A G-Quadruplex Structure in the Promoter Region of CLIC4 Functions as a Regulatory Element for Gene Expression
Abstract
1. Introduction
2. Results
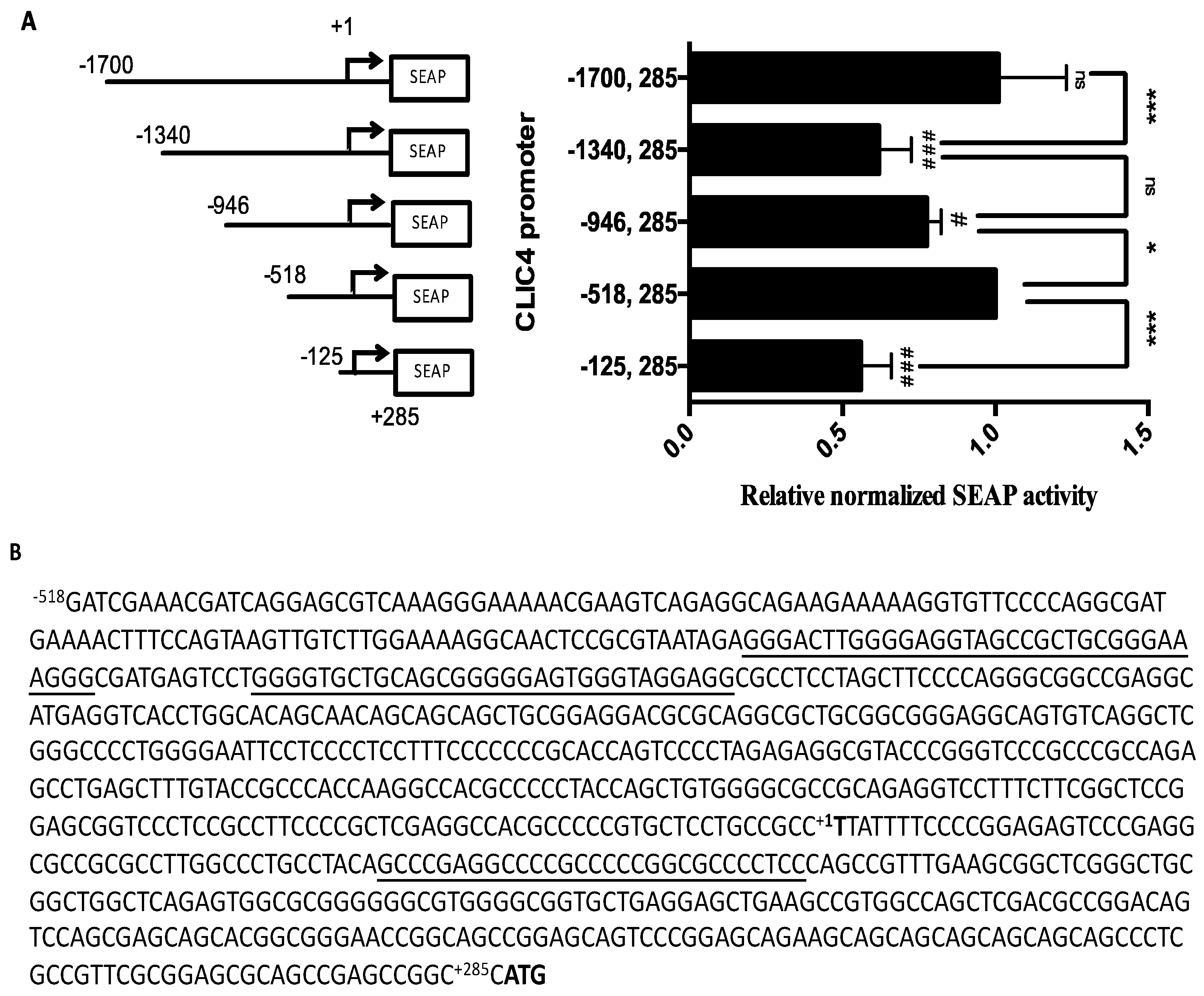
2.1. CLIC4 Promoter Analysis
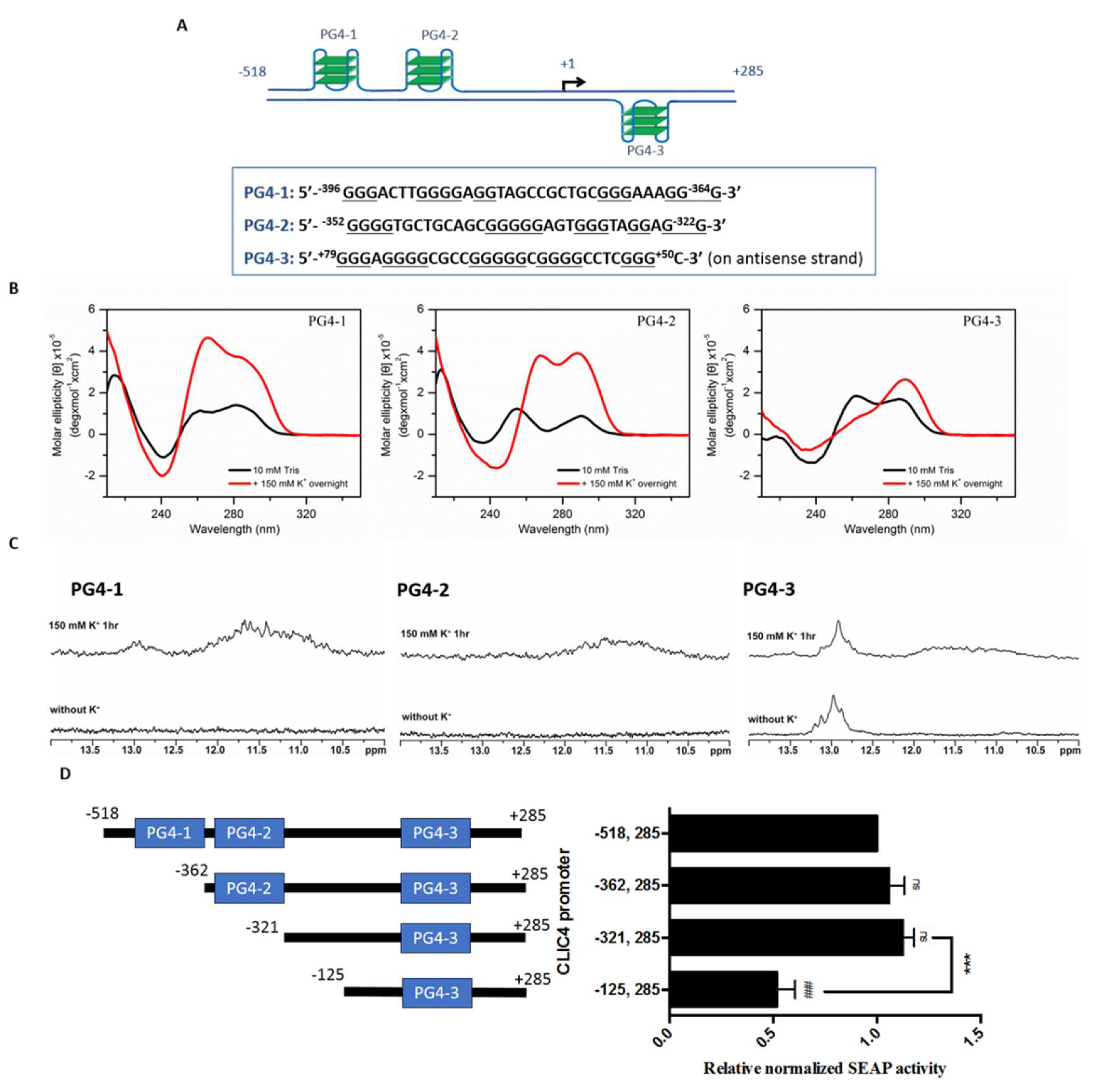
2.2. Putative G-Quadruplexes in the CLIC4 Promoter
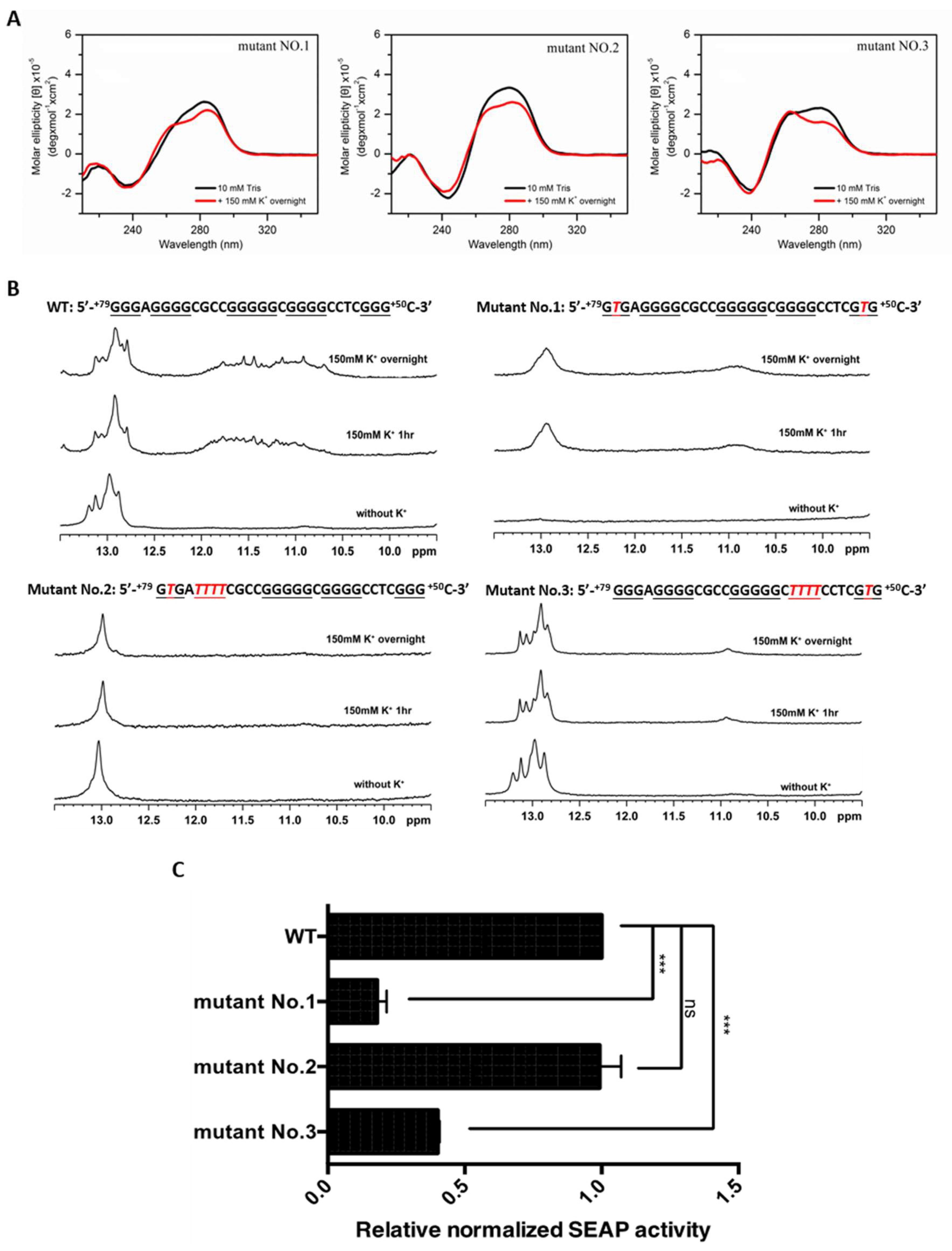
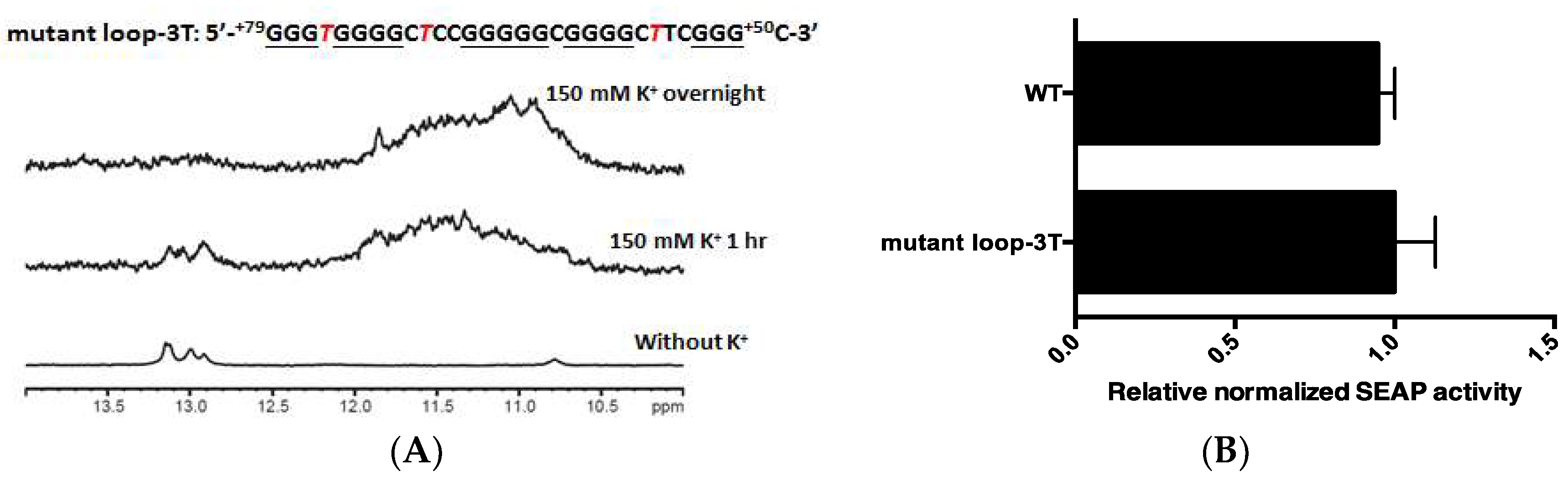
2.3. PG4-3 Is Involved in Regulating CLIC4 Transcription
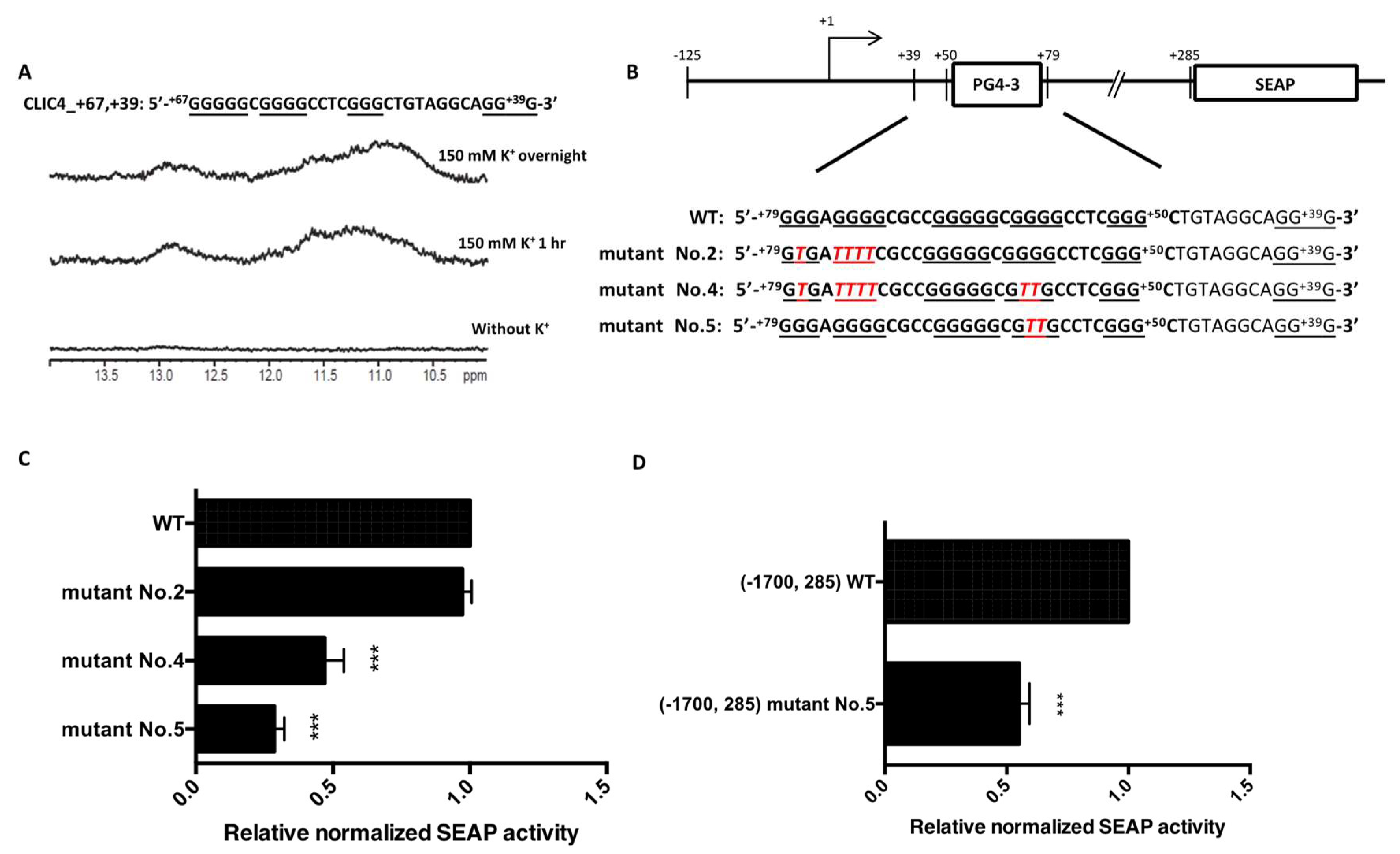
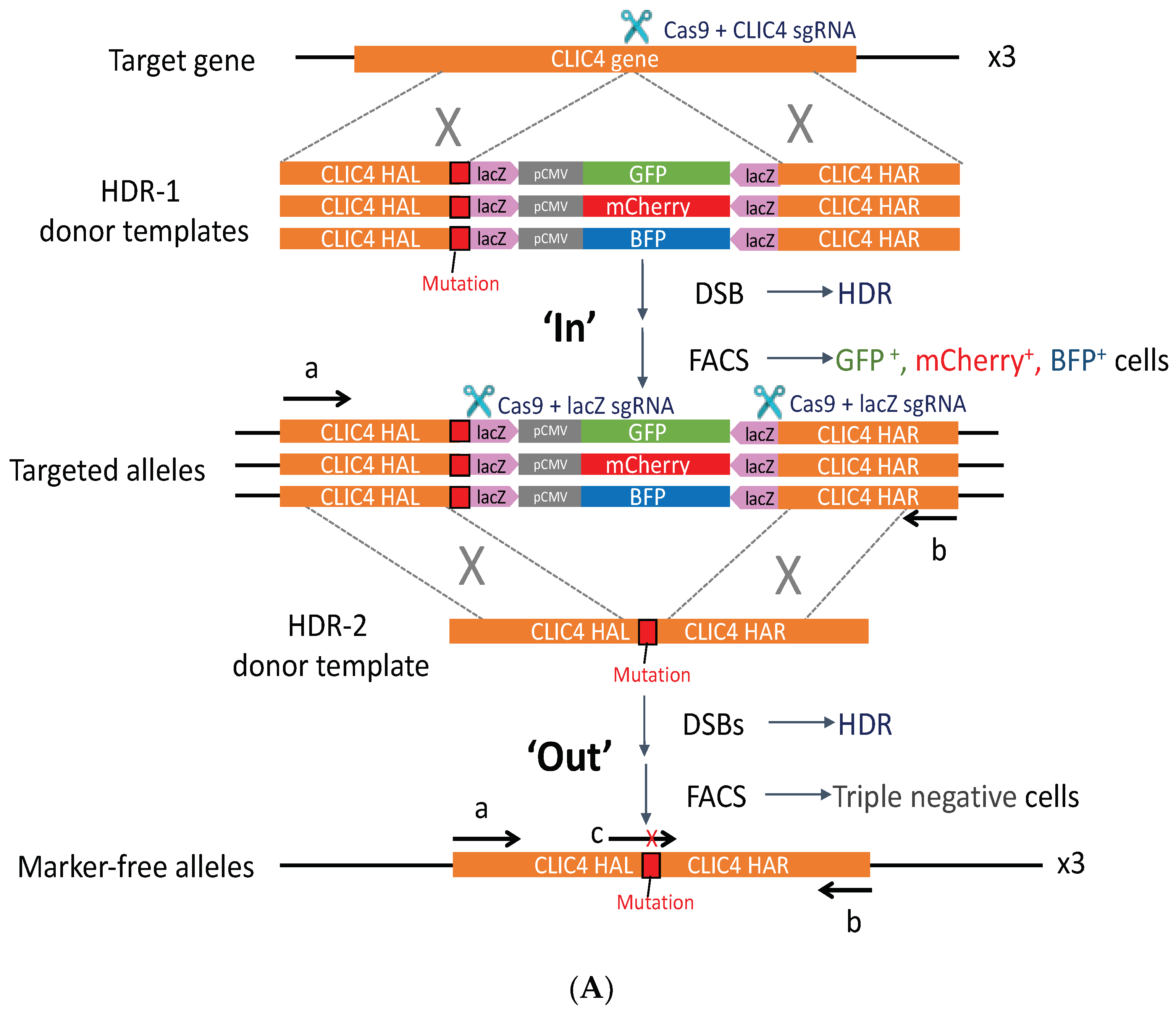
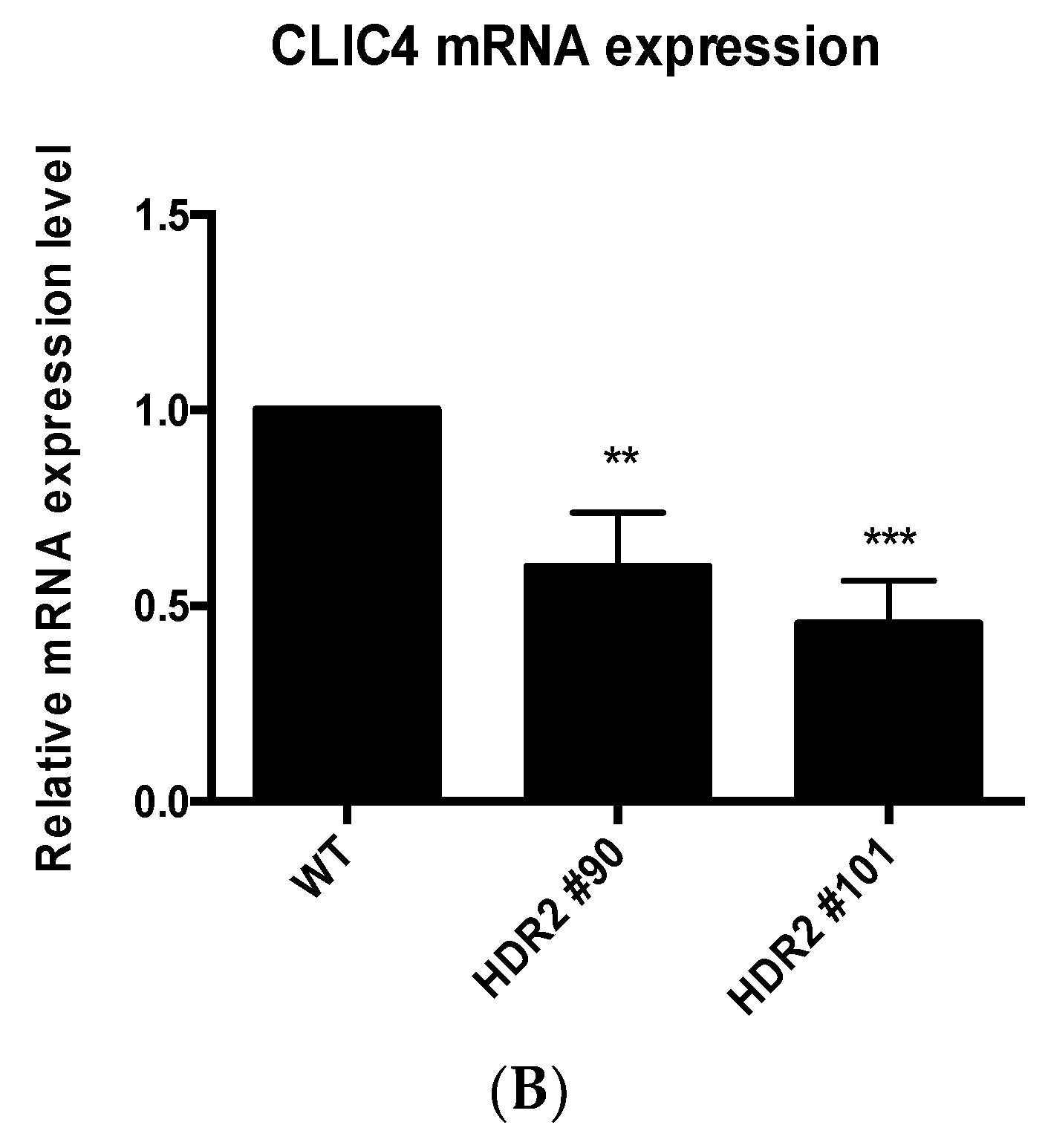
2.4. PG4-3 Acts as a Regulatory Element in the CLIC4 Promoter Region of the Cell Chromosome
3. Discussion
4. Materials and Methods
4.1. Cell Culturing
4.2. Circular Dichroism (CD) Spectroscopy
4.3. NMR
4.4. Reporter Assay
4.5. CRISPR/Cas9
4.6. Real-Time PCR Analysis
4.7. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Berryman, M.A.; Goldenring, J.R. CLIC4 is enriched at cell-cell junctions and colocalizes with AKAP350 at the centrosome and midbody of cultured mammalian cells. Cell Motil. Cytoskel. 2003, 56, 159–172. [Google Scholar] [CrossRef] [PubMed]
- Suh, K.S.; Mutoh, M.; Nagashima, K.; Fernandez-Salas, E.; Edwards, L.E.; Hayes, D.D.; Crutchley, J.M.; Marin, K.G.; Dumont, R.A.; Levy, J.M.; et al. The organellular chloride channel protein CLIC4/mtCLIC translocates to the nucleus in response to cellular stress and accelerates apoptosis. J. Biol. Chem. 2004, 279, 4632–4641. [Google Scholar] [CrossRef] [PubMed]
- Littler, D.R.; Harrop, S.J.; Goodchild, S.C.; Phang, J.M.; Mynott, A.V.; Jiang, L.; Valenzuela, S.M.; Mazzanti, M.; Brown, L.J.; Breit, S.N.; et al. The enigma of the CLIC proteins: Ion channels, redox proteins, enzymes, scaffolding proteins? FEBS Lett. 2010, 584, 2093–2101. [Google Scholar] [CrossRef] [PubMed]
- Shukla, A.; Malik, M.; Cataisson, C.; Ho, Y.; Friesen, T.; Suh, K.S.; Yuspa, S.H. TGF-beta signalling is regulated by Schnurri-2-dependent nuclear translocation of CLIC4 and consequent stabilization of phospho-Smad2 and 3. Nat. Cell Biol. 2009, 11, 777–784. [Google Scholar] [CrossRef] [PubMed]
- Suh, K.S.; Crutchley, J.M.; Koochek, A.; Ryscavage, A.; Bhat, K.; Tanaka, T.; Oshima, A.; Fitzgerald, P.; Yuspa, S.H. Reciprocal modifications of CLIC4 in tumor epithelium and stroma mark malignant progression of multiple human cancers. Clin. Cancer Res. 2007, 13, 121–131. [Google Scholar] [CrossRef] [PubMed]
- Suh, K.S.; Malik, M.; Shukla, A.; Ryscavage, A.; Wright, L.; Jividen, K.; Crutchley, J.M.; Dumont, R.A.; Fernandez-Salas, E.; Webster, J.D.; et al. CLIC4 is a tumor suppressor for cutaneous squamous cell cancer. Carcinogenesis 2012, 33, 986–995. [Google Scholar] [CrossRef] [PubMed]
- Ronnov-Jessen, L.; Villadsen, R.; Edwards, J.C.; Petersen, O.W. Differential expression of a chloride intracellular channel gene, CLIC4, in transforming growth factor-beta1-mediated conversion of fibroblasts to myofibroblasts. Am. J. Pathol. 2002, 161, 471–480. [Google Scholar] [CrossRef]
- Todeschini, A.L.; Georges, A.; Veitia, R.A. Transcription factors: Specific DNA binding and specific gene regulation. Trends Genet. 2014, 30, 211–219. [Google Scholar] [CrossRef] [PubMed]
- Gibney, E.R.; Nolan, C.M. Epigenetics and gene expression. Heredity 2010, 105, 4–13. [Google Scholar] [CrossRef] [PubMed]
- Edwards, J.C. A novel p64-related Cl−channel: Subcellular distribution and nephron segment-specific expression. Am. J. Physiol. 1999, 276, F398–F408. [Google Scholar] [CrossRef] [PubMed]
- Chuang, J.Z.; Milner, T.A.; Zhu, M.; Sung, C.H. A 29 kDa intracellular chloride channel p64H1 is associated with large dense-core vesicles in rat hippocampal neurons. J. Neurosci. 1999, 19, 2919–2928. [Google Scholar] [CrossRef] [PubMed]
- Boyer, L.A.; Lee, T.I.; Cole, M.F.; Johnstone, S.E.; Levine, S.S.; Zucker, J.P.; Guenther, M.G.; Kumar, R.M.; Murray, H.L.; Jenner, R.G.; et al. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell 2005, 122, 947–956. [Google Scholar] [CrossRef] [PubMed]
- Fernandez-Salas, E.; Suh, K.S.; Speransky, V.V.; Bowers, W.L.; Levy, J.M.; Adams, T.; Pathak, K.R.; Edwards, L.E.; Hayes, D.D.; Cheng, C.; et al. mtCLIC/CLIC4, an organellular chloride channel protein, is increased by DNA damage and participates in the apoptotic response to p53. Mol. Cell. Biol. 2002, 22, 3610–3620. [Google Scholar] [CrossRef] [PubMed]
- Shiio, Y.; Suh, K.S.; Lee, H.; Yuspa, S.H.; Eisenman, R.N.; Aebersold, R. Quantitative proteomic analysis of myc-induced apoptosis: A direct role for Myc induction of the mitochondrial chloride ion channel, mtCLIC/CLIC4. J. Biol. Chem. 2006, 281, 2750–2756. [Google Scholar] [CrossRef] [PubMed]
- Suh, K.S.; Mutoh, M.; Mutoh, T.; Li, L.; Ryscavage, A.; Crutchley, J.M.; Dumont, R.A.; Cheng, C.; Yuspa, S.H. CLIC4 mediates and is required for Ca2+-induced keratinocyte differentiation. J. Cell Sci. 2007, 120, 2631–2640. [Google Scholar] [CrossRef] [PubMed]
- Burge, S.; Parkinson, G.N.; Hazel, P.; Todd, A.K.; Neidle, S. Quadruplex DNA: Sequence, topology and structure. Nucleic Acids Res. 2006, 34, 5402–5415. [Google Scholar] [CrossRef] [PubMed]
- Gellert, M.; Lipsett, M.N.; Davies, D.R. Helix formation by guanylic acid. Proc. Natl. Acad. Sci. USA 1962, 48, 2013–2018. [Google Scholar] [CrossRef] [PubMed]
- Henderson, E.; Hardin, C.C.; Walk, S.K.; Tinoco, I.; Blackburn, E.H. Telomeric DNA oligonucleotides form novel intramolecular structures containing guanine·guanine base pairs. Cell 1987, 51, 899–908. [Google Scholar] [CrossRef]
- Biffi, G.; Tannahill, D.; McCafferty, J.; Balasubramanian, S. Quantitative visualization of DNA G-quadruplex structures in human cells. Nat. Chem. 2013, 5, 182–186. [Google Scholar] [CrossRef] [PubMed]
- Tseng, T.Y.; Chien, C.H.; Chu, J.F.; Huang, W.C.; Lin, M.Y.; Chang, C.C.; Chang, T.C. Fluorescent probe for visualizing guanine-quadruplex DNA by fluorescence lifetime imaging microscopy. J. Biomed. Opt. 2013, 18, 101309. [Google Scholar] [CrossRef] [PubMed]
- Du, Z.; Zhao, Y.; Li, N. Genome-wide colonization of gene regulatory elements by G4 DNA motifs. Nucleic Acids Res. 2009, 37, 6784–6798. [Google Scholar] [CrossRef] [PubMed]
- Qin, Y.; Hurley, L.H. Structures, folding patterns, and functions of intramolecular DNA G-quadruplexes found in eukaryotic promoter regions. Biochimie 2008, 90, 1149–1171. [Google Scholar] [CrossRef] [PubMed]
- Yang, D.; Hurley, L.H. Structure of the biologically relevant G-quadruplex in the c-MYC promoter. Nucleosides Nucleotides Nucleic Acids 2006, 25, 951–968. [Google Scholar] [CrossRef] [PubMed]
- Paramasivam, M.; Membrino, A.; Cogoi, S.; Fukuda, H.; Nakagama, H.; Xodo, L.E. Protein hnRNP A1 and its derivative Up1 unfold quadruplex DNA in the human KRAS promoter: Implications for transcription. Nucleic Acids Res. 2009, 37, 2841–2853. [Google Scholar] [CrossRef] [PubMed]
- Qin, Y.; Rezler, E.M.; Gokhale, V.; Sun, D.; Hurley, L.H. Characterization of the G-quadruplexes in the duplex nuclease hypersensitive element of the PDGF-A promoter and modulation of PDGF-A promoter activity by TMPyP4. Nucleic Acids Res. 2007, 35, 7698–7713. [Google Scholar] [CrossRef] [PubMed]
- Dai, J.; Dexheimer, T.S.; Chen, D.; Carver, M.; Ambrus, A.; Jones, R.A.; Yang, D. An intramolecular G-quadruplex structure with mixed parallel/antiparallel G-strands formed in the human BCL-2 promoter region in solution. J. Am. Chem. Soc. 2006, 128, 1096–1098. [Google Scholar] [CrossRef] [PubMed]
- Bochman, M.L.; Paeschke, K.; Zakian, V.A. DNA secondary structures: Stability and function of G-quadruplex structures. Nat. Rev. Genet. 2012, 13, 770–780. [Google Scholar] [CrossRef] [PubMed]
- Kikin, O.; D’Antonio, L.; Bagga, P.S. QGRS Mapper: A web-based server for predicting G-quadruplexes in nucleotide sequences. Nucleic Acids Res. 2006, 34, W676–W682. [Google Scholar] [CrossRef] [PubMed]
- Cogoi, S.; Shchekotikhin, A.E.; Xodo, L.E. HRAS is silenced by two neighboring G-quadruplexes and activated by MAZ, a zinc-finger transcription factor with DNA unfolding property. Nucleic Acids Res. 2014, 42, 8379–8388. [Google Scholar] [CrossRef] [PubMed]
- Raiber, E.A.; Kranaster, R.; Lam, E.; Nikan, M.; Balasubramanian, S. A non-canonical DNA structure is a binding motif for the transcription factor SP1 in vitro. Nucleic Acids Res. 2012, 40, 1499–1508. [Google Scholar] [CrossRef] [PubMed]
- Xi, L.; Schmidt, J.C.; Zaug, A.J.; Ascarrunz, D.R.; Cech, T.R. A novel two-step genome editing strategy with CRISPR-Cas9 provides new insights into telomerase action and TERT gene expression. Genome Biol. 2015, 16, 231. [Google Scholar] [CrossRef] [PubMed]
- Hansel-Hertsch, R.; Beraldi, D.; Lensing, S.V.; Marsico, G.; Zyner, K.; Parry, A.; Di Antonio, M.; Pike, J.; Kimura, H.; Narita, M.; et al. G-quadruplex structures mark human regulatory chromatin. Nat. Genet. 2016, 48, 1267–1272. [Google Scholar] [CrossRef] [PubMed]
- Mladenova, V.; Mladenov, E.; Russev, G. Organization of Plasmid DNA into Nucleosome-Like Structures after Transfection in Eukaryotic Cells. Biotechnol. Biotechnol. Equip. 2009, 23, 1044–1047. [Google Scholar] [CrossRef]
- Fleming, A.M.; Zhou, J.; Wallace, S.S.; Burrows, C.J. A Role for the Fifth G-Track in G-Quadruplex Forming Oncogene Promoter Sequences during Oxidative Stress: Do These “Spare Tires” Have an Evolved Function? ACS Cent. Sci. 2015, 1, 226–233. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Fleming, A.M.; Averill, A.M.; Burrows, C.J.; Wallace, S.S. The NEIL glycosylases remove oxidized guanine lesions from telomeric and promoter quadruplex DNA structures. Nucleic Acids Res. 2015, 43, 4039–4054. [Google Scholar] [CrossRef] [PubMed]
- Fleming, A.M.; Ding, Y.; Burrows, C.J. Oxidative DNA damage is epigenetic by regulating gene transcription via base excision repair. Proc. Natl. Acad. Sci. USA 2017, 114, 2604–2609. [Google Scholar] [CrossRef] [PubMed]
- Chiang, P.C.; Chou, R.H.; Chien, H.F.; Tsai, T.; Chen, C.T. Chloride intracellular channel 4 involves in the reduced invasiveness of cancer cells treated by photodynamic therapy. Lasers Surg. Med. 2013, 45, 38–47. [Google Scholar] [CrossRef] [PubMed]
- Yao, Q.; Qu, X.; Yang, Q.; Wei, M.; Kong, B. CLIC4 mediates TGF-beta1-induced fibroblast-to-myofibroblast transdifferentiation in ovarian cancer. Oncol. Rep. 2009, 22, 541–548. [Google Scholar] [PubMed]
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Huang, M.-C.; Chu, I.-T.; Wang, Z.-F.; Lin, S.; Chang, T.-C.; Chen, C.-T. A G-Quadruplex Structure in the Promoter Region of CLIC4 Functions as a Regulatory Element for Gene Expression. Int. J. Mol. Sci. 2018, 19, 2678. https://doi.org/10.3390/ijms19092678
Huang M-C, Chu I-T, Wang Z-F, Lin S, Chang T-C, Chen C-T. A G-Quadruplex Structure in the Promoter Region of CLIC4 Functions as a Regulatory Element for Gene Expression. International Journal of Molecular Sciences. 2018; 19(9):2678. https://doi.org/10.3390/ijms19092678
Chicago/Turabian StyleHuang, Mu-Ching, I-Te Chu, Zi-Fu Wang, Steven Lin, Ta-Chau Chang, and Chin-Tin Chen. 2018. "A G-Quadruplex Structure in the Promoter Region of CLIC4 Functions as a Regulatory Element for Gene Expression" International Journal of Molecular Sciences 19, no. 9: 2678. https://doi.org/10.3390/ijms19092678
APA StyleHuang, M.-C., Chu, I.-T., Wang, Z.-F., Lin, S., Chang, T.-C., & Chen, C.-T. (2018). A G-Quadruplex Structure in the Promoter Region of CLIC4 Functions as a Regulatory Element for Gene Expression. International Journal of Molecular Sciences, 19(9), 2678. https://doi.org/10.3390/ijms19092678



