Molecular Regulation of Nitrate Responses in Plants
Abstract
1. Introduction
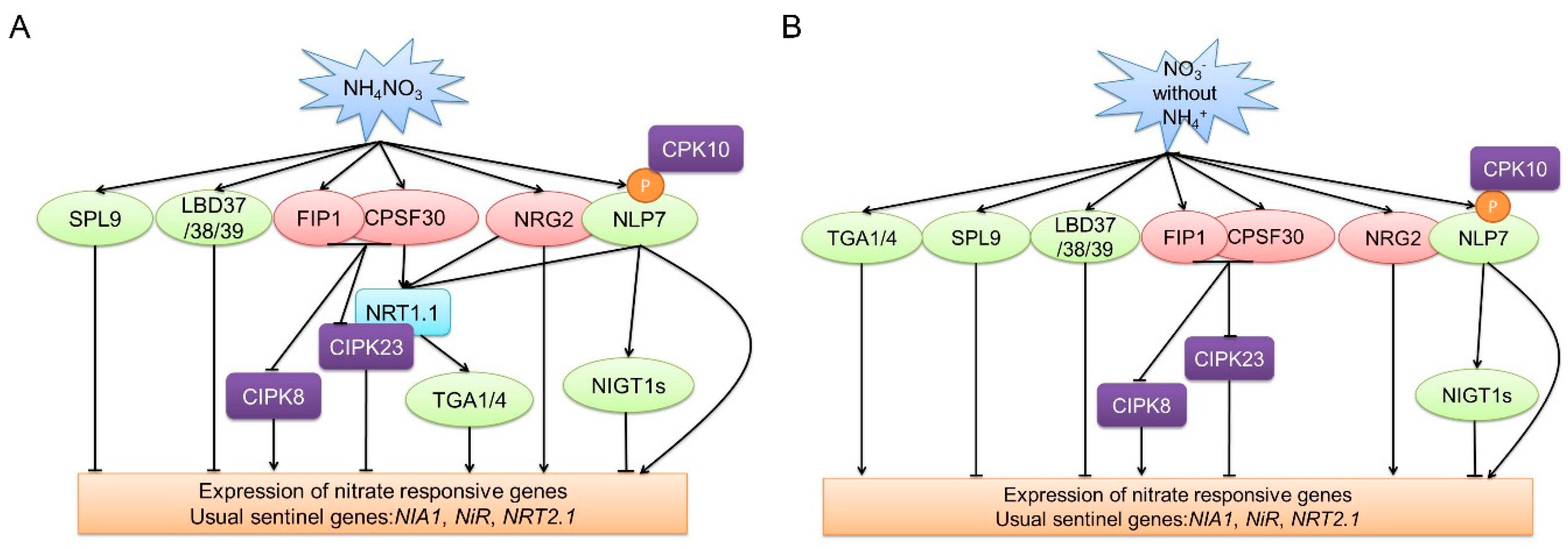
2. Short-Term Nitrate Signaling: The Primary Nitrate Response
2.1. Nitrate Sensor
2.2. Transcription Factors
2.3. Protein Kinases
2.4. Other Factors, Including NRG2, CPSF30, and FIP1
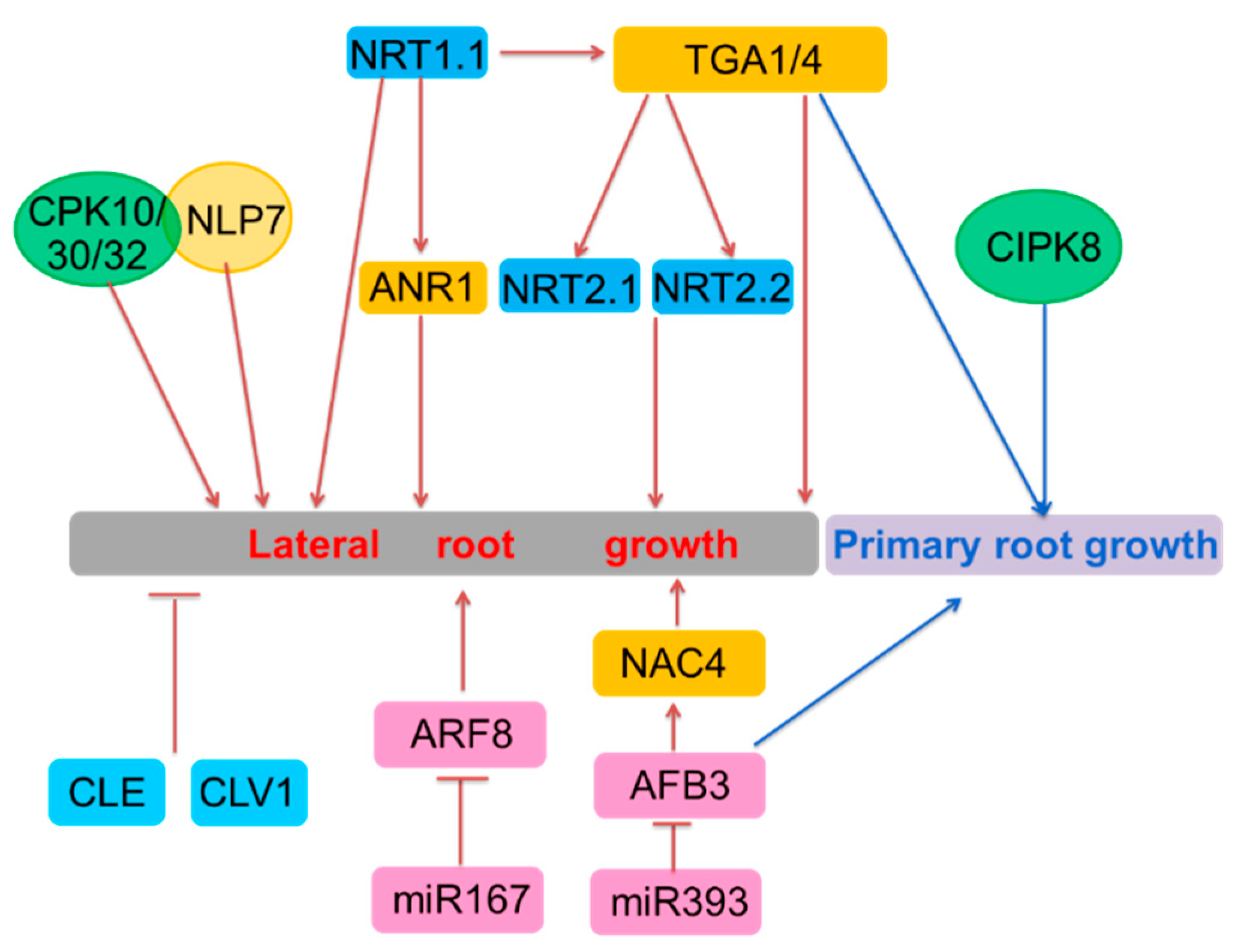
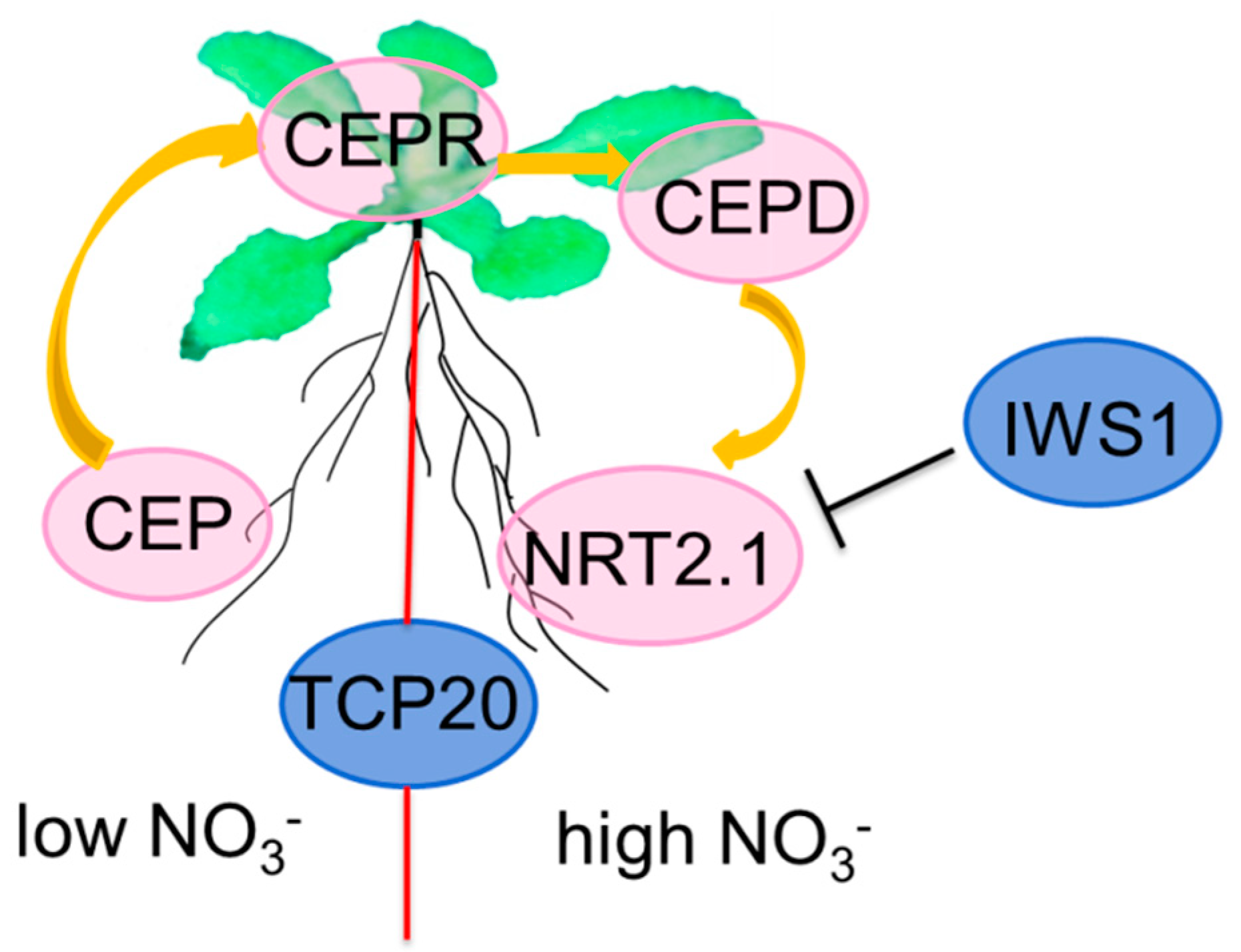
3. Long-Term Nitrate Signaling
4. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Crawford, N.M.; Glass, A.D. Molecular and physiological aspects of nitrate uptake in plants. Trends Plant Sci. 1998, 3, 389–395. [Google Scholar] [CrossRef]
- Broadley, M.; Brown, P.; Buerkert, A.; Cakmak, I.; Cooper, J.; Eichert, T.; Engels, C.; Fernández, V.; Kirkby, E.; George, E. Marschner’s Mineral Nutrition of Higher Plants, 3rd ed.; Academic Press: Cambridge, MA, USA, 2012; Volume 68. [Google Scholar]
- Krouk, G.; Crawford, N.M.; Coruzzi, G.M.; Tsay, Y.-F. Nitrate signaling: Adaptation to fluctuating environments. Curr. Opin. Plant Biol. 2010, 13, 265–272. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.; Guegler, K.; LaBrie, S.T.; Crawford, N.M. Genomic analysis of a nutrient response in Arabidopsis reveals diverse expression patterns and novel metabolic and potential regulatory genes induced by nitrate. Plant Cell 2000, 12, 1491–1509. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.; Okamoto, M.; Xing, X.; Crawford, N.M. Microarray analysis of the nitrate response in Arabidopsis roots and shoots reveals over 1000 rapidly responding genes and new linkages to glucose, trehalose-6-phosphate, iron, and sulfate metabolism. Plant Physiol. 2003, 132, 556–567. [Google Scholar] [CrossRef] [PubMed]
- Scheible, W.-R.; Morcuende, R.; Czechowski, T.; Fritz, C.; Osuna, D.; Palacios-Rojas, N.; Schindelasch, D.; Thimm, O.; Udvardi, M.K.; Stitt, M. Genome-wide reprogramming of primary and secondary metabolism, protein synthesis, cellular growth processes, and the regulatory infrastructure of Arabidopsis in response to nitrogen. Plant Physiol. 2004, 136, 2483–2499. [Google Scholar] [CrossRef] [PubMed]
- Gutiérrez, R.A.; Lejay, L.V.; Dean, A.; Chiaromonte, F.; Shasha, D.E.; Coruzzi, G.M. Qualitative network models and genome-wide expression data define carbon/nitrogen-responsive molecular machines in Arabidopsis. Genome Biol. 2007, 8, R7. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.; Xing, X.; Crawford, N. Nitrite acts as a transcriptome signal at micromolar concentrations in Arabidopsis roots. Plant Physiol. 2007, 145, 1735–1745. [Google Scholar] [CrossRef] [PubMed]
- Roenneberg, T.; Rehman, J. Nitrate, a nonphotic signal for the 3circadian system. FASEB J. 1996, 10, 1443–1447. [Google Scholar] [CrossRef] [PubMed]
- Stitt, M. Nitrate regulation of metabolism and growth. Curr. Opin. Plant Biol. 1999, 2, 178–186. [Google Scholar] [CrossRef]
- Alboresi, A.; Gestin, C.; Leydecker, M.T.; Bedu, M.; Meyer, C.; Truong, H.N. Nitrate, a signal relieving seed dormancy in Arabidopsis. Plant Cell Environ. 2005, 28, 500–512. [Google Scholar] [CrossRef] [PubMed]
- Walch-Liu, P.; Ivanov, I.I.; Filleur, S.; Gan, Y.; Remans, T.; Forde, B.G. Nitrogen regulation of root branching. Ann. Bot. 2006, 97, 875–881. [Google Scholar] [CrossRef] [PubMed]
- Wilkinson, S.; Bacon, M.A.; Davies, W.J. Nitrate signalling to stomata and growing leaves: Interactions with soil drying, ABA, and xylem sap pH in maize. J. Exp. Bot. 2007, 58, 1705–1716. [Google Scholar] [CrossRef] [PubMed]
- Krouk, G.; Lacombe, B.; Bielach, A.; Perrine-Walker, F.; Malinska, K.; Mounier, E.; Hoyerova, K.; Tillard, P.; Leon, S.; Ljung, K. Nitrate-regulated auxin transport by NRT1. 1 defines a mechanism for nutrient sensing in plants. Dev. Cell 2010, 18, 927–937. [Google Scholar] [CrossRef] [PubMed]
- Medici, A.; Krouk, G. The primary nitrate response: A multifaceted signalling pathway. J. Exp. Bot. 2014, 65, 5567–5576. [Google Scholar] [CrossRef] [PubMed]
- Cavicchioli, R.; Chiang, R.C.; Kalman, L.V.; Gunsalus, R.P. Role of the periplasmic domain of the Escherichia coli NARX sensor-transmitter protein in nitrate-dependent signal transduction and gene regulation. Mol. Microbiol. 1996, 21, 901–911. [Google Scholar] [CrossRef] [PubMed]
- Chiang, R.C.; Cavicchioli, R.; Gunsalus, R.P. ‘Locked-on’ and ‘locked-off’ signal transduction mutations in the periplasmic domain of the Escherichia coli NARQ and NARX sensors affect nitrate-and nitrite-dependent regulation by narl and narp. Mol. Microbiol. 1997, 24, 1049–1060. [Google Scholar] [CrossRef] [PubMed]
- Fu, Y.; Kneesi, J.Y.; Marzluf, G.A. Isolation of Nit-4, the minor nitrogen regulatory gene which mediates nitrate induction in Neurospora crassa. J. Bacteriol. 1989, 171, 4067–4070. [Google Scholar] [CrossRef] [PubMed]
- Burger, G.; Tilburn, J.; Scazzocchio, C. Molecular cloning and functional characterization of the pathway-specific regulatory gene NirA, which controls nitrate assimilation in Aspergillus nidulans. Mol. Cell. Biol. 1991, 11, 795–802. [Google Scholar] [CrossRef] [PubMed]
- And, N.M.; Arst, H.N., Jr. The molecular genetics of nitrate assimilation in fungi and plants. Ann. Rev. Genet. 1993, 27, 115–146. [Google Scholar]
- Léran, S.; Varala, K.; Boyer, J.-C.; Chiurazzi, M.; Crawford, N.; Daniel-Vedele, F.; David, L.; Dickstein, R.; Fernandez, E.; Forde, B. A unified nomenclature of nitrate transporter 1/peptide transporter family members in plants. Trends Plant Sci. 2014, 19, 5–9. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.; Liu, D.; Crawford, N.M. The Arabidopsis CHL1 protein plays a major role in high-affinity nitrate uptake. Proc. Natl. Acad. Sci. USA 1998, 95, 15134–15139. [Google Scholar] [CrossRef] [PubMed]
- Liu, K.-H.; Huang, C.-Y.; Tsay, Y.-F. CHL1 is a dual-affinity nitrate transporter of Arabidopsis involved in multiple phases of nitrate uptake. Plant Cell 1999, 11, 865–874. [Google Scholar] [CrossRef] [PubMed]
- Liu, K.H.; Tsay, Y.F. Switching between the two action modes of the dual-affinity nitrate transporter CHL1 by phosphorylation. EMBO J. 2003, 22, 1005–1013. [Google Scholar] [CrossRef] [PubMed]
- Tsay, Y.F.; Schroeder, J.I.; Feldmann, K.A.; Crawford, N.M. The herbicide sensitivity gene CHL1 of Arabidopsis encodes a nitrate-inducible nitrate transporter. Cell 1993, 72, 705–713. [Google Scholar] [CrossRef]
- Remans, T.; Nacry, P.; Pervent, M.; Filleur, S.; Diatloff, E.; Mounier, E.; Tillard, P.; Forde, B.G.; Gojon, A. The Arabidopsis NRT1.1 transporter participates in the signaling pathway triggering root colonization of nitrate-rich patches. Proc. Natl. Acad. Sci. USA 2006, 103, 19206–19211. [Google Scholar] [CrossRef] [PubMed]
- Walch-Liu, P.; Forde, B. Nitrate signalling mediated by the NRT1.1 nitrate transporter antagonises l-glutamate-induced changes in root architecture. Plant J. 2010, 54, 820–828. [Google Scholar] [CrossRef] [PubMed]
- Ho, C.-H.; Lin, S.-H.; Hu, H.-C.; Tsay, Y.-F. CHL1 functions as a nitrate sensor in plants. Cell 2009, 138, 1184–1194. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.; Xing, X.; Wang, Y.; Tran, A.; Crawford, N.M. A genetic screen for nitrate regulatory mutants captures the nitrate transporter gene NRT1.1. Plant Physiol. 2009, 151, 472. [Google Scholar] [CrossRef] [PubMed]
- Bouguyon, E.; Brun, F.; Meynard, D.; Kubeš, M.; Pervent, M.; Leran, S.; Lacombe, B.; Krouk, G.; Guiderdoni, E.; Zažímalová, E. Multiple mechanisms of nitrate sensing by Arabidopsis nitrate transceptor NRT1.1. Nat. Plants 2015, 1, 15015. [Google Scholar] [CrossRef] [PubMed]
- Hu, B.; Wang, W.; Ou, S.; Tang, J.; Li, H.; Che, R.; Zhang, Z.; Chai, X.; Wang, H.; Wang, Y.; et al. Variation in NRT1.1b contributes to nitrate-use divergence between rice subspecies. Nat. Genet. 2015, 47, 834. [Google Scholar] [CrossRef] [PubMed]
- Schauser, L.; Roussis, A.; Stiller, J.; Stougaard, J. A plant regulator controlling development of symbiotic root nodules. Nature 1999, 402, 191–195. [Google Scholar] [CrossRef] [PubMed]
- Borisov, A.Y.; Madsen, L.H.; Tsyganov, V.E.; Umehara, Y.; Voroshilova, V.A.; Batagov, A.O.; Sandal, N.; Mortensen, A.; Schauser, L.; Ellis, N. The SYM35 gene required for root nodule development in pea is an ortholog of NIN from Lotus japonicus. Plant Physiol. 2003, 131, 1009–1017. [Google Scholar] [CrossRef] [PubMed]
- Schauser, L.; Wieloch, W.; Stougaard, J. Evolution of NIN-like proteins in Arabidopsis, rice, and Lotus japonicus. J. Mol. Evol. 2005, 60, 229–237. [Google Scholar] [CrossRef] [PubMed]
- Castaings, L.; Camargo, A.; Pocholle, D.; Gaudon, V.; Texier, Y.; Boutet-Mercey, S.; Taconnat, L.; Renou, J.P.; Daniel-Vedele, F.; Fernandez, E. The nodule inception-like protein 7 modulates nitrate sensing and metabolism in Arabidopsis. Plant J. 2009, 57, 426–435. [Google Scholar] [CrossRef] [PubMed]
- Camargo, A.; Llamas, Á.; Schnell, R.A.; Higuera, J.J.; González-Ballester, D.; Lefebvre, P.A.; Fernández, E.; Galván, A. Nitrate signaling by the regulatory gene NIT2 in CHLamydomonas. Plant Cell 2007, 19, 3491–3503. [Google Scholar] [CrossRef] [PubMed]
- Yu, L.-H.; Wu, J.; Tang, H.; Yuan, Y.; Wang, S.-M.; Wang, Y.-P.; Zhu, Q.-S.; Li, S.-G.; Xiang, C.-B. Overexpression of Arabidopsis NLP7 improves plant growth under both nitrogen-limiting and-sufficient conditions by enhancing nitrogen and carbon assimilation. Sci. Rep. 2016, 6, 27795. [Google Scholar] [CrossRef] [PubMed]
- Marchive, C.; Roudier, F.; Castaings, L.; Brehaut, V.; Blondet, E.; Colot, V.; Meyer, C.; Krapp, A. Nuclear retention of the transcription factor NLP7 orchestrates the early response to nitrate in plants. Nat. Commun. 2013, 4, 1713. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Zhang, W.; Yang, Y.; Li, Z.; Li, N.; Qi, S.; Crawford, N.M.; Wang, Y. The Arabidopsis NLP7 gene regulates nitrate signaling via NRT1.1–dependent pathway in the presence of ammonium. Sci. Rep. 2018, 8, 1487. [Google Scholar] [CrossRef] [PubMed]
- O’Brien, J.A.; Vega, A.; Bouguyon, E.; Krouk, G.; Gojon, A.; Coruzzi, G.; Gutiérrez, R.A. Nitrate transport, sensing, and responses in plants. Mol. Plant 2016, 9, 837–856. [Google Scholar] [CrossRef] [PubMed]
- Konishi, M.; Yanagisawa, S. Arabidopsis NIN-like transcription factors have a central role in nitrate signalling. Nat. Commun. 2013, 4, 1617. [Google Scholar] [CrossRef] [PubMed]
- Liu, K.-H.; Niu, Y.; Konishi, M.; Wu, Y.; Du, H.; Sun Chung, H.; Li, L.; Boudsocq, M.; McCormack, M.; Maekawa, S.; et al. Discovery of nitrate–CPK–NLP signalling in central nutrient–growth networks. Nature 2017, 545, 311–316. [Google Scholar] [CrossRef] [PubMed]
- Cao, H.; Qi, S.; Sun, M.; Li, Z.; Yang, Y.; Crawford, N.M.; Wang, Y. Overexpression of the maize ZMNLP6 and ZMNLP8 can complement the Arabidopsis nitrate regulatory mutant NLP7 by restoring nitrate signaling and assimilation. Front. Plant Sci. 2017, 8, 1703. [Google Scholar] [CrossRef] [PubMed]
- Rubin, G.; Tohge, T.; Matsuda, F.; Saito, K.; Scheible, W.-R. Members of the LBD family of transcription factors repress anthocyanin synthesis and affect additional nitrogen responses in Arabidopsis. Plant Cell 2009, 21, 3567–3584. [Google Scholar] [CrossRef] [PubMed]
- Vidal, E.A.; Álvarez, J.M.; Moyano, T.C.; Gutiérrez, R.A. Transcriptional networks in the nitrate response of Arabidopsis thaliana. Curr. Opin. Plant Biol. 2015, 27, 125–132. [Google Scholar] [CrossRef] [PubMed]
- Krouk, G.; Mirowski, P.; LeCun, Y.; Shasha, D.E.; Coruzzi, G.M. Predictive network modeling of the high-resolution dynamic plant transcriptome in response to nitrate. Genome Biol. 2010, 11, 1. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.-W.; Czech, B.; Weigel, D. miR156-regulated SPL transcription factors define an endogenous flowering pathway in Arabidopsis thaliana. Cell 2009, 138, 738–749. [Google Scholar] [CrossRef] [PubMed]
- Alvarez, J.M.; Riveras, E.; Vidal, E.A.; Gras, D.E.; Contreras-López, O.; Tamayo, K.P.; Aceituno, F.; Gómez, I.; Ruffel, S.; Lejay, L. Systems approach identifies TGA1 and TGA4 transcription factors as important regulatory components of the nitrate response of Arabidopsis thaliana roots. Plant J. 2014, 80, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Maeda, Y.; Konishi, M.; Kiba, T.; Sakuraba, Y.; Sawaki, N.; Kurai, T.; Ueda, Y.; Sakakibara, H.; Yanagisawa, S. A NIGT1-centred transcriptional cascade regulates nitrate signalling and incorporates phosphorus starvation signals in Arabidopsis. Nat. Commun. 2018, 9, 1376. [Google Scholar] [CrossRef] [PubMed]
- Medici, A.; Marshallcolon, A.; Ronzier, E.; Szponarski, W.; Wang, R.; Gojon, A.; Crawford, N.M.; Ruffel, S.; Coruzzi, G.M.; Krouk, G. ATNIGT1/HRS1 integrates nitrate and phosphate signals at the Arabidopsis root tip. Nat. Commun. 2015, 6, 6274. [Google Scholar] [CrossRef] [PubMed]
- Hu, H.C.; Wang, Y.Y.; Tsay, Y.F. ATCIPK8, a CBL-interacting protein kinase, regulates the low-affinity phase of the primary nitrate response. Plant J. 2009, 57, 264–278. [Google Scholar] [CrossRef] [PubMed]
- Xu, N.; Wang, R.; Zhao, L.; Zhang, C.; Li, Z.; Lei, Z.; Liu, F.; Guan, P.; Chu, Z.; Crawford, N.M. The Arabidopsis NRG2 protein mediates nitrate signaling and interacts with and regulates key nitrate regulators. Plant Cell 2016, 28, 485–504. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Wang, R.; Gao, Y.; Wang, C.; Zhao, L.; Xu, N.; Chen, K.E.; Qi, S.; Zhang, M.; Tsay, Y.F. The Arabidopsis CPSF30-l gene plays an essential role in nitrate signaling and regulates the nitrate transceptor gene NRT1. 1. New Phytol. 2017, 216, 1205–1222. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Zhang, W.; Li, Z.; Li, Z.; Bi, Y.; Crawford, N.M.; Wang, Y. FIP1 plays an important role in nitrate signaling and regulates CIPK8 and CIPK23 expression in Arabidopsis. Front. Plant Sci. 2018, 9. [Google Scholar] [CrossRef] [PubMed]
- Delaney, K.J.; Xu, R.; Zhang, J.; Li, Q.Q.; Yun, K.-Y.; Falcone, D.L.; Hunt, A.G. Calmodulin interacts with and regulates the RNA-binding activity of an Arabidopsis polyadenylation factor subunit. Plant Physiol. 2006, 140, 1507–1521. [Google Scholar] [CrossRef] [PubMed]
- Addepalli, B.; Hunt, A.G. A novel endonuclease activity associated with the Arabidopsis ortholog of the 30-KDA subunit of cleavage and polyadenylation specificity factor. Nucleic Acids Res. 2007, 35, 4453–4463. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Addepalli, B.; Yun, K.-Y.; Hunt, A.G.; Xu, R.; Rao, S.; Li, Q.Q.; Falcone, D.L. A polyadenylation factor subunit implicated in regulating oxidative signaling in Arabidopsis thaliana. PLoS ONE 2008, 3, e2410. [Google Scholar] [CrossRef] [PubMed]
- Guan, P.; Ripoll, J.-J.; Wang, R.; Vuong, L.; Bailey-Steinitz, L.J.; Ye, D.; Crawford, N.M. Interacting TCP and NLP transcription factors control plant responses to nitrate availability. Proc. Natl. Acad. Sci. USA 2017, 114, 2419–2424. [Google Scholar] [CrossRef] [PubMed]
- Hunt, A.G.; Xu, R.; Addepalli, B.; Rao, S.; Forbes, K.P.; Meeks, L.R.; Xing, D.; Mo, M.; Zhao, H.; Bandyopadhyay, A. Arabidopsis mRNA polyadenylation machinery: Comprehensive analysis of protein-protein interactions and gene expression profiling. BMC Genom. 2008, 9, 220. [Google Scholar] [CrossRef] [PubMed]
- Riveras, E.; Alvarez, J.M.; Vidal, E.A.; Oses, C.; Vega, A.; Gutiérrez, R.A. The calcium ion is a second messenger in the nitrate signaling pathway of Arabidopsis. Plant Physiol. 2015, 169, 1397–1404. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Forde, B.G. An Arabidopsis MADs box gene that controls nutrient-induced changes in root architecture. Science 1998, 279, 407. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Forde, B.G. Regulation of Arabidopsis root development by nitrate availability. J. Exp. Bot. 2000, 51, 51. [Google Scholar] [CrossRef] [PubMed]
- Remans, T.; Nacry, P.; Pervent, M.; Girin, T.; Tillard, P.; Lepetit, M.; Gojon, A. A central role for the nitrate transporter NRT2.1 in the integrated morphological and physiological responses of the root system to nitrogen limitation in Arabidopsis. Plant Physiol. 2006, 140, 909–921. [Google Scholar] [PubMed]
- Gan, Y.; Bernreiter, A.; Filleur, S.; Abram, B.; Forde, B.G. Overexpressing the ANR1 MADs-box gene in transgenic plants provides new insights into its role in the nitrate regulation of root development. Plant Cell Physiol. 2012, 53, 1003–1016. [Google Scholar] [CrossRef] [PubMed]
- Bouguyon, E.; Perrine-Walker, F.; Pervent, M.; Rochette, J.; Cuesta, C.; Benkova, E.; Martinière, A.; Bach, L.; Krouk, G.; Gojon, A.; et al. Nitrate controls root development through posttranscriptional regulation of the NRT1.1/NPF6.3 transporter/sensor. Plant Physiol. 2016, 172, 1237–1248. [Google Scholar] [PubMed]
- Zhang, H.; Jennings, A.; Barlow, P.W.; Forde, B.G. Dual pathways for regulation of root branching by nitrate. Proc. Natl. Acad. Sci. USA 1999, 96, 6529–6534. [Google Scholar] [CrossRef] [PubMed]
- Signora, L.; De Smet, I.; Foyer, C.H.; Zhang, H. ABA plays a central role in mediating the regulatory effects of nitrate on root branching in Arabidopsis. Plant J. 2001, 28, 655–662. [Google Scholar] [CrossRef] [PubMed]
- Cerezo, M.; Tillard, P.; Filleur, S.; Muños, S.; Daniel-Vedele, F.; Gojon, A. Major alterations of the regulation of root NO3− uptake are associated with the mutation of NRT2.1 and NRT2.2 genes in Arabidopsis. Plant Physiol. 2001, 127, 262–271. [Google Scholar] [CrossRef] [PubMed]
- Filleur, S.; Dorbe, M.-F.; Cerezo, M.; Orsel, M.; Granier, F.; Gojon, A.; Daniel-Vedele, F. An Arabidopsis t-DNA mutant affected in NRT2 genes is impaired in nitrate uptake. FEBS Lett. 2001, 489, 220–224. [Google Scholar] [CrossRef]
- Lejay, L.; Tillard, P.; Lepetit, M.; Olive, F.D.; Filleur, S.; Daniel-Vedele, F.; Gojon, A. Molecular and functional regulation of two NO3− uptake systems by N- and C-status of Arabidopsis plants. Plant J. 1999, 18, 509–519. [Google Scholar] [CrossRef] [PubMed]
- Girin, T.; El-Kafafi, E.-S.; Widiez, T.; Erban, A.; Hubberten, H.-M.; Kopka, J.; Hoefgen, R.; Gojon, A.; Lepetit, M. Identification of Arabidopsis mutants impaired in the systemic regulation of root nitrate uptake by the nitrogen status of the plant. Plant Physiol. 2010, 153, 1250–1260. [Google Scholar] [CrossRef] [PubMed]
- Malamy, J.E.; Ryan, K.S. Environmental regulation of lateral root initiation in Arabidopsis. Plant Physiol. 2001, 127, 899–909. [Google Scholar] [CrossRef] [PubMed]
- Little, D.Y.; Rao, H.; Oliva, S.; Daniel-Vedele, F.; Krapp, A.; Malamy, J.E. The putative high-affinity nitrate transporter NRT2.1 represses lateral root initiation in response to nutritional cues. Proc. Natl. Acad. Sci. USA 2005, 102, 13693–13698. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Wang, Y.; Okamoto, M.; Crawford, N.M.; Siddiqi, M.Y.; Glass, A.D.M. Dissection of the ATNRT2.1:ATNRT2.2 inducible high-affinity nitrate transporter gene cluster. Plant Physiol. 2007, 143, 425–433. [Google Scholar] [CrossRef] [PubMed]
- Gifford, M.L.; Dean, A.; Gutierrez, R.A.; Coruzzi, G.M.; Birnbaum, K.D. Cell-specific nitrogen responses mediate developmental plasticity. Proc. Natl. Acad. Sci. USA 2008, 105, 803–808. [Google Scholar] [CrossRef] [PubMed]
- Wu, M.-F.; Tian, Q.; Reed, J.W. Arabidopsis microRNA167 controls patterns of ARF6 and ARF8 expression, and regulates both female and male reproduction. Development 2006, 133, 4211–4218. [Google Scholar] [CrossRef] [PubMed]
- Vidal, E.A.; Araus, V.; Lu, C.; Parry, G.; Green, P.J.; Coruzzi, G.M.; Gutiérrez, R.A. Nitrate-responsive MIR393/AFB3 regulatory module controls root system architecture in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 2010, 107, 4477–4482. [Google Scholar] [CrossRef] [PubMed]
- Vidal, E.A.; Moyano, T.C.; Riveras, E.; Contreras-López, O.; Gutiérrez, R.A. Systems approaches map regulatory networks downstream of the auxin receptor AFB3 in the nitrate response of Arabidopsis thaliana roots. Proc. Natl. Acad. Sci. USA 2013, 110, 12840–12845. [Google Scholar] [CrossRef] [PubMed]
- Vidal, E.A.; Álvarez, J.M.; Gutiérrez, R.A. Nitrate regulation of AFB3 and NAC4 gene expression in Arabidopsis roots depends on NRT1. 1 nitrate transport function. Plant Signal. Behav. 2014, 9, e28501. [Google Scholar] [CrossRef] [PubMed]
- Araya, T.; Miyamoto, M.; Wibowo, J.; Suzuki, A.; Kojima, S.; Tsuchiya, Y.N.; Sawa, S.; Fukuda, H.; von Wirén, N.; Takahashi, H. CLE-clavata1 peptide-receptor signaling module regulates the expansion of plant root systems in a nitrogen-dependent manner. Proc. Natl. Acad. Sci. USA 2014, 111, 2029–2034. [Google Scholar] [CrossRef] [PubMed]
- Gansel, X.; Muños, S.; Tillard, P.; Gojon, A. Differential regulation of the NO3− and NH4+ transporter genes ATNRT2.1 and ATAMT1.1 in Arabidopsis: Relation with long-distance and local controls by N status of the plant. Plant J. 2001, 26, 143–155. [Google Scholar] [CrossRef] [PubMed]
- Tabata, R.; Sumida, K.; Yoshii, T.; Ohyama, K.; Shinohara, H.; Matsubayashi, Y. Perception of root-derived peptides by shoot LRR-RKS mediates systemic N-demand signaling. Science 2014, 346, 343–346. [Google Scholar] [CrossRef] [PubMed]
- Ohkubo, Y.; Tanaka, M.; Tabata, R.; Ogawa-Ohnishi, M.; Matsubayashi, Y. Shoot-to-root mobile polypeptides involved in systemic regulation of nitrogen acquisition. Nat. Plants 2017, 3, 17029. [Google Scholar] [CrossRef] [PubMed]
- Widiez, T.; El Kafafi, E.S.; Girin, T.; Berr, A.; Ruffel, S.; Krouk, G.; Vayssières, A.; Shen, W.-H.; Coruzzi, G.M.; Gojon, A.; et al. High nitrogen insensitive 9 (HNI9)-mediated systemic repression of root NO3− uptake is associated with changes in histone methylation. Proc. Natl. Acad. Sci. USA 2011, 108, 13329–13334. [Google Scholar] [CrossRef] [PubMed]
- Guan, P.; Wang, R.; Nacry, P.; Breton, G.; Kay, S.A.; Pruneda-Paz, J.L.; Davani, A.; Crawford, N.M. Nitrate foraging by Arabidopsis roots is mediated by the transcription factor TCP20 through the systemic signaling pathway. Proc. Natl. Acad. Sci. USA 2014, 111, 15267–15272. [Google Scholar] [CrossRef] [PubMed]
| The Group of the Genes | Gene | Gene Family | Target Genes | Roles in Nitrate Signaling | Nitrate Responsive | Identification Methodology |
|---|---|---|---|---|---|---|
| Genes involved in short-term nitrate signaling/primary nitrate response | NLP6 | RWP-PK | NRT2.1, NRT2.2, and NIA | Involved in primary nitrate response (positive) | No | Binding to NRE |
| LBD37/38/39 | LBD | NRT2.1, NRT2.2, NIA1, and NIA2 | Involved in primary nitrate response (negative) | Yes | Nitrate-responsive transcription factor | |
| SPL9 | SPL | NiR, NIA2, and NRT1.1 (potential) | Potential regulatory hub (negative) | Yes | Integrated systems biology approach | |
| NIGT1s | NIGT | NRT2.1 | Involved in primary nitrate response (negative) | Yes | Homologues of OsNIGT1 | |
| CIPK23 | CBL-interacting protein kinases | Phosphorylating NRT1.1 | Involved in primary nitrate response (negative) | Yes | Downregulated in chl1 | |
| NRG2 | Nitrate regulatory gene 2 | Regulating NRT1.1 and interacting with NLP7 | Involved in primary nitrate response (positive) | No | Forward genetics screening | |
| CPSF30 | Polyadenylation specificity factor | Affecting NRT1.1 mRNA 3’UTR alternative polyadenylation | Involved in primary nitrate response (positive) | No | Forward genetics screening | |
| FIP1 | Factor interacting with poly(A) polymerase 1 | Interacting with CPSF30, regulating CIPK8 and CIPK23, and affecting 3’UTR alternative polyadenylation of NRT1.1 | Involved in primary nitrate response (positive) | No | Interaction with CPSF30 | |
| Genes involved in both short-term and long-term nitrate signaling | NRT1.1/NPF6.3 | NPF | Regulating CIPK8, CIPK23, TGA1/4 | Involved in primary nitrate response (positive) and regulating lateral root growth | Yes | Forward genetics screening |
| TGA1/4 | bZIP | NRT2.1 and NRT2.2 | Involved in primary nitrate response (positive) and regulating lateral root emergency and primary root growth | Yes | Integrated systems biology approach | |
| CIPK8 | CBL-interacting protein kinases | Unknown | Involved in primary nitrate response (positive) and regulating primary root growth | Yes | Downregulated in chl1-5 | |
| NLP7 | RWP-PK | NRT2.1, NiR, NRT2.2, and NIA | Involved in primary nitrate response (positive) and regulating lateral root density and primary root growth | No | Homologous to NIN protein in legumes and binding to NRE | |
| CPK10 | Subgroup III Ca2+-sensor protein kinase | Phosphorylating NLP7 | Involved in primary nitrate response (positive) and regulating lateral root primordia density and lateral root elongation | Yes | Induced by nitrate | |
| Genes involved in long-term nitrate signaling | ANR1 | MADS-box | Regulating NRT1.1 | Regulating lateral root growth under high nitrate | Yes | Isolated in a screen for nitrate-responsive genes in roots |
| NRT2.1 | NPF | Unknown | Regulating lateral root initiation under the conditions of high C/N ratio and lateral root growth under limited nitrogen | Yes | Forward genetics screening | |
| ARF8 | Auxin response factors | Unknown | Induced in the pericycle and lateral root cap and marginally repressed in the stele in response to nitrogen | Yes | Cell-specific response to nitrogen | |
| miR167 | microRNA | Regulating the expression of ARF8 | Controlling the lateral root growth in response to nitrogen with ARF8 | Yes | Regulator of ARF8 | |
| miR393 | microRNA | Specifically cleaving AFB3 | miR393/AFB3 controlled the lateral root growth and primary root growth | Yes | 454 sequencing technology | |
| AFB3 | Auxin receptor | Unknown | Induced by nitrate and involved in the regulation of nitrate in primary and lateral root growth | Target of miR393 | ||
| NAC4 | NAM/ATAF/CUC | OBP4 | Regulating lateral roots induction | Yes | Integrated systems biology approach | |
| CLE | CLAVATA3/ESR-related | CLV1 | Regulating lateral roots elongation | Induced under N-deficiency | Upregulated by N deficiency | |
| CLV1 | XI LRR-RLKs | Feedback regulation of CLE | Regulating lateral roots emergence and length | unknown | Bound by CLE | |
| CEP | CEP | Unknown | Root-derived ascending signals to the shoot | unknown | Originating from N-starved roots | |
| HIN9/IWS1 | Component of RNAPII complexes | NRT2.1 | Involved in the transduction of N systemic signal | No | Forward genetics Screening | |
| TCP20 | TCP | NRT1.1, NIA, NRT2.1, and NiR | Regulating lateral root elongation under high nitrate | No | Binding to the promoter of NIA1 and NRT2.1 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhao, L.; Liu, F.; Crawford, N.M.; Wang, Y. Molecular Regulation of Nitrate Responses in Plants. Int. J. Mol. Sci. 2018, 19, 2039. https://doi.org/10.3390/ijms19072039
Zhao L, Liu F, Crawford NM, Wang Y. Molecular Regulation of Nitrate Responses in Plants. International Journal of Molecular Sciences. 2018; 19(7):2039. https://doi.org/10.3390/ijms19072039
Chicago/Turabian StyleZhao, Lufei, Fei Liu, Nigel M. Crawford, and Yong Wang. 2018. "Molecular Regulation of Nitrate Responses in Plants" International Journal of Molecular Sciences 19, no. 7: 2039. https://doi.org/10.3390/ijms19072039
APA StyleZhao, L., Liu, F., Crawford, N. M., & Wang, Y. (2018). Molecular Regulation of Nitrate Responses in Plants. International Journal of Molecular Sciences, 19(7), 2039. https://doi.org/10.3390/ijms19072039