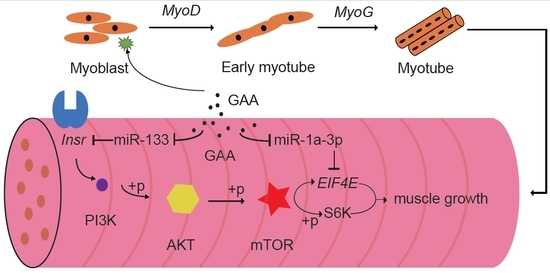
Guanidinoacetic Acid Regulates Myogenic Differentiation and Muscle Growth Through miR-133a-3p and miR-1a-3p Co-mediated Akt/mTOR/S6K Signaling Pathway
Abstract
1. Introduction
2. Results
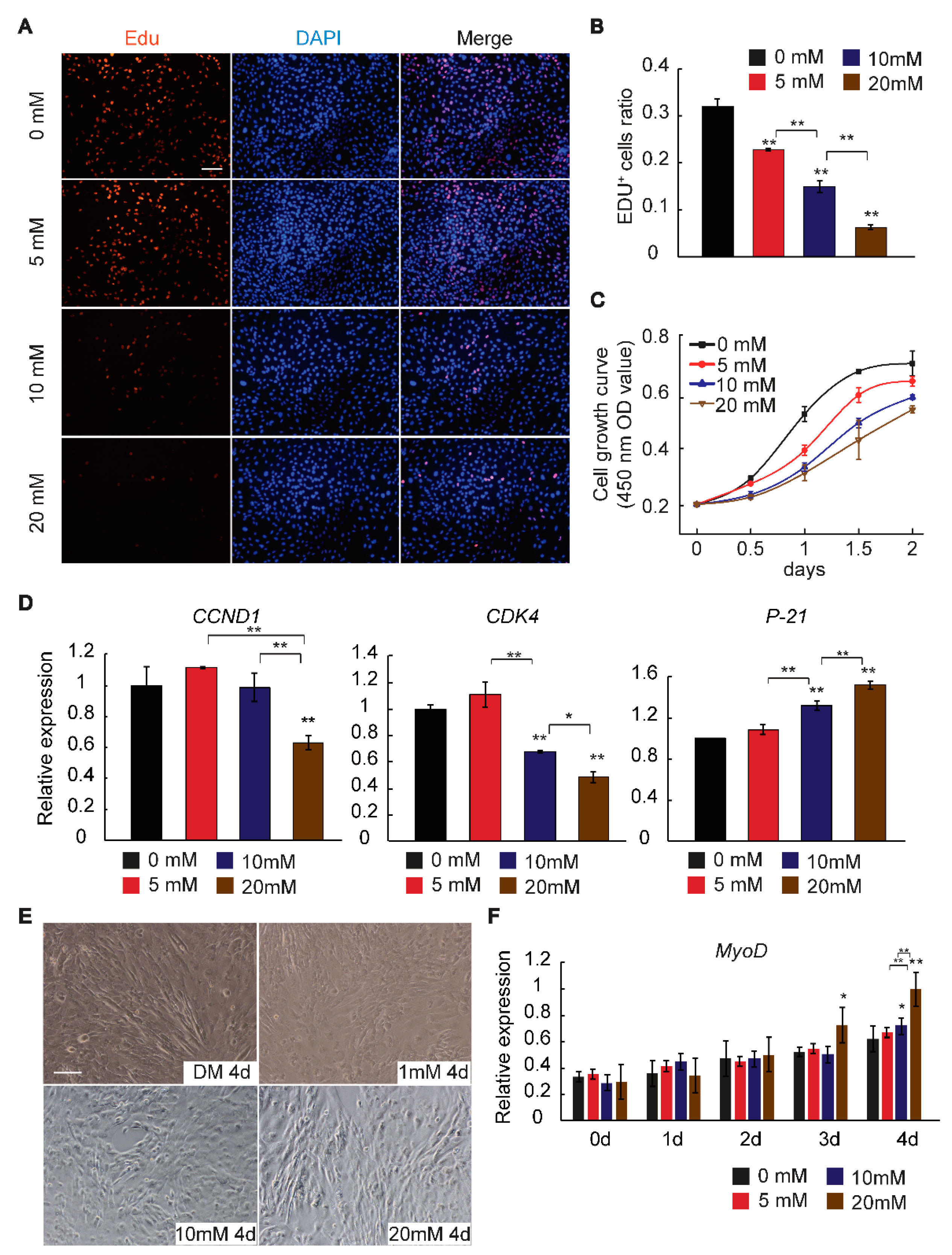
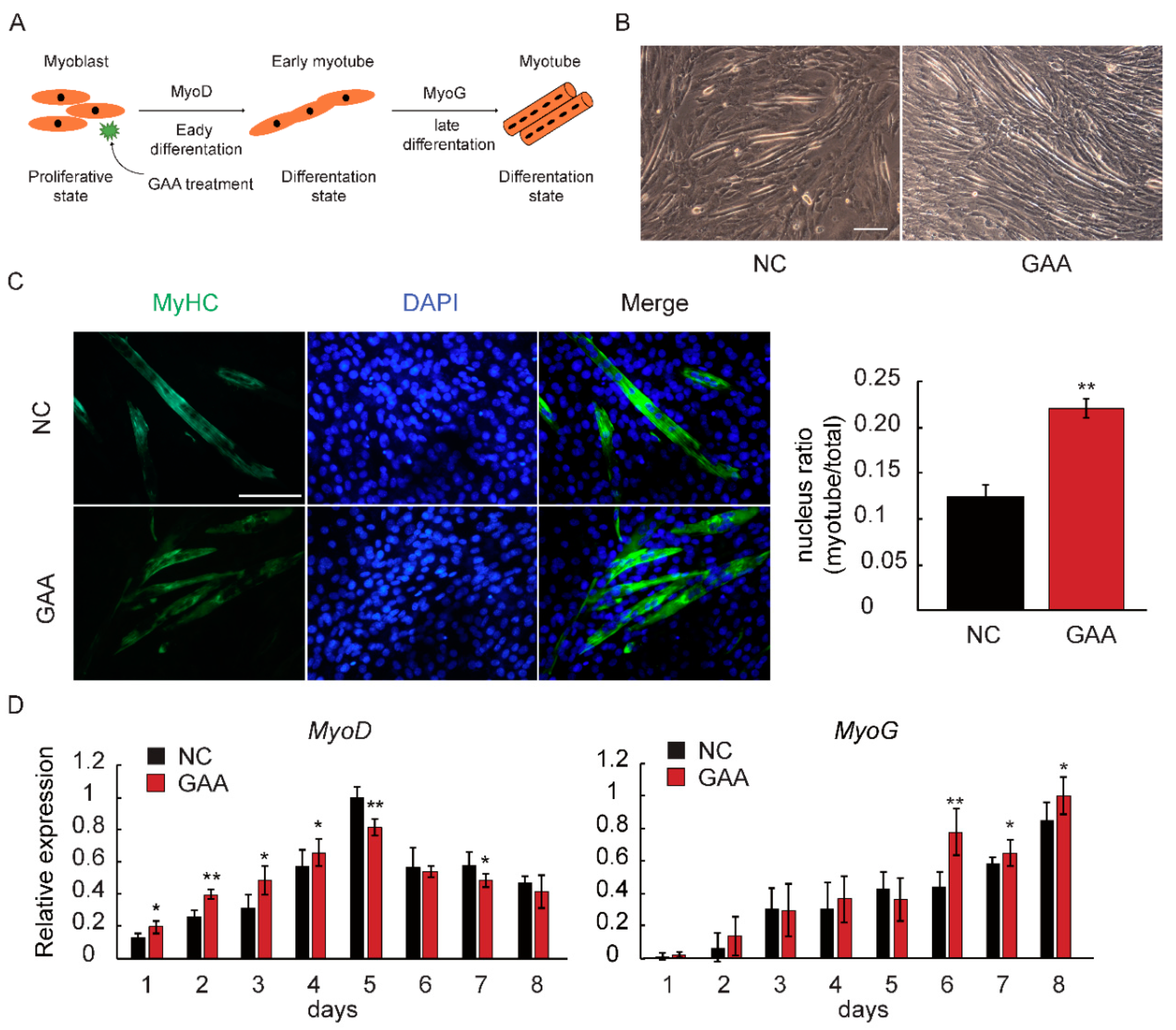
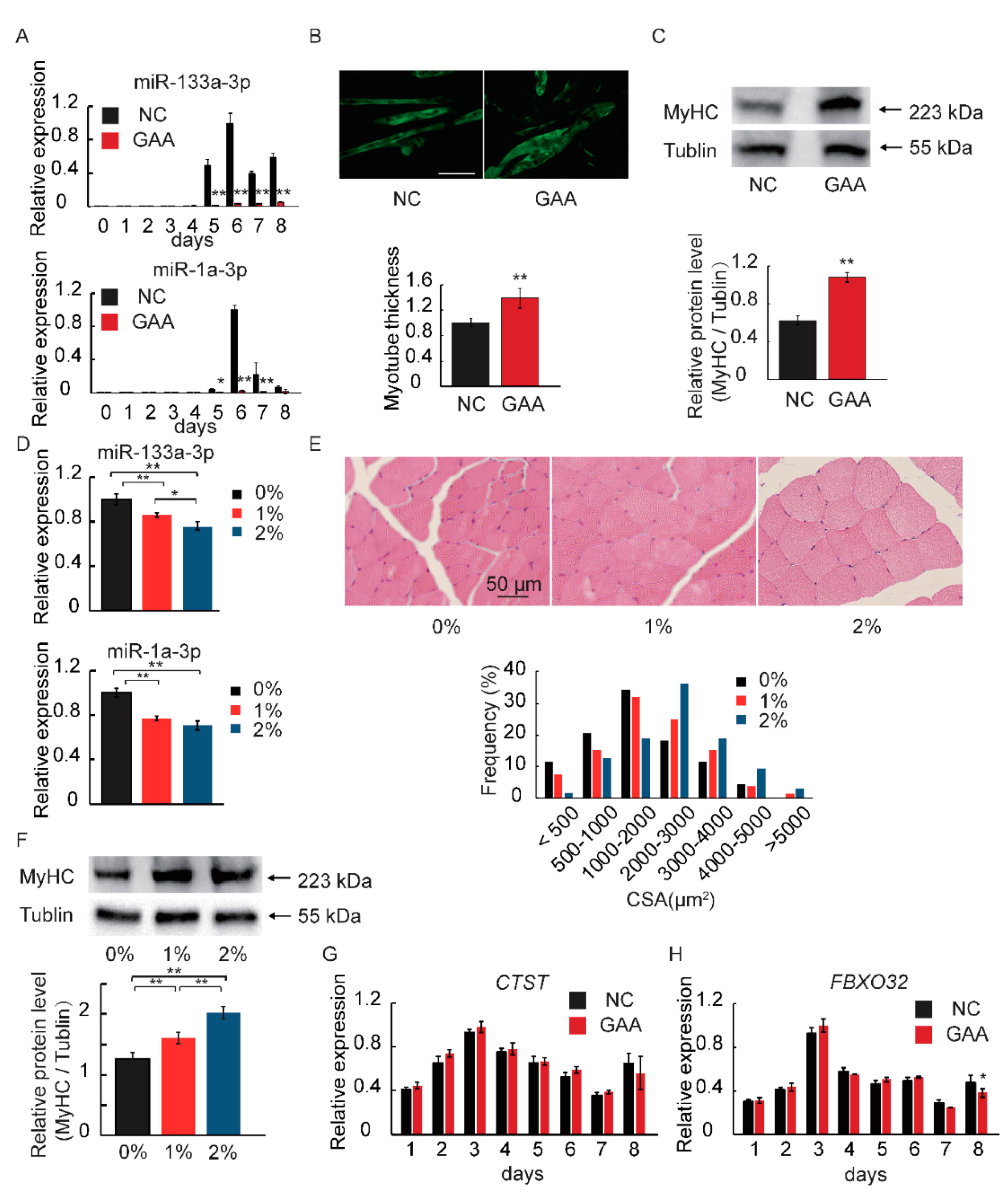
2.1. GAA Affects Myogenic Differentiation of Myoblast in Dose-Dependent Manner
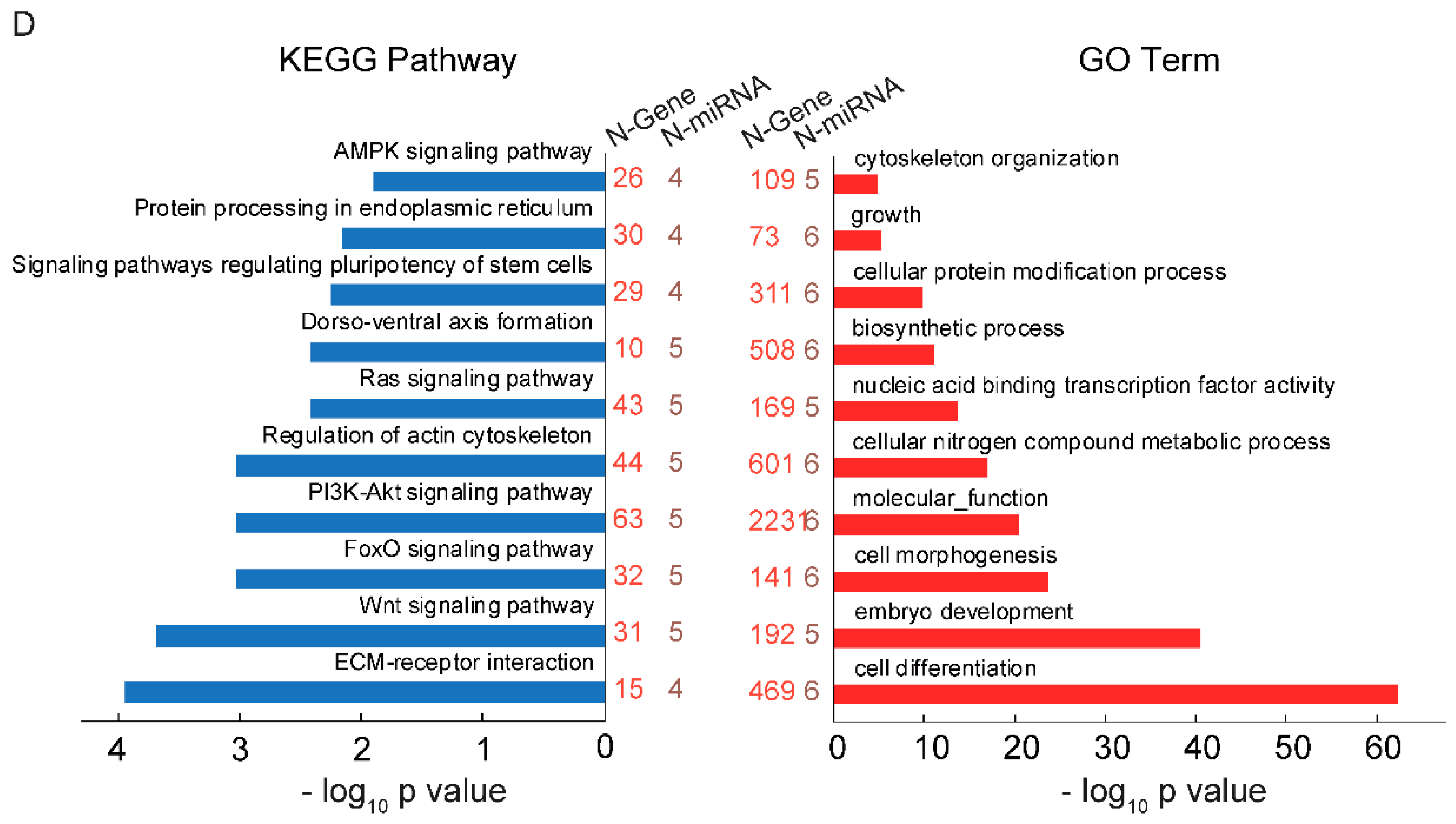
2.2. GAA Modulates Myogenesis through MicroRNA Regulatory Network
2.3. MiR-133a-3p and MiR-1a-3p Expression is Decreased during Muscle Growth
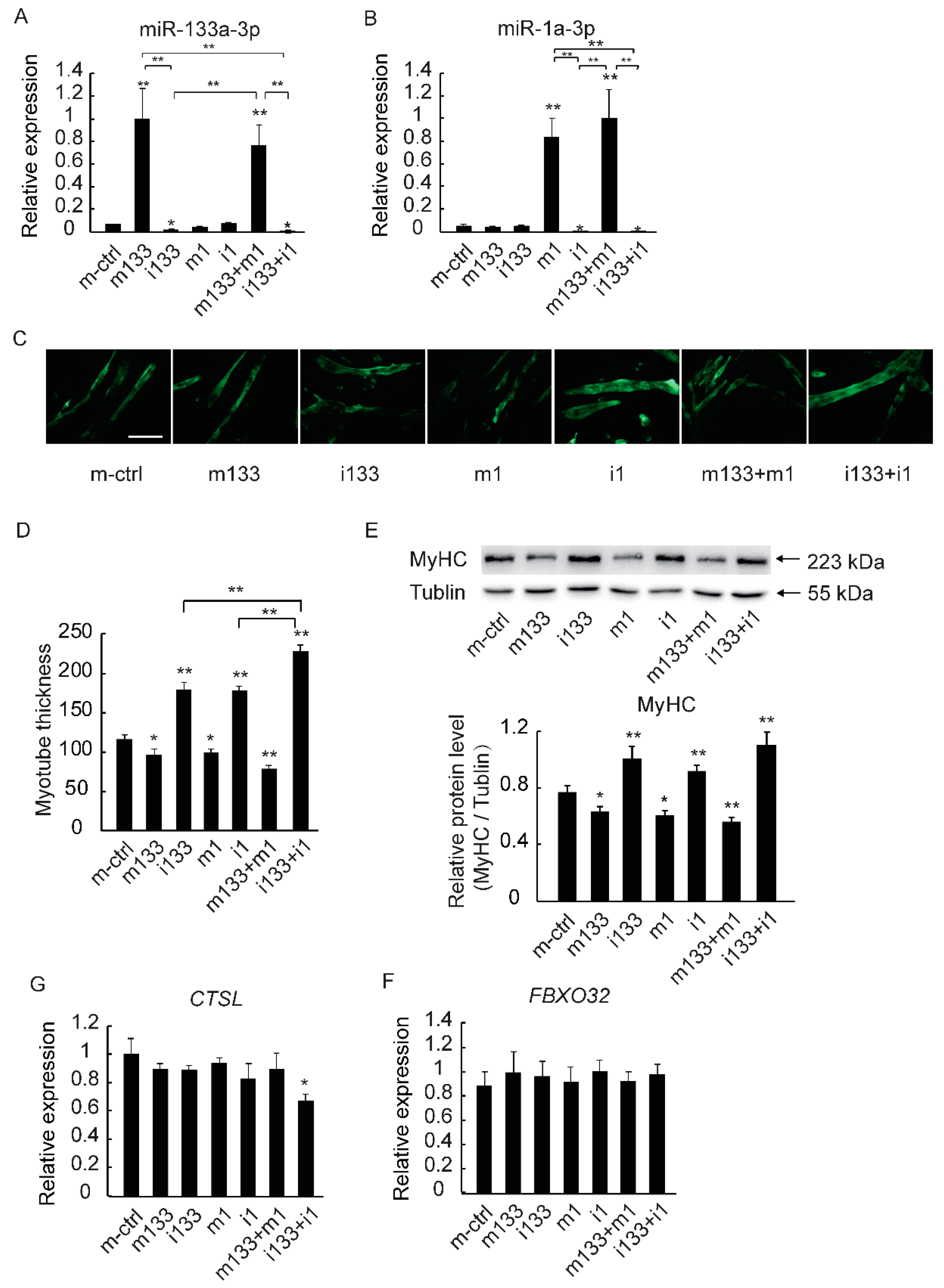
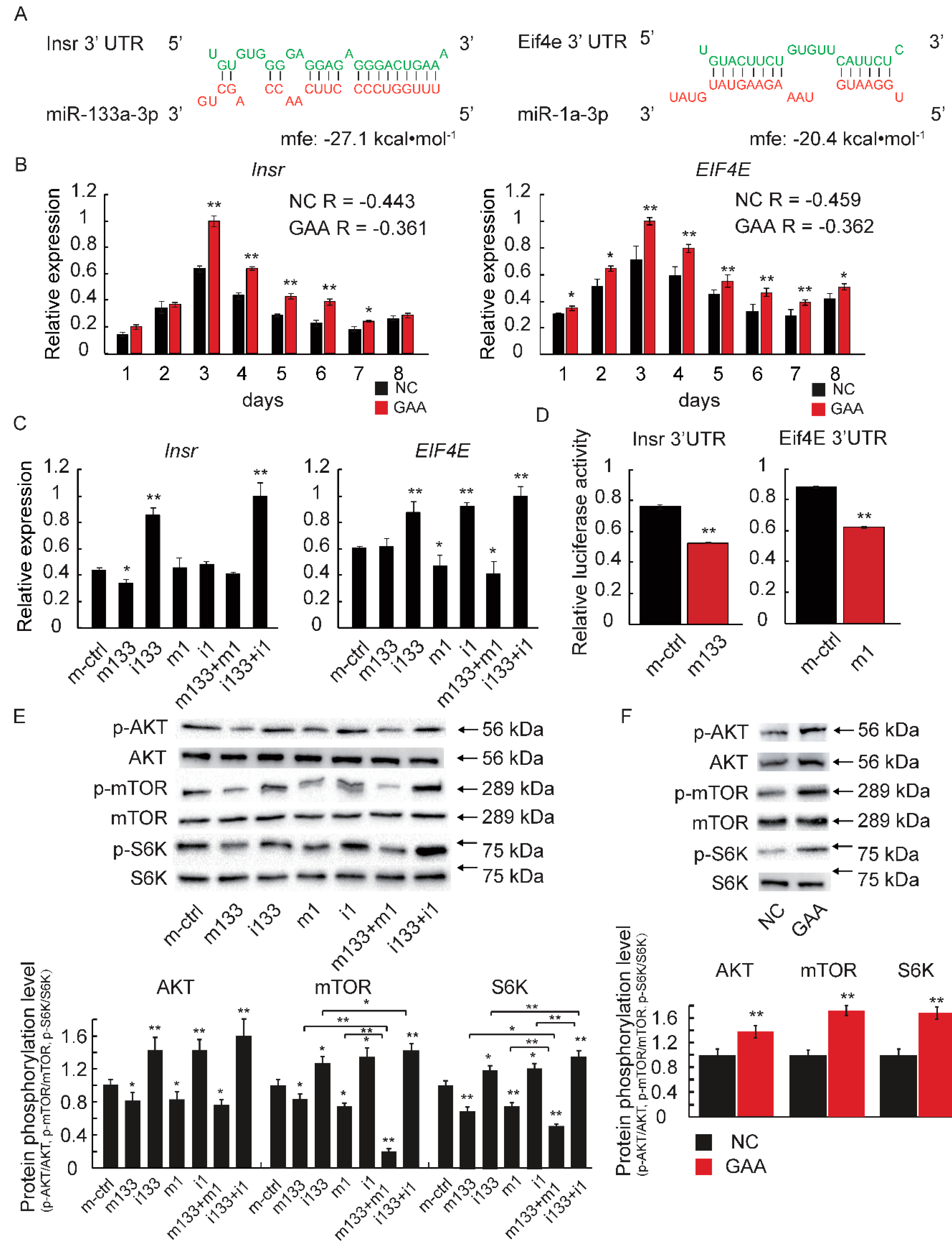
2.4. The Downregulation of MiR-133a-3p and MiR-1a-3p Promotes Muscle Growth by Activating Akt/mTOR/S6K Signaling
3. Discussion
4. Materials and Methods
4.1. Regents and Antibodies
4.2. Cell Culture and Treatment
4.3. Animals and Muscle Collection
4.4. RNA Extraction, Reverse Transcription, and qRT-PCR
4.5. Cell Proliferation Ratio Assay
4.6. Western Blot Assay
4.7. Immunocytochemistry
4.8. HE Staining and Skeletal Muscle Cross-Sectional Area Statistics
4.9. Small RNA Library Construction and Sequencing
4.10. Analysis of High-Throughput Sequencing Data
4.11. Differential Expression Analysis of MiRNAs
4.12. Prediction and Functional Enrichment Analysis of DE MiRNA Target Genes
4.13. Bioinformatic Analyses and Dual-Luciferase Reporter Assay
4.14. MiRNA Transfection
4.15. Creatine and Creatinine Content Assay
4.16. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Montesano, A.; Luzi, L.; Senesi, P.; Mazzocchi, N.; Terruzzi, I. Resveratrol promotes myogenesis and hypertrophy in murine myoblasts. J. Transl. Med. 2013, 11, 310. [Google Scholar] [CrossRef] [PubMed]
- Parr, M.K.; Zhao, P.; Haupt, O.; Ngueu, S.T.; Hengevoss, J.; Fritzemeier, K.H.; Piechotta, M.; Schlörer, N.; Muhn, P.; Zheng, W.Y. Estrogen receptor beta is involved in skeletal muscle hypertrophy induced by the phytoecdysteroid ecdysterone. Mol. Nutr. Food Res. 2014, 58, 1861–1872. [Google Scholar] [CrossRef] [PubMed]
- Kunkel, S.D.; Suneja, M.; Ebert, S.M.; Bongers, K.S.; Fox, D.K.; Malmberg, S.E.; Alipour, F.; Shields, R.K.; Adams, C.M. Mrna expression signatures of human skeletal muscle atrophy identify a natural compound that increases muscle mass. Cell Metab. 2011, 13, 627–638. [Google Scholar] [CrossRef] [PubMed]
- Ostojic, S.M.; Niess, B.; Stojanovic, M.; Obrenovic, M. Creatine metabolism and safety profiles after six-week oral guanidinoacetic acid administration in healthy humans. Int. Med. Sci. 2013, 10, 141–147. [Google Scholar] [CrossRef] [PubMed]
- Tsubakihara, Y.; Suzuki, A.; Hayashi, T.; Shoji, T.; Togawa, M.; Okada, N. The effect of guanidinoacetic acid supplementation in patients with chronic renal failure. In Guanidino Compounds in Biology and Medicine; Mori, A., Ishida, M., Clark, J.F., Eds.; Blackwell Science: Hoboken, NJ, USA, 1999; Volume 3, pp. 139–144. [Google Scholar]
- Ostojic, S.M.; Hoffman, J.R.; Stojanovic, M.; Drid, P. 28-day guanidinoacetic acid supplementation improves clinical outcomes in patients with chronic fatigue syndrome. Med. Sci. Sport Exerc. 2015, 48, S47. [Google Scholar]
- Ringel, J.; Lemme, A.; Redshaw, M.S.; Damme, K. The effects of supplemental guanidino acetic acid as a precursor of creatine in vegetable broiler diets on performance and carcass parameters. Poult. Sci. 2008, 87, 72. [Google Scholar]
- Michiels, J.; Maertens, L.; Buyse, J.; Lemme, A.; Rademacher, M.; Dierick, N.A.; De, S.S. Supplementation of guanidinoacetic acid to broiler diets: Effects on performance, carcass characteristics, meat quality, and energy metabolism. Poult. Sci. 2012, 91, 402–412. [Google Scholar] [CrossRef] [PubMed]
- Mcbreairty, L.E.; Robinson, J.L.; Furlong, K.R.; Brunton, J.A.; Bertolo, R.F. Guanidinoacetate is more effective than creatine at enhancing tissue creatine stores while consequently limiting methionine availability in yucatan miniature pigs. PLoS ONE 2014, 10, e0131563. [Google Scholar] [CrossRef] [PubMed]
- Ambros, V. The c. Elegans heterochronic gene lin-4 encodes small rnas with antisense complementarity to lin-14. Cell 1993, 75, 843–854. [Google Scholar]
- Pasquinelli, A.E.; Reinhart, B.J.; Slack, F.; Martindale, M.Q.; Kuroda, M.I.; Maller, B.; Hayward, D.C.; Ball, E.E.; Degnan, B.; Müller, P. Conservation of the sequence and temporal expression of let-7 heterochronic regulatory RNA. Nature 2000, 408, 86–89. [Google Scholar] [CrossRef] [PubMed]
- Xu, P.; Guo, M.; Hay, B.A. Micrornas and the regulation of cell death. Trends Genet. 2004, 20, 617–624. [Google Scholar] [CrossRef] [PubMed]
- Cheng, A.M.; Byrom, M.W.; Shelton, J.; Ford, L.P. Antisense inhibition of human miRNAs and indications for an involvement of miRNA in cell growth and apoptosis. Nucleic Acids Res. 2005, 33, 1290–1297. [Google Scholar] [CrossRef] [PubMed]
- O’Rourke, J.R.; Georges, S.A.; Seay, H.R.; Tapscott, S.J.; Mcmanus, M.T.; Goldhamer, D.J.; Swanson, M.S.; Harfe, B.D. Essential role for dicer during skeletal muscle development. Dev. Biol. 2007, 311, 359–368. [Google Scholar] [CrossRef] [PubMed]
- Horak, M.; Novak, J.; Bienertovavasku, J. Muscle-specific microRNAs in skeletal muscle development. Dev. Biol. 2016, 410, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Samal, E.; Srivastava, D. Serum response factor regulates a muscle-specific microRNA that targets hand2 during cardiogenesis. Nature 2005, 436, 214–220. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.F.; Mandel, E.M.; Thomson, J.M.; Wu, Q.; Callis, T.E.; Hammond, S.M.; Conlon, F.L.; Wang, D.Z. The role of microRNA-1 and microrna-133 in skeletal muscle proliferation and differentiation. Nat. Genet. 2006, 38, 228–233. [Google Scholar] [CrossRef] [PubMed]
- Crist, C.G.; Montarras, D.; Pallafacchina, G.; Rocancourt, D.; Cumano, A.; Conway, S.J.; Buckingham, M. Muscle stem cell behavior is modified by microRNA-27 regulation of pax3 expression. Proc. Natl. Acad. Sci. USA 2009, 106, 13383–13387. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.F.; Tao, Y.; Li, J.; Deng, Z.; Yan, Z.; Xiao, X.; Wang, D.Z. Microrna-1 and microrna-206 regulate skeletal muscle satellite cell proliferation and differentiation by repressing pax7. J. Cell Biol. 2010, 190, 867–879. [Google Scholar] [CrossRef] [PubMed]
- Dey, B.K.; Gagan, J.; Dutta, A. miR-206 and -486 induce myoblast differentiation by downregulating Pax7. Mol. Cell. Biol. 2011, 31, 203–214. [Google Scholar] [CrossRef] [PubMed]
- Mccormick, R.; Goljanek-Whysall, K. MicroRNA dysregulation in aging and pathologies of the skeletal muscle. Int. Rev. Cell Mol. Biol. 2017, 334, 265–308. [Google Scholar] [PubMed]
- Hitachi, K.; Tsuchida, K. Role of microRNAs in skeletal muscle hypertrophy. Front. Physiol. 2013, 4, 408. [Google Scholar] [CrossRef] [PubMed]
- Rao, P.K.; Kumar, R.M.; Farkhondeh, M.; Baskerville, S.; Lodish, H.F. Myogenic factors that regulate expression of muscle-specific microRNAs. Proc. Natl. Acad. Sci. USA 2006, 103, 8721–8726. [Google Scholar] [CrossRef] [PubMed]
- Rosenberg, M.I.; Georges, S.A.; Asawachaicharn, A.; Analau, E.; Tapscott, S.J. Myod inhibits Fstl1 and utrn expression by inducing transcription of miR-206. J. Cell Biol. 2006, 175, 77–85. [Google Scholar] [CrossRef] [PubMed]
- Liu, N.; Williams, A.H.; Kim, Y.; Mcanally, J.; Bezprozvannaya, S.; Sutherland, L.B.; Richardson, J.A.; Basselduby, R.; Olson, E.N. An intragenic MEF2-dependent enhancer directs muscle-specific expression of micrornas 1 and 133. Proc. Natl. Acad. Sci. USA 2007, 104, 20844–20849. [Google Scholar] [CrossRef] [PubMed]
- Gan, M.; Yang, D.; Fan, Y.; Du, J.; Shen, L.; Li, Q.; Jiang, Y.; Tang, G.; Li, M.; Wang, J. Bidirectional regulation of genistein on the proliferation and differentiation of C2C12 myoblasts. Xenobiotica 2017, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Latimer, M.; Sabin, N.; Le, C.A.; Seiliez, I.; Biga, P.; Gabillard, J.C. miR-210 expression is associated with methionine-induced differentiation of trout satellite cells. J. Exp. Biol. 2017, 220, 2932–2938. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Li, J.; Zhang, L.; Yu, C.; Lin, M.; Gao, F.; Zhou, G.; Zhang, Y.; Fan, Y.; Nuldnali, L. Effects of dietary energy sources on post mortem glycolysis, meat quality and muscle fibre type transformation of finishing pigs. PLoS ONE 2015, 10, e0131958. [Google Scholar] [CrossRef] [PubMed]
- Ostojic, S.M.; Stojanovic, M.D.; Hoffman, J.R. Six-week oral guanidinoacetic acid administration improves muscular performance in healthy volunteers. J. Investig. Med. 2015, 63, 942–946. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.S.; Shi, B.M.; Shan, A.S.; Zhang, Y.Y. Effects of guanidinoacetic acid on growth performance, meat quality and antioxidation in growing-finishing pigs. J. Anim. Vet. Adv. 2012, 11, 631–636. [Google Scholar] [CrossRef]
- Bates, S.; Bonetta, L.; Macallan, D.; Parry, D. Cdk6 (PLSTIRE) and CDK4 (PSK-J3) are a distinct subset of the cyclin-dependent kinases that associate with cyclin d1. Oncogene 1994, 9, 71–79. [Google Scholar] [PubMed]
- Datar, S.A.; Jacobs, H.W.; Af, D.L.C.; Lehner, C.F.; Edgar, B.A. The drosophila cyclin D-Cdk4 complex promotes cellular growth. EMBO J. 2014, 19, 4543–4554. [Google Scholar] [CrossRef] [PubMed]
- Halevy, O.; Novitch, B.G.; Spicer, D.B.; Skapek, S.X.; Rhee, J.; Hannon, G.J.; Beach, D.; Lassar, A.B. Correlation of terminal cell cycle arrest of skeletal muscle with induction of p21 by myod. Science 1995, 267, 1018–1021. [Google Scholar] [CrossRef] [PubMed]
- Ferri, P.; Barbieri, E.; Burattini, S.; Guescini, M.; D’Emilio, A.; Biagiotti, L.; Del, G.P.; De, L.A.; Stocchi, V.; Falcieri, E. Expression and subcellular localization of myogenic regulatory factors during the differentiation of skeletal muscle c2c12 myoblasts. J. Cell. Biochem. 2009, 108, 1302–1317. [Google Scholar] [CrossRef] [PubMed]
- Perry, R.L.; Rudnick, M.A. Molecular mechanisms regulating myogenic determination and differentiation. Front. Biosci. 2000, 5, D750–D767. [Google Scholar] [CrossRef] [PubMed]
- Williams, A.H.; Liu, N.; Rooij, E.V.; Olson, E.N. MicroRNA control of muscle development and disease. Curr. Opin. Cell. Biol. 2009, 21, 461–469. [Google Scholar] [CrossRef] [PubMed]
- Gracia, A.; Fernández-Quintela, A.; Miranda, J.; Eseberri, I.; González, M.; Portillo, M.P. Are mirna-103, miRNA-107 and miRNA-122 involved in the prevention of liver steatosis induced by resveratrol? Nutrients 2017, 9, 360. [Google Scholar] [CrossRef] [PubMed]
- Lim, E.; Lim, J.Y.; Kim, E.; Kim, Y.S.; Shin, J.H.; Seok, P.R.; Jung, S.; Yoo, S.H.; Kim, Y. Xylobiose, an alternative sweetener, ameliorates diabetes-related metabolic changes by regulating hepatic lipogenesis and miR-122a/33a in db/db mice. Nutrients 2016, 8, 791. [Google Scholar] [CrossRef] [PubMed]
- Lokireddy, S.; Bonala, S. Myostatin induces degradation of sarcomeric proteins through a Smad3 signaling mechanism during skeletal muscle wasting. Mol. Endocrinol. 2015, 29, 153. [Google Scholar] [CrossRef]
- Yamazaki, Y.; Kamei, Y.; Sugita, S.; Akaike, F.; Kanai, S.; Miura, S.; Hirata, Y.; Troen, B.R.; Kitamura, T.; Nishino, I. The cathepsin l gene is a direct target of foxo1 in skeletal muscle. Biochem. J. 2010, 427, 171–178. [Google Scholar] [CrossRef] [PubMed]
- Van Pilsum, J.F. Evidence for a dual role of creatine in the regulation of kidney transamidinase activities in the rat. J. Nutr. 1971, 101, 1085–1091. [Google Scholar] [CrossRef] [PubMed]
- Edison, E.E.; Brosnan, M.E.; Meyer, C.; Brosnan, J.T. Creatine synthesis: Production of guanidinoacetate by the rat and human kidney in vivo. Am. J. Physiol. Ren. 2007, 293, F1799–F1804. [Google Scholar] [CrossRef] [PubMed]
- Ostojic, S.M.; Ostojic, J. Human skeletal muscle contains no detectable guanidinoacetic acid. Appl. Physiol. Nutr. Metab. 2018, 43, 647–649. [Google Scholar] [CrossRef] [PubMed]
- Wyss, M.; Kaddurahdaouk, R. Creatine and creatinine metabolism. Physiol. Rev. 2000, 80, 1107–1213. [Google Scholar] [CrossRef] [PubMed]
- Daly, M.M. Guanidinoacetate methyltransferase activity in tissues and cultured cells. Arch. Biochem. Biophys. 1985, 236, 576–584. [Google Scholar] [CrossRef]
- Egerman, M.A.; Glass, D.J. Signaling pathways controlling skeletal muscle mass. Crit. Rev. Biochem. Mol. 2014, 49, 59–68. [Google Scholar] [CrossRef] [PubMed]
- Larsson, L. Histochemical characteristics of human skeletal muscle during aging. Acta Physiol. Scand. 2010, 117, 469–471. [Google Scholar] [CrossRef] [PubMed]
- Engel, W.K. Selective and nonselective susceptibility of muscle fiber types: A new approach to human neuromuscular diseases. Arch. Neurol. 1970, 22, 97–117. [Google Scholar] [CrossRef] [PubMed]
- Webster, C.; Silberstein, L.; Hays, A.P.; Blau, H.M. Fast muscle fibers are preferentially affected in duchenne muscular dystrophy. Cell 1988, 52, 503–513. [Google Scholar] [CrossRef]
- Weintraub, H. The MyoD family and myogenesis: Redundancy, networks, and thresholds. Cell 1993, 75, 1241–1244. [Google Scholar] [CrossRef]
- Mccarthy, J.; Esser, K. Microrna-1 and microrna-133a expression are decreased during skeletal muscle hypertrophy. J. Appl. Physiol. 2007, 102, 306–313. [Google Scholar] [CrossRef] [PubMed]
- Izumiya, Y.; Hopkins, T.; Morris, C.; Sato, K.; Zeng, L.; Viereck, J.; Hamilton, J.A.; Ouchi, N.; Lebrasseur, N.K.; Walsh, K. Fast/glycolytic muscle fiber growth reduces fat mass and improves metabolic parameters in obese mice. Cell. Metab. 2008, 7, 159–172. [Google Scholar] [CrossRef] [PubMed]
- Miyata, K.; Miyata, T.; Nakabayashi, K.; Okamura, K.; Naito, M.; Kawai, T.; Takada, S.; Kato, K.; Miyamoto, S.; Hata, K. DNA methylation analysis of human myoblasts during in vitro myogenic differentiation: De novo methylation of promoters of muscle-related genes and its involvement in transcriptional down-regulation. Hum. Mol. Genet. 2015, 24, 410–423. [Google Scholar] [CrossRef] [PubMed]
- Lee, E.J.; Malik, A.; Pokharel, S.; Ahmad, S.; Mir, B.A.; Cho, K.H.; Kim, J.; Kong, J.C.; Lee, D.M.; Chung, K.Y. Identification of genes differentially expressed in myogenin knock-down bovine muscle satellite cells during differentiation through RNA sequencing analysis. PLoS ONE 2014, 9, e92447. [Google Scholar] [CrossRef] [PubMed]
- Tan, L.C.H. Regulation of the follistatin gene by RSPO-LGR4 signaling via activation of the WNT/beta-catenin pathway in skeletal myogenesis. Mol. Cell. Biol. 2015, 34, 752–764. [Google Scholar]
- Xu, Q.; Wu, Z. The insulin-like growth factor-phosphatidylinositol 3-kinase-Akt signaling pathway regulates myogenin expression in normal myogenic cells but not in rhabdomyosarcoma-derived rd cells. J. Biol. Chem. 2000, 275, 36750–36757. [Google Scholar] [CrossRef] [PubMed]
- Long, Y.C.; Cheng, Z.; Copps, K.D.; White, M.F. Insulin receptor substrates Irs1 and Irs2 coordinate skeletal muscle growth and metabolism via the Akt and AMPK pathways. Mol. Cell. Biol. 2011, 31, 430–441. [Google Scholar] [CrossRef] [PubMed]
- Mounier, R.; Théret, M.; Lantier, L.; Foretz, M.; Viollet, B. Expanding roles for AMPK in skeletal muscle plasticity. Trends Endocrinol. Metab. 2015, 26, 275–286. [Google Scholar] [CrossRef] [PubMed]
- De Deyn, P.P.; Macdonald, R.L. Guanidino compounds that are increased in cerebrospinal fluid and brain of uremic patients inhibit GABA and glycine responses on mouse neurons in cell culture. Ann. Neurol. 1990, 28, 627–633. [Google Scholar] [CrossRef] [PubMed]
- Do, D.N.; Strathe, A.B.; Ostersen, T.; Jensen, J.; Mark, T.; Kadarmideen, H.N. Genome-wide association study reveals genetic architecture of eating behavior in pigs and its implications for humans obesity by comparative mapping. PLoS ONE 2013, 8, e71509. [Google Scholar] [CrossRef] [PubMed]
- Do, D.N.; Ostersen, T.; Strathe, A.B.; Mark, T.; Jensen, J.; Kadarmideen, H.N. Genome-wide association and systems genetic analyses of residual feed intake, daily feed consumption, backfat and weight gain in pigs. BMC Genet. 2014, 15, 27–42. [Google Scholar] [CrossRef] [PubMed]
- Cai, X.; Zhu, C.; Xu, Y.; Jing, Y.; Yuan, Y.; Wang, L.; Wang, S.; Zhu, X.; Gao, P.; Zhang, Y. Alpha-ketoglutarate promotes skeletal muscle hypertrophy and protein synthesis through Akt/mTOR signaling pathways. Sci. Rep. 2016, 6, 26802. [Google Scholar] [CrossRef] [PubMed]
- Senesi, P.; Luzi, L.; Montesano, A.; Mazzocchi, N.; Terruzzi, I. Betaine supplement enhances skeletal muscle differentiation in murine myoblasts via IGF-1 signaling activation. J. Transl. Med. 2013, 11, 174. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.Y.; Go, G.Y.; Vuong, T.A.; Kim, J.W.; Lee, S.; Jo, A.; An, J.M.; Kim, S.N.; Seo, D.W.; Kim, J.S. Black ginseng activates Akt signaling, thereby enhancing myoblast differentiation and myotube growth. J. Ginseng Res. 2018, 42, 116–121. [Google Scholar] [CrossRef] [PubMed]
- Hwang, S.Y.; Kang, Y.J.; Sung, B.; Kim, M.; Kim, D.H.; Lee, Y.; Yoo, M.A.; Kim, C.M.; Chung, H.Y.; Kim, N.D. Folic acid promotes the myogenic differentiation of C2C12 murine myoblasts through the Akt signaling pathway. Int. J. Med. 2015, 36, 1073–1080. [Google Scholar] [CrossRef] [PubMed]
- O’Neill, B.T.; Lauritzen, H.P.; Hirshman, M.F.; Smyth, G.; Goodyear, L.J.; Kahn, C.R. Differential role of insulin/IGF-1 receptor signaling in muscle growth and glucose homeostasis. Cell. Rep. 2015, 11, 1220–1235. [Google Scholar] [CrossRef] [PubMed]
- Aynsley-Green, A.; Alberti, K.G. In vivo stimulation of insulin secretion by guanidine derivatives in the rat. Horm. Metab. Res. 1974, 6, 115–120. [Google Scholar] [CrossRef] [PubMed]
- Umeki, D.; Ohnuki, Y.; Mototani, Y.; Shiozawa, K.; Fujita, T.; Nakamura, Y.; Saeki, Y.; Okumura, S. Effects of chronic Akt/mTOR inhibition by rapamycin on mechanical overload-induced hypertrophy and myosin heavy chain transition in masseter muscle. J. Pharmacol. Sci. 2013, 122, 278–288. [Google Scholar] [CrossRef] [PubMed]
- Gao, H.; Ao, M.; Wang, H.; Yu, L. Rapamycin represses myotube hypertrophy and preserves viability of C2C12 cells during myogenesis in vitro. Transplantation 2014, 98, 139–147. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Liu, W.; Nie, Y.; Qaher, M.; Horton, H.E.; Yue, F.; Asakura, A.; Kuang, S. Loss of myod promotes fate transdifferentiation of myoblasts into brown adipocytes. Ebiomedicine 2017, 16, 212–223. [Google Scholar] [CrossRef] [PubMed]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. Edger: A bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef] [PubMed]
- Agarwal, V.; Bell, G.W.; Nam, J.W.; Bartel, D.P. Predicting effective microRNA target sites in mammalian mrnas. Life Sci. 2015, 4, e05005. [Google Scholar] [CrossRef] [PubMed]
- Huang, D.; Sherman, B.; Lempicki, R. Systematic and integrative analysis of large gene lists using david bioinformatics resources. Nat. Protoc. 2009, 4, 44–57. [Google Scholar] [CrossRef] [PubMed]
- Wong, N.; Wang, X. Mirdb: An online resource for microrna target prediction and functional annotations. Nucleic Acids Res. 2015, 43, 146–152. [Google Scholar] [CrossRef] [PubMed]
- Rehmsmeier, M.; Steffen, P.; Hochsmann, M.; Giegerich, R. Fast and effective prediction of microrna/target duplexes. RNA 2004, 10, 1507–1517. [Google Scholar] [CrossRef] [PubMed]

© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, Y.; Ma, J.; Qiu, W.; Zhang, J.; Feng, S.; Zhou, X.; Wang, X.; Jin, L.; Long, K.; Liu, L.; et al. Guanidinoacetic Acid Regulates Myogenic Differentiation and Muscle Growth Through miR-133a-3p and miR-1a-3p Co-mediated Akt/mTOR/S6K Signaling Pathway. Int. J. Mol. Sci. 2018, 19, 2837. https://doi.org/10.3390/ijms19092837
Wang Y, Ma J, Qiu W, Zhang J, Feng S, Zhou X, Wang X, Jin L, Long K, Liu L, et al. Guanidinoacetic Acid Regulates Myogenic Differentiation and Muscle Growth Through miR-133a-3p and miR-1a-3p Co-mediated Akt/mTOR/S6K Signaling Pathway. International Journal of Molecular Sciences. 2018; 19(9):2837. https://doi.org/10.3390/ijms19092837
Chicago/Turabian StyleWang, Yujie, Jideng Ma, Wanling Qiu, Jinwei Zhang, Siyuan Feng, Xiankun Zhou, Xun Wang, Long Jin, Keren Long, Lingyan Liu, and et al. 2018. "Guanidinoacetic Acid Regulates Myogenic Differentiation and Muscle Growth Through miR-133a-3p and miR-1a-3p Co-mediated Akt/mTOR/S6K Signaling Pathway" International Journal of Molecular Sciences 19, no. 9: 2837. https://doi.org/10.3390/ijms19092837
APA StyleWang, Y., Ma, J., Qiu, W., Zhang, J., Feng, S., Zhou, X., Wang, X., Jin, L., Long, K., Liu, L., Xiao, W., Tang, Q., Zhu, L., Jiang, Y., Li, X., & Li, M. (2018). Guanidinoacetic Acid Regulates Myogenic Differentiation and Muscle Growth Through miR-133a-3p and miR-1a-3p Co-mediated Akt/mTOR/S6K Signaling Pathway. International Journal of Molecular Sciences, 19(9), 2837. https://doi.org/10.3390/ijms19092837