Diagnostic and Prognostic Implications of FGFR3high/Ki67high Papillary Bladder Cancers
Abstract
1. Introduction
2. Results
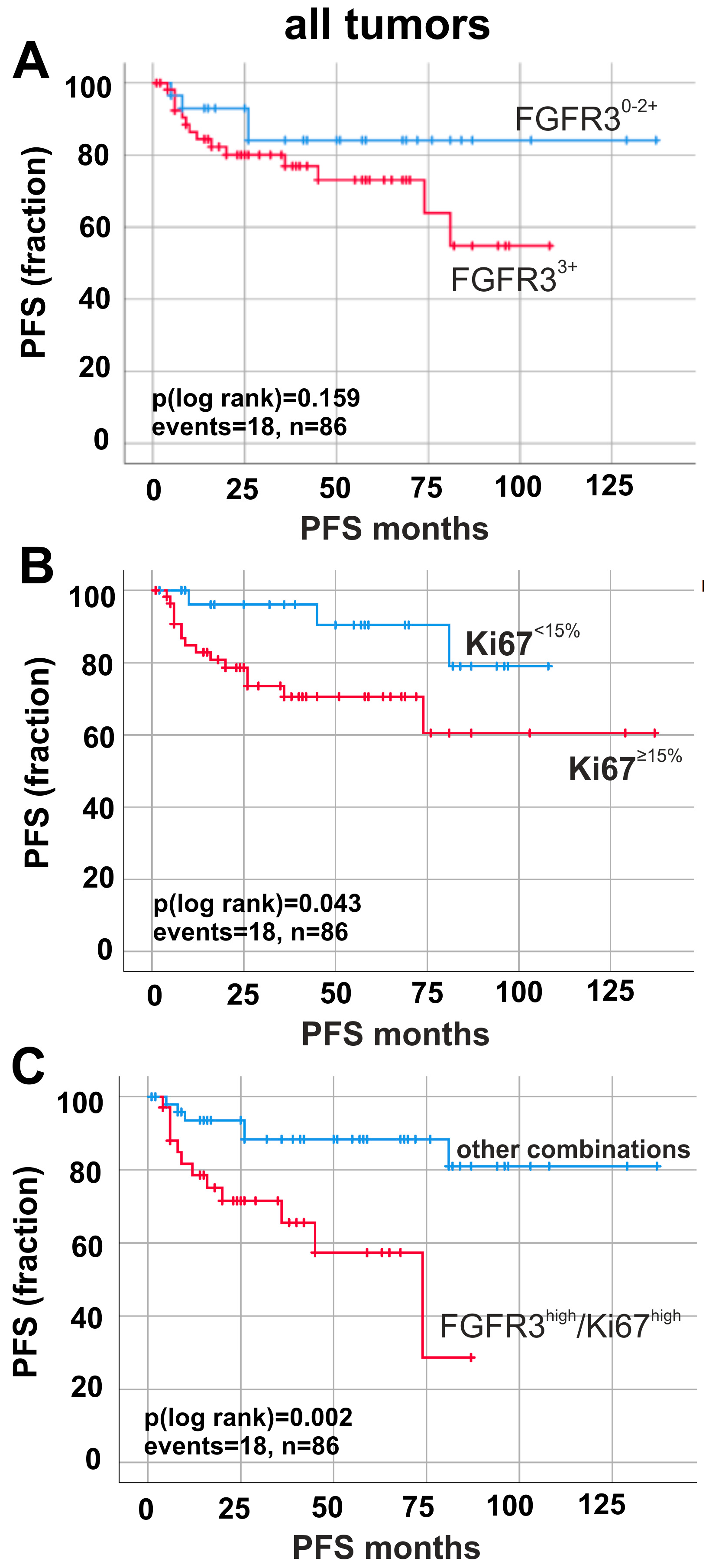
2.1. Immunohistochemical Combination of Ki67-Index and FGFR3 Levels Defines Worse Patients’ Outcome
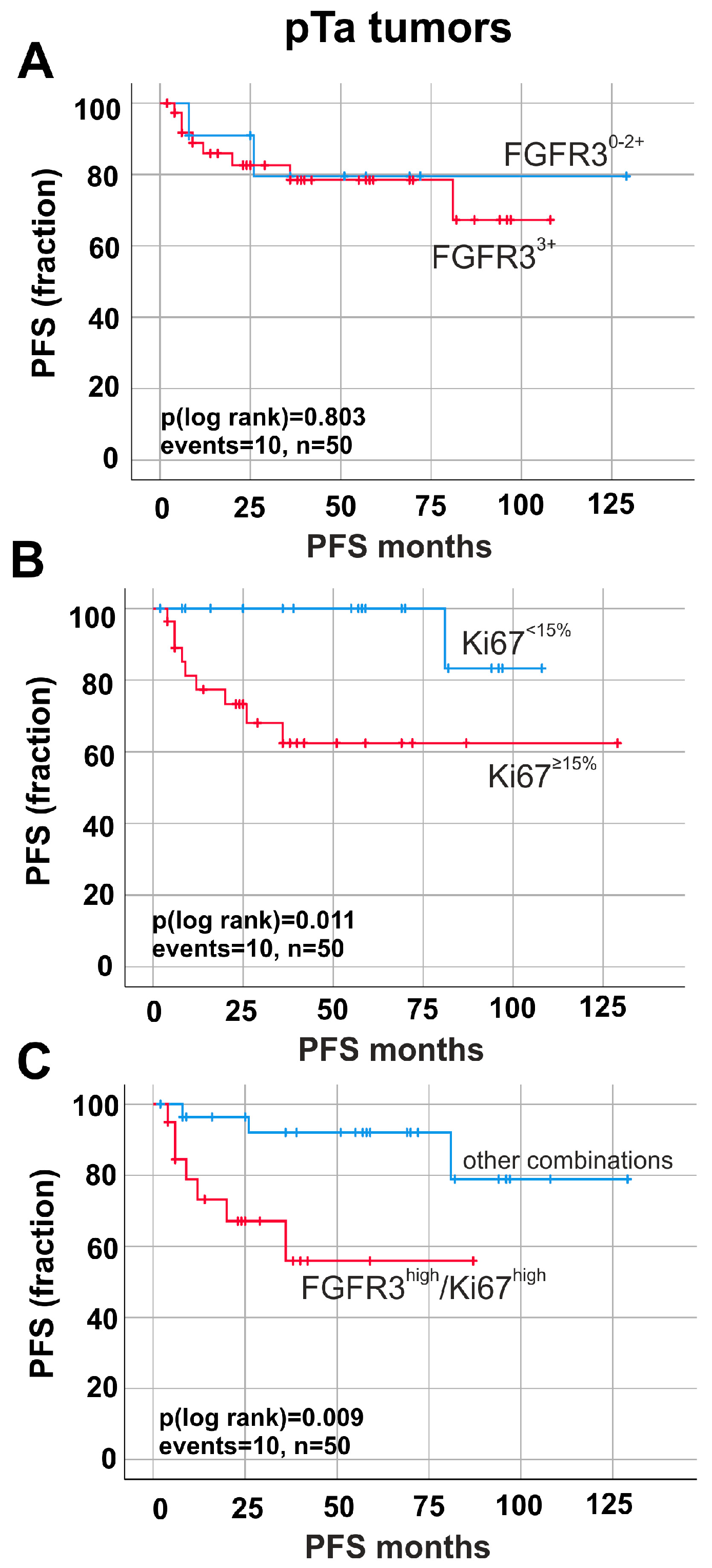
2.2. Prognostic Impact of Ki67-Index and FGFR3 Overexpression in Papillary Non-Invasive (pTa) Tumors
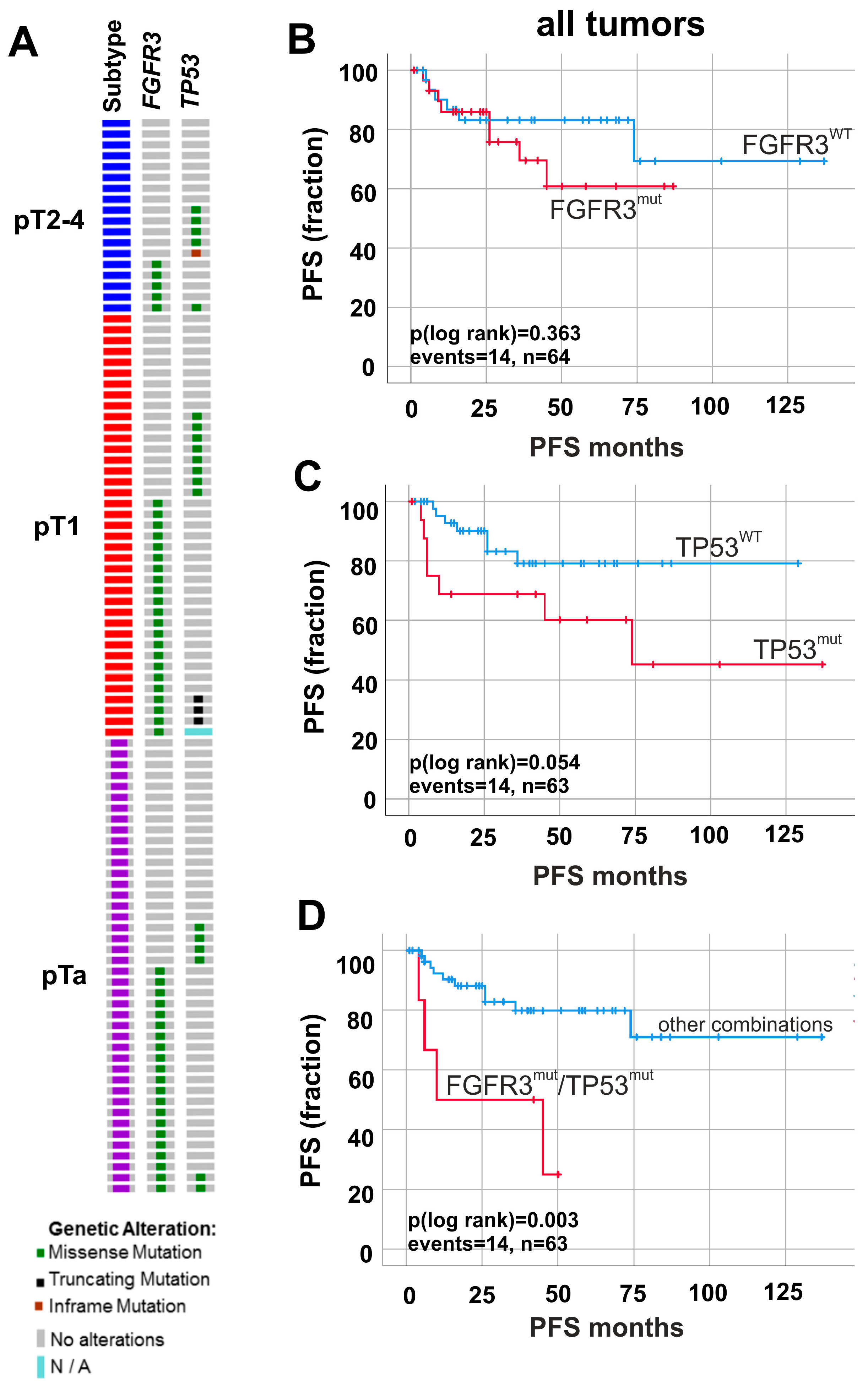
2.3. Altered Molecular FGFR3/TP53 Status Predicts Worse Patients’ Survival
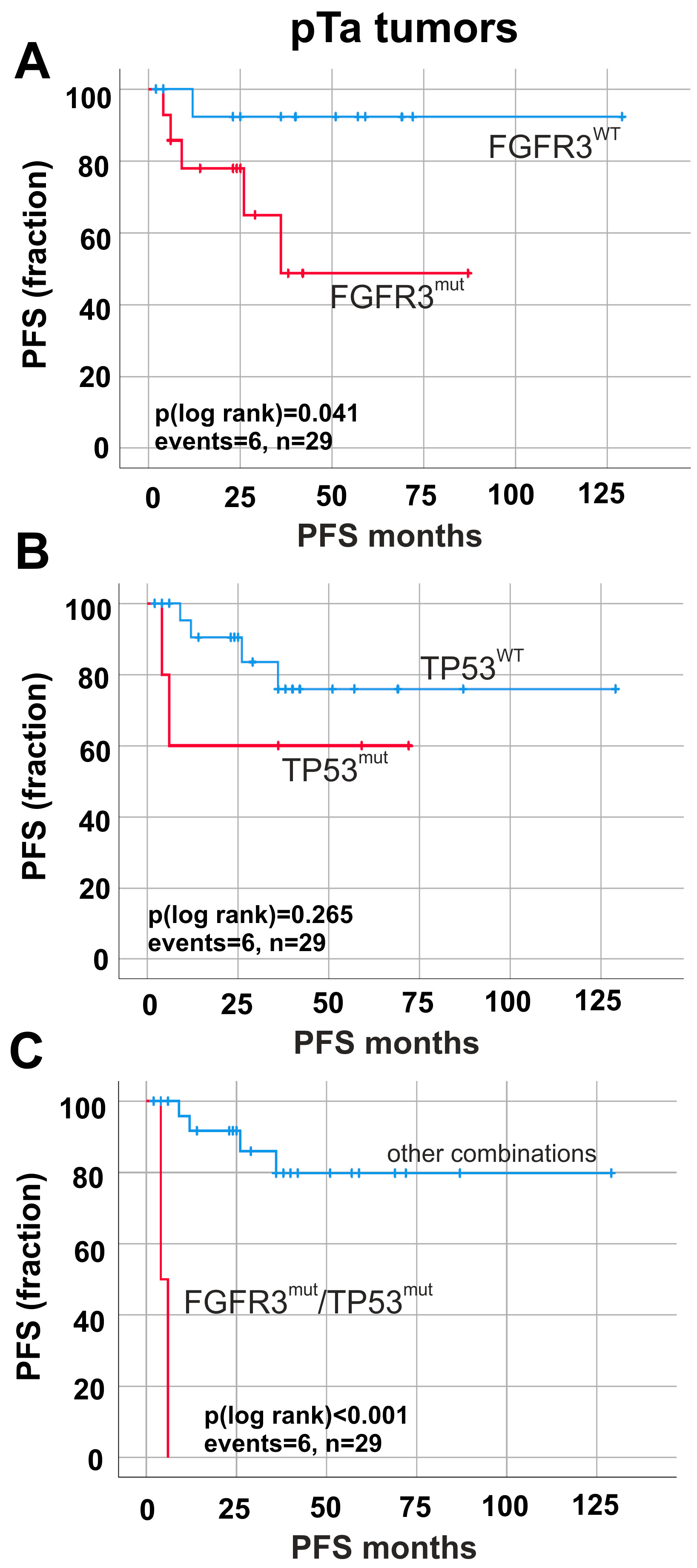
2.4. Prognostic Impact of FGFR3 and TP53 Mutations in Papillary Non-Invasive (pTa) Tumors
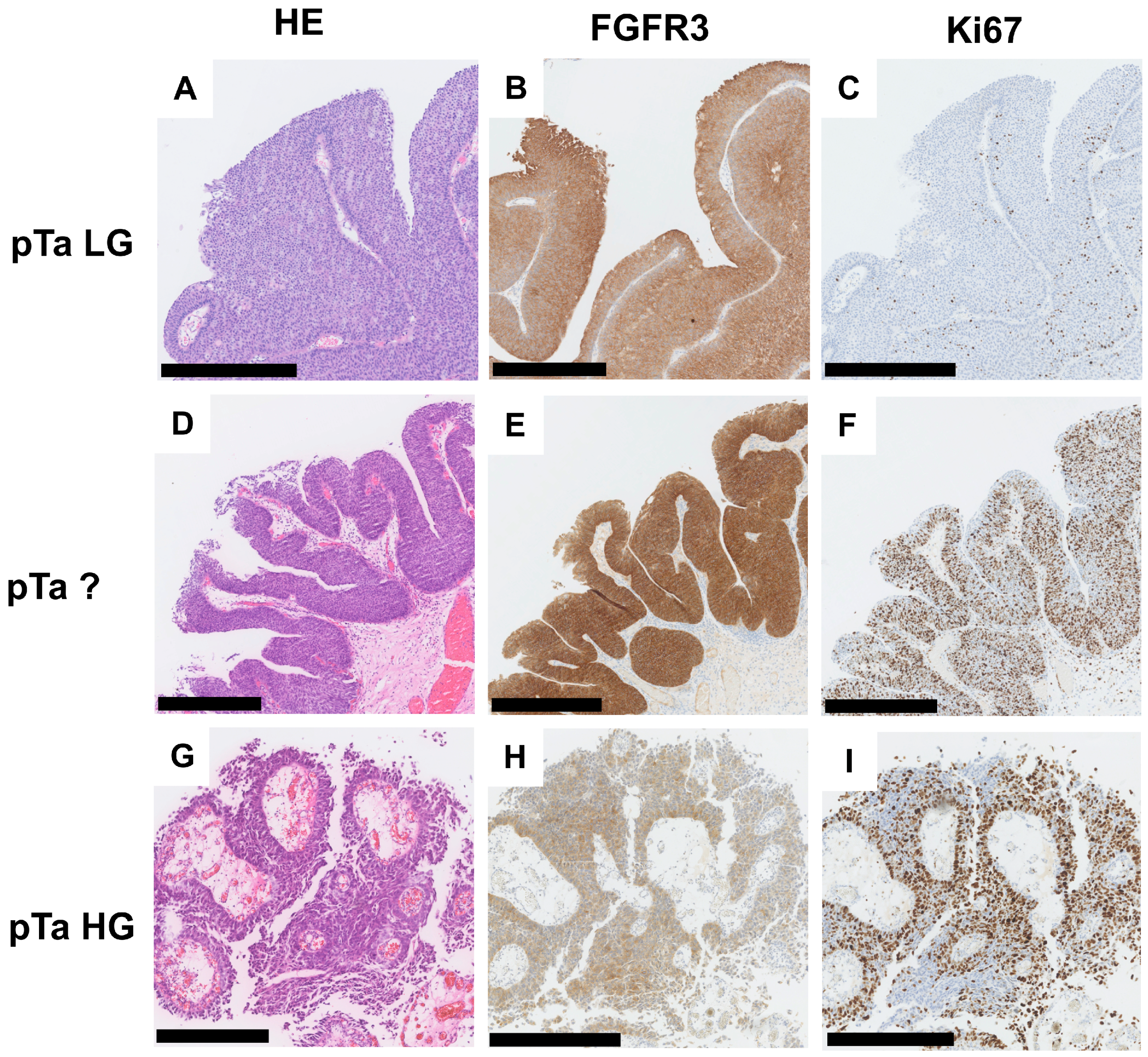
2.5. FGFR3high/Ki67high Tumors Define a Subset of pTa Tumors Including Lesions with Molecular FGFR3 Pathway Activation
3. Discussion
4. Materials and Methods
4.1. Patient Samples, Tissue Microarrays and DNA
4.2. Immunohistochemistry
4.3. Fluorescence In Situ Hybridization
4.4. Sanger Sequencing
4.5. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer statistics. CA Cancer J. Clin. 2018, 68, 7–30. [Google Scholar] [CrossRef]
- Van Rhijn, B.W.; Lurkin, I.; Radvanyi, F.; Kirkels, W.J.; Van der Kwast, T.H.; Zwarthoff, E.C. The fibroblast growth factor receptor 3 (FGFR3) mutation is a strong indicator of superficial bladder cancer with low recurrence rate. Cancer Res. 2001, 61, 1265–1268. [Google Scholar] [PubMed]
- Billerey, C.; Chopin, D.; Aubriot-Lorton, M.H.; Ricol, D.; Gil Diez de Medina, S.; van Rhijn, B.; Bralet, M.P.; Lefrere-Belda, M.A.; Lahaye, J.B.; Abbou, C.C.; et al. Frequent FGFR3 mutations in papillary non-invasive bladder (pTa) tumors. Am. J. Pathol. 2001, 158, 1955–1959. [Google Scholar] [CrossRef]
- Hernandez, S.; Lopez-Knowles, E.; Lloreta, J.; Kogevinas, M.; Jaramillo, R.; Amoros, A.; Tardón, A.; García-Closas, R.; Serra, C.; Carrato, A.; et al. FGFR3 and Tp53 mutations in T1G3 transitional bladder carcinomas: Independent distribution and lack of association with prognosis. Clin. Cancer. Res. 2005, 11, 5444–5450. [Google Scholar] [CrossRef] [PubMed]
- Neuzillet, Y.; van Rhijn, B.W.; Prigoda, N.L.; Bapat, B.; Liu, L.; Bostrom, P.J.; Fleshner, N.E.; Gallie, B.L.; Zlotta, A.R.; Jewett, M.A.; et al. FGFR3 mutations, but not FGFR3 expression and FGFR3 copy-number variations, are associated with favourable non-muscle invasive bladder cancer. Virchows. Arch. 2014, 465, 207–213. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.R. Urothelial tumorigenesis: A tale of divergent pathways. Nat. Rev. Cancer. 2005, 5, 713–725. [Google Scholar] [CrossRef] [PubMed]
- Knowles, M.A.; Hurst, C.D. Molecular biology of bladder cancer: New insights into pathogenesis and clinical diversity. Nat. Rev. Cancer 2015, 15, 25–41. [Google Scholar] [CrossRef] [PubMed]
- Brierley, J.; Gospodarowicz, M.K.; Wittekind, C. TNM Classification of Malignant Tumours, 8th ed.; John Wiley & Sons Inc.: Chichester, UK; Hoboken, NJ, USA, 2017; ISBN 9781119263579. [Google Scholar]
- Montironi, R.; Lopez-Beltran, A.; Scarpelli, M.; Mazzucchelli, R.; Cheng, L. Morphological classification and definition of benign, preneoplastic and non-invasive neoplastic lesions of the urinary bladder. Histopathology 2008, 53, 621–633. [Google Scholar] [CrossRef] [PubMed]
- Moch, H.; Humphrey, P.A.; Ulbright, T.M.; Reuter, V.E. International Agency for Research on Cancer. WHO Classification of Tumours of the Urinary System and Male Genital Organs, 4th ed.; International Agency for Research on Cancer: Lyon, France, 2016; ISBN 9789283224372. [Google Scholar]
- Cina, S.J.; Lancaster-Weiss, K.J.; Lecksell, K.; Epstein, J.I. Correlation of Ki-67 and p53 with the new World Health Organization/International Society of Urological Pathology Classification System for Urothelial Neoplasia. Arch. Pathol. Lab. Med. 2001, 125, 646–651. [Google Scholar] [CrossRef] [PubMed]
- Hentic, O.; Couvelard, A.; Rebours, V.; Zappa, M.; Dokmak, S.; Hammel, P.; Maire, F.; O’Toole, D.; Lévy, P.; Sauvanet, A.; et al. Ki-67 index, tumor differentiation, and extent of liver involvement are independent prognostic factors in patients with liver metastases of digestive endocrine carcinomas. Endocr. Relat. Cancer 2011, 18, 51–59. [Google Scholar] [CrossRef] [PubMed]
- Bertz, S.; Otto, W.; Denzinger, S.; Wieland, W.F.; Burger, M.; Stohr, R.; Link, S.; Hofstädter, F.; Hartmann, A. Combination of CK20 and Ki-67 immunostaining analysis predicts recurrence, progression, and cancer-specific survival in pT1 urothelial bladder cancer. Eur. Urol. 2014, 65, 218–226. [Google Scholar] [CrossRef] [PubMed]
- Harnden, P.; Mahmood, N.; Southgate, J. Expression of cytokeratin 20 redefines urothelial papillomas of the bladder. Lancet 1999, 353, 974–977. [Google Scholar] [CrossRef]
- Hurst, C.D.; Alder, O.; Platt, F.M.; Droop, A.; Stead, L.F.; Burns, J.E.; Burghel, G.J.; Jain, S.; Klimczak, L.J.; Lindsay, H.; et al. Genomic Subtypes of Non-invasive Bladder Cancer with Distinct Metabolic Profile and Female Gender Bias in KDM6A Mutation Frequency. Cancer Cell 2017, 32, 701–715. [Google Scholar] [CrossRef] [PubMed]
- Tomlinson, D.C.; Baldo, O.; Harnden, P.; Knowles, M.A. FGFR3 protein expression and its relationship to mutation status and prognostic variables in bladder cancer. J. Pathol. 2007, 213, 91–98. [Google Scholar] [CrossRef] [PubMed]
- Van Rhijn, B.W.; van der Kwast, T.H.; Vis, A.N.; Kirkels, W.J.; Boeve, E.R.; Jobsis, A.C.; Zwarthoff, E.C. FGFR3 and P53 characterize alternative genetic pathways in the pathogenesis of urothelial cell carcinoma. Cancer. Res. 2004, 64, 1911–1914. [Google Scholar] [CrossRef] [PubMed]
- Bakkar, A.A.; Wallerand, H.; Radvanyi, F.; Lahaye, J.B.; Pissard, S.; Lecerf, L.; Kouyoumdjian, J.C.; Abbou, C.C.; Pairon, J.C.; Jaurand, M.C.; et al. FGFR3 and TP53 gene mutations define two distinct pathways in urothelial cell carcinoma of the bladder. Cancer. Res. 2003, 63, 8108–8112. [Google Scholar] [PubMed]
- Hernandez, S.; Lopez-Knowles, E.; Lloreta, J.; Kogevinas, M.; Amoros, A.; Tardon, A.; Carrato, A.; Serra, C.; Malats, N.; Real, F.X. Prospective study of FGFR3 mutations as a prognostic factor in nonmuscle invasive urothelial bladder carcinomas. J. Clin. Oncol. 2006, 24, 3664–3671. [Google Scholar] [CrossRef] [PubMed]
- Junker, K.; van Oers, J.M.; Zwarthoff, E.C.; Kania, I.; Schubert, J.; Hartmann, A. Fibroblast growth factor receptor 3 mutations in bladder tumors correlate with low frequency of chromosome alterations. Neoplasia 2008, 10, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Van Rhijn, B.W.; Zuiverloon, T.C.; Vis, A.N.; Radvanyi, F.; van Leenders, G.J.; Ooms, B.C.; Kirkels, W.J.; Lockwood, G.A.; Boevé, E.R.; Jöbsis, A.C.; et al. Molecular grade (FGFR3/MIB-1) and EORTC risk scores are predictive in primary non-muscle-invasive bladder cancer. Eur. Urol. 2010, 58, 433–441. [Google Scholar] [CrossRef] [PubMed]
- Sigal, A.; Rotter, V. Oncogenic mutations of the p53 tumor suppressor: The demons of the guardian of the genome. Cancer. Res. 2000, 60, 6788–6793. [Google Scholar] [PubMed]
- Zuckerman, V.; Wolyniec, K.; Sionov, R.V.; Haupt, S.; Haupt, Y. Tumour suppression by p53: The importance of apoptosis and cellular senescence. J. Pathol. 2009, 219, 3–15. [Google Scholar] [CrossRef] [PubMed]
- Oren, M.; Rotter, V. Mutant p53 gain-of-function in cancer. Cold Spring Harb. Perspect Biol. 2010, 2, a001107. [Google Scholar] [CrossRef] [PubMed]
- Rivlin, N.; Brosh, R.; Oren, M.; Rotter, V. Mutations in the p53 Tumor Suppressor Gene: Important Milestones at the Various Steps of Tumorigenesis. Genes Cancer 2011, 2, 466–474. [Google Scholar] [CrossRef] [PubMed]
- Weinstein, J.N.; Akbani, R.; Broom, B.M.; Wang, W.; Verhaak, R.G.; McConkey, D.; Lerner, S.; Morgan, M.; Creighton, C.J.; Smith, C.; et al. Comprehensive molecular characterization of urothelial bladder carcinoma. Nature 2014, 507, 315–322. [Google Scholar] [CrossRef]
- Hedegaard, J.; Lamy, P.; Nordentoft, I.; Algaba, F.; Hoyer, S.; Ulhoi, B.P.; Vang, S.; Reinert, T.; Hermann, G.G.; Mogensen, K.; et al. Comprehensive Transcriptional Analysis of Early-Stage Urothelial Carcinoma. Cancer Cell 2016, 30, 27–42. [Google Scholar] [CrossRef] [PubMed]
- Sung, J.Y.; Sun, J.M.; Chang, J.B.; Seo, S.I.; Soo, J.S.; Moo, L.H.; Yong, C.H.; Young, K.S.; Choi, Y.L.; Young, K.G. FGFR3 overexpression is prognostic of adverse outcome for muscle-invasive bladder carcinoma treated with adjuvant chemotherapy. Urol. Oncol. 2014, 32, e23–e31. [Google Scholar] [CrossRef]
- Gaisa, N.T.; Graham, T.A.; McDonald, S.A.; Canadillas-Lopez, S.; Poulsom, R.; Heidenreich, A.; Jakse, G.; Tadrous, P.J.; Knuechel, R.; Wright, N.A. The human urothelium consists of multiple clonal units, each maintained by a stem cell. J Pathol. 2011, 225, 163–171. [Google Scholar] [CrossRef] [PubMed]
- Fischbach, A.; Rogler, A.; Erber, R.; Stoehr, R.; Poulsom, R.; Heidenreich, A.; Schneevoigt, B.S.; Hauke, S.; Hartmann, A.; Knuechel, R.; et al. Fibroblast growth factor receptor (FGFR) gene amplifications are rare events in bladder cancer. Histopathology 2015, 66, 639–649. [Google Scholar] [CrossRef] [PubMed]
- Molitor, M.; Junker, K.; Eltze, E.; Toma, M.; Denzinger, S.; Siegert, S.; Knuechel, R.; Gaisa, N.T. Comparison of structural genetics of non-schistosoma-associated squamous cell carcinoma of the urinary bladder. Int. J. Clin. Exp. Pathol. 2015, 8, 8143–8158. [Google Scholar] [PubMed]
- Gaisa, N.T.; Braunschweig, T.; Reimer, N.; Bornemann, J.; Eltze, E.; Siegert, S.; Toma, M.; Villa, L.; Hartmann, A.; Knuechel, R. Different immunohistochemical and ultrastructural phenotypes of squamous differentiation in bladder cancer. Virchows. Arch. 2011, 458, 301–312. [Google Scholar] [CrossRef] [PubMed]
- Baldia, P.H.; Maurer, A.; Heide, T.; Rose, M.; Stoehr, R.; Hartmann, A.; Williams, S.V.; Knowles, M.A.; Knuechel, R.; Gaisa, N.T. Fibroblast growth factor receptor (FGFR) alterations in squamous differentiated bladder cancer: A putative therapeutic target for a small subgroup. Oncotarget 2016, 7, 71429–71439. [Google Scholar] [CrossRef] [PubMed]
| FGFR3 Expression a | |||||
|---|---|---|---|---|---|
| n | 0–2 | 3 | p-Value b | Spearman ρ | |
| Parameter: | |||||
| Age at diagnosis | |||||
| <70 years | 67 | 20 | 47 | 0.946 | 0.006 |
| ≥70 years | 75 | 22 | 53 | ||
| Gender | |||||
| female | 31 | 10 | 21 | 0.372 | 0.031 |
| male | 111 | 32 | 79 | ||
| Histological tumor grade | |||||
| low grade | 49 | 3 | 46 | <0.001 | −0.373 |
| high grade | 93 | 39 | 54 | ||
| Tumor stage | |||||
| pTa | 82 | 14 | 68 | <0.001 | −0.320 |
| pT1–pT4 | 60 | 28 | 32 | ||
| Ki67 Expression a | |||||
|---|---|---|---|---|---|
| n | <15% | ≥15% | p-Value b | Spearman ρ | |
| Parameter: | |||||
| Age at diagnosis | |||||
| <70 years | 67 | 31 | 36 | 0.083 | 0.146 |
| ≥70 years | 75 | 24 | 51 | ||
| Gender | |||||
| female | 31 | 14 | 17 | 0.408 | 0.070 |
| male | 111 | 41 | 70 | ||
| Histological tumor grade | |||||
| low grade | 49 | 34 | 15 | <0.001 | 0.457 |
| high grade | 93 | 21 | 72 | ||
| Tumor stage | |||||
| pTa | 82 | 37 | 45 | 0.069 | 0.175 |
| pT1–pT4 | 60 | 18 | 42 | ||
| Variable | HR | p-Value | 95%CI | |
|---|---|---|---|---|
| Lower | Upper | |||
| FGFR3 high/Ki67high | 3.943 | 0.019 | 1.247 | 12.466 |
| pT status | 0.957 | 0.941 | 0.295 | 3.105 |
| Tumor grade | 0.846 | 0.823 | 0.196 | 3.653 |
| Keratin 5/6 | 0.482 | 0.280 | 0.128 | 1.812 |
| Keratin 20 | 0.424 | 0.115 | 0.146 | 1.232 |
| Age | 1.773 | 0.347 | 0.537 | 5.847 |
| Variable | HR | p-Value | 95%CI | |
|---|---|---|---|---|
| Lower | Upper | |||
| FGFR3mut/TP53mut | 6.563 | 0.006 | 1.694 | 25.425 |
| pT status | 1.179 | 0.821 | 0.284 | 4.896 |
| Tumor grade | 0.241 | 0.138 | 0.037 | 1.580 |
| Keratin 5/6 | 0.714 | 0.621 | 0.188 | 2.712 |
| Keratin 20 | 0.872 | 0.814 | 0.279 | 2.730 |
| Age | 1.41 | 0.584 | 0.412 | 2.809 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Geelvink, M.; Babmorad, A.; Maurer, A.; Stöhr, R.; Grimm, T.; Bach, C.; Knuechel, R.; Rose, M.; Gaisa, N.T. Diagnostic and Prognostic Implications of FGFR3high/Ki67high Papillary Bladder Cancers. Int. J. Mol. Sci. 2018, 19, 2548. https://doi.org/10.3390/ijms19092548
Geelvink M, Babmorad A, Maurer A, Stöhr R, Grimm T, Bach C, Knuechel R, Rose M, Gaisa NT. Diagnostic and Prognostic Implications of FGFR3high/Ki67high Papillary Bladder Cancers. International Journal of Molecular Sciences. 2018; 19(9):2548. https://doi.org/10.3390/ijms19092548
Chicago/Turabian StyleGeelvink, Mirja, Armin Babmorad, Angela Maurer, Robert Stöhr, Tobias Grimm, Christian Bach, Ruth Knuechel, Michael Rose, and Nadine T. Gaisa. 2018. "Diagnostic and Prognostic Implications of FGFR3high/Ki67high Papillary Bladder Cancers" International Journal of Molecular Sciences 19, no. 9: 2548. https://doi.org/10.3390/ijms19092548
APA StyleGeelvink, M., Babmorad, A., Maurer, A., Stöhr, R., Grimm, T., Bach, C., Knuechel, R., Rose, M., & Gaisa, N. T. (2018). Diagnostic and Prognostic Implications of FGFR3high/Ki67high Papillary Bladder Cancers. International Journal of Molecular Sciences, 19(9), 2548. https://doi.org/10.3390/ijms19092548






