Parkinson’s disease and other synucleinopathies. Acta Neuropathol. Commun. 2013, 1, 79. [Google Scholar] [CrossRef] [PubMed]
- Kang, C.H.; Jung, W.Y.; Kang, Y.H.; Kim, J.Y.; Kim, D.G.; Jeong, J.C.; Baek, D.W.; Jin, J.B.; Lee, J.Y.; Kim, M.O.; et al. AtBAG6, a novel calmodulin-binding protein, induces programmed cell death in yeast and plants. Cell Death Differ. 2006, 13, 84–95. [Google Scholar] [CrossRef] [PubMed]
- Cheong, N.E.; Choi, Y.O.; Lee, K.O.; Kim, W.Y.; Jung, B.G.; Chi, Y.H.; Jeong, J.S.; Kim, K.; Cho, M.J.; Lee, S.Y. Molecular cloning, expression, and functional characterization of a 2Cys-peroxiredoxin in Chinese cabbage. Plant Mol. Biol. 1999, 40, 825–834. [Google Scholar] [CrossRef] [PubMed]
- Chae, H.Z.; Chung, S.J.; Rhee, S.G. Thioredoxin-dependent peroxide reductase from yeast. J. Biol. Chem. 1994, 269, 27670–27678. [Google Scholar]
- Bollag, G.; McCormick, F. Intrinsic and GTPase-activating protein-stimulated Ras GTPase assays. Methods Enzymol. 1995, 255, 161–170. [Google Scholar] [PubMed]
- Seo, H.S.; Jeong, J.Y.; Nahm, M.Y.; Kim, S.W.; Lee, S.Y.; Bahk, J.D. The effect of pH and various cations on the GTP hydrolysis of rice heterotrimeric G-protein alpha subunit expressed in Escherichia coli. J. Biochem. Mol. Biol. 2003, 36, 196–200. [Google Scholar] [PubMed]
- Rohrbough, J.G.; Breci, L.; Merchant, N.; Miller, S.; Haynes, P.A. Verification of single-peptide protein identifications by the application of complementary database search algorithms. J. Biomol. Tech. 2006, 17, 327–332. [Google Scholar] [PubMed]
- Oliveros, J.C. VENNY. An. Interactive Tool for Comparing Lists with Venn Diagrams; CNB_CSIC: Madrid, Spain, 2007. [Google Scholar]


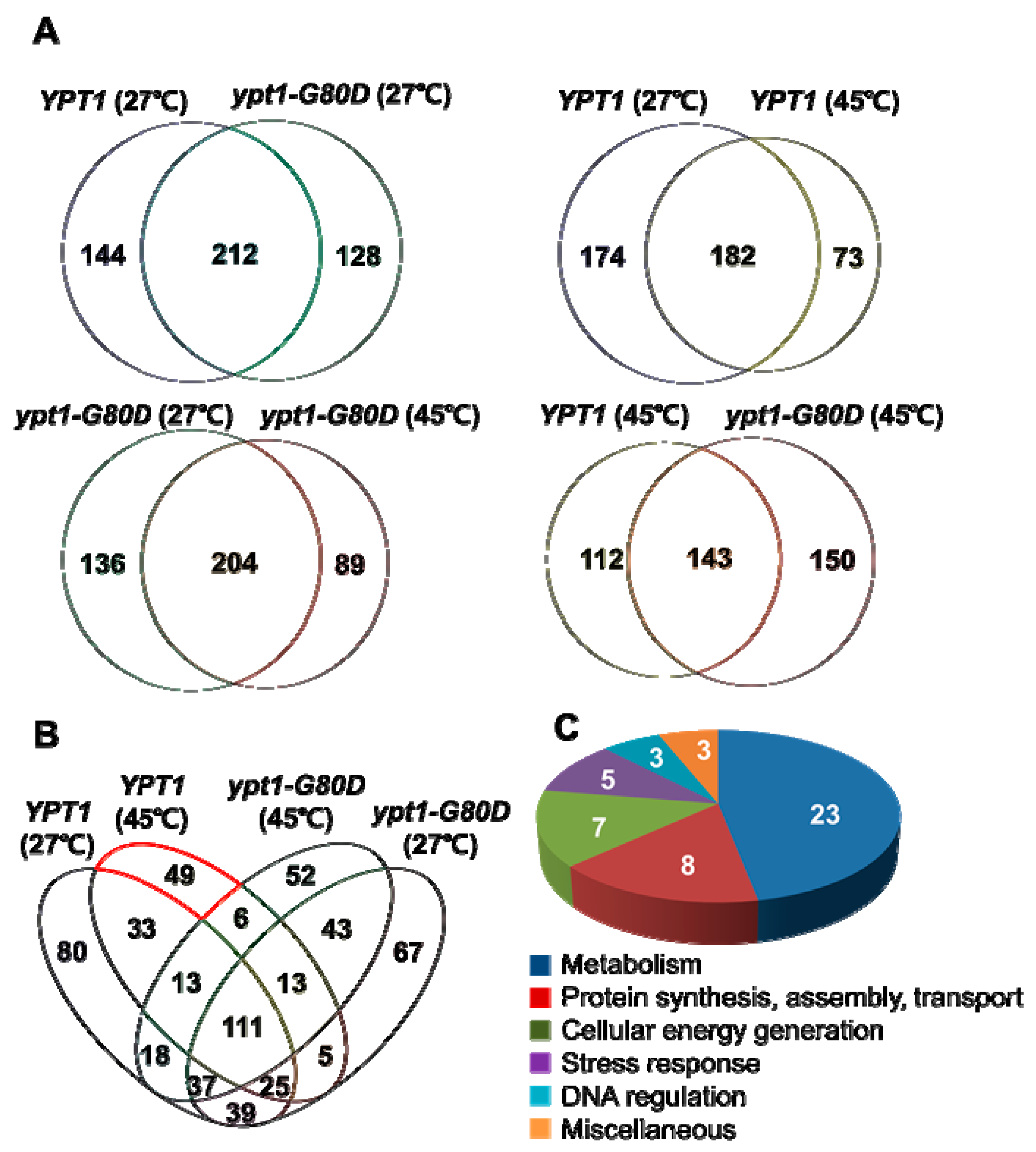
| Groups (# proteins) | Putative Ypt1p-Regulatory Proteins under Heat-Shock |
|---|---|
| Metabolism (23) | Inorganic pyrophosphatase, Glucose-6-phosphate isomerase, Ach1p, YKR097Wp-like protein, Thr4p, 6-phosphogluconate dehydrogenase, Alpha-ketoglutarate dehydrogenase, Carboxypeptidase Y inhibitor, Ses1p, YGL202Wp-like protein, Transaldolase, IDH2 (Isocitric dehydrogenase 2), Putative uncharacterized protein (A6ZLW7), Pbi2p, IDP2 (Isocitrate dehydrogenase), Branched chain a.a. aminotransferase, YGR124Wp-like protein, Bts1p, Enolase, Ypr1p, Erg10p, Cys4p, K7_Ade17bp |
| Protein synthesis, assembly, and transport (8) | Eft2p, Om45p, Ses1p, Tropomyosin-1, YFL037Wp-like protein, KAP123 (Karyopherin beta 4), YBL050Wp-like protein, Rpl7bp |
| Cellular energy generation (7) | ATP synthase subunit beta, Porin, Rhr2p, ADK1 (Adenylate kinase), Sdh1p, Atp16p, YBR039Wp-like protein, Cytochrome b-c1 complex subunit 7 |
| Stress response (5) | Ssa1p, Ssa3p, Prx1p, SIP18, YML128Cp-like protein |
| DNA regulation (3) | REV3 (DNA polymerase), YPL016Wp-like protein, Mbp1p |
| Miscellaneous (3) | Putative uncharacterized protein (A6ZYB0), SOF1 (Conserved protein), Tma19p |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kang, C.H.; Park, J.H.; Lee, E.S.; Paeng, S.K.; Chae, H.B.; Chi, Y.H.; Lee, S.Y. Exploring Novel Functions of the Small GTPase Ypt1p under Heat-Shock by Characterizing a Temperature-Sensitive Mutant Yeast Strain, ypt1-G80D. Int. J. Mol. Sci. 2019, 20, 132. https://doi.org/10.3390/ijms20010132
Kang CH, Park JH, Lee ES, Paeng SK, Chae HB, Chi YH, Lee SY. Exploring Novel Functions of the Small GTPase Ypt1p under Heat-Shock by Characterizing a Temperature-Sensitive Mutant Yeast Strain, ypt1-G80D. International Journal of Molecular Sciences. 2019; 20(1):132. https://doi.org/10.3390/ijms20010132
Chicago/Turabian StyleKang, Chang Ho, Joung Hun Park, Eun Seon Lee, Seol Ki Paeng, Ho Byoung Chae, Yong Hun Chi, and Sang Yeol Lee. 2019. "Exploring Novel Functions of the Small GTPase Ypt1p under Heat-Shock by Characterizing a Temperature-Sensitive Mutant Yeast Strain, ypt1-G80D" International Journal of Molecular Sciences 20, no. 1: 132. https://doi.org/10.3390/ijms20010132
APA StyleKang, C. H., Park, J. H., Lee, E. S., Paeng, S. K., Chae, H. B., Chi, Y. H., & Lee, S. Y. (2019). Exploring Novel Functions of the Small GTPase Ypt1p under Heat-Shock by Characterizing a Temperature-Sensitive Mutant Yeast Strain, ypt1-G80D. International Journal of Molecular Sciences, 20(1), 132. https://doi.org/10.3390/ijms20010132





