miR-196a Is Able to Restore the Aggressive Phenotype of Annexin A1 Knock-Out in Pancreatic Cancer Cells by CRISPR/Cas9 Genome Editing
Abstract
:1. Introduction
2. Results
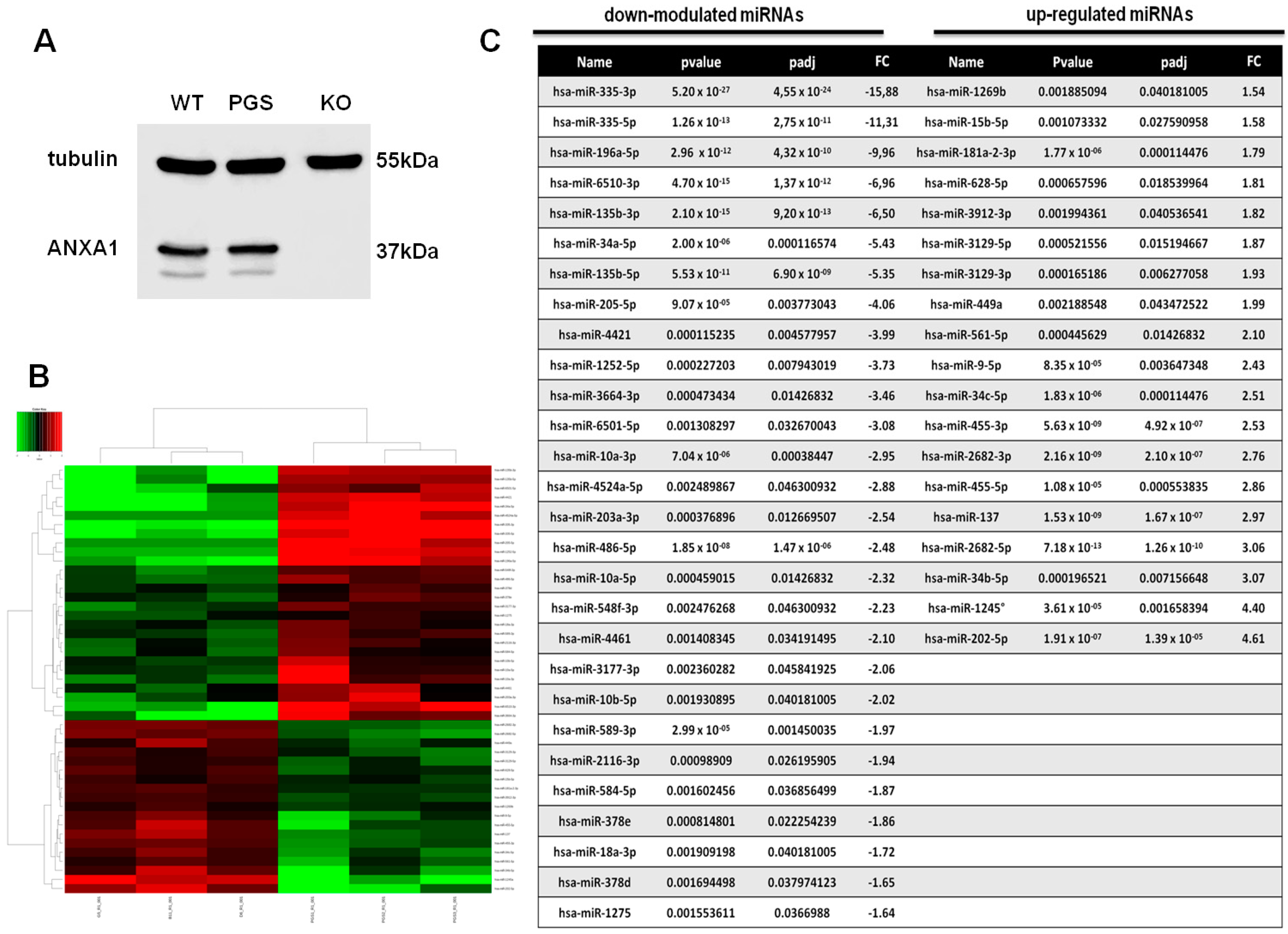
2.1. Differential miRNA Expression Profile in PGS and ANXA1 KO MIA PaCa-2 Cells
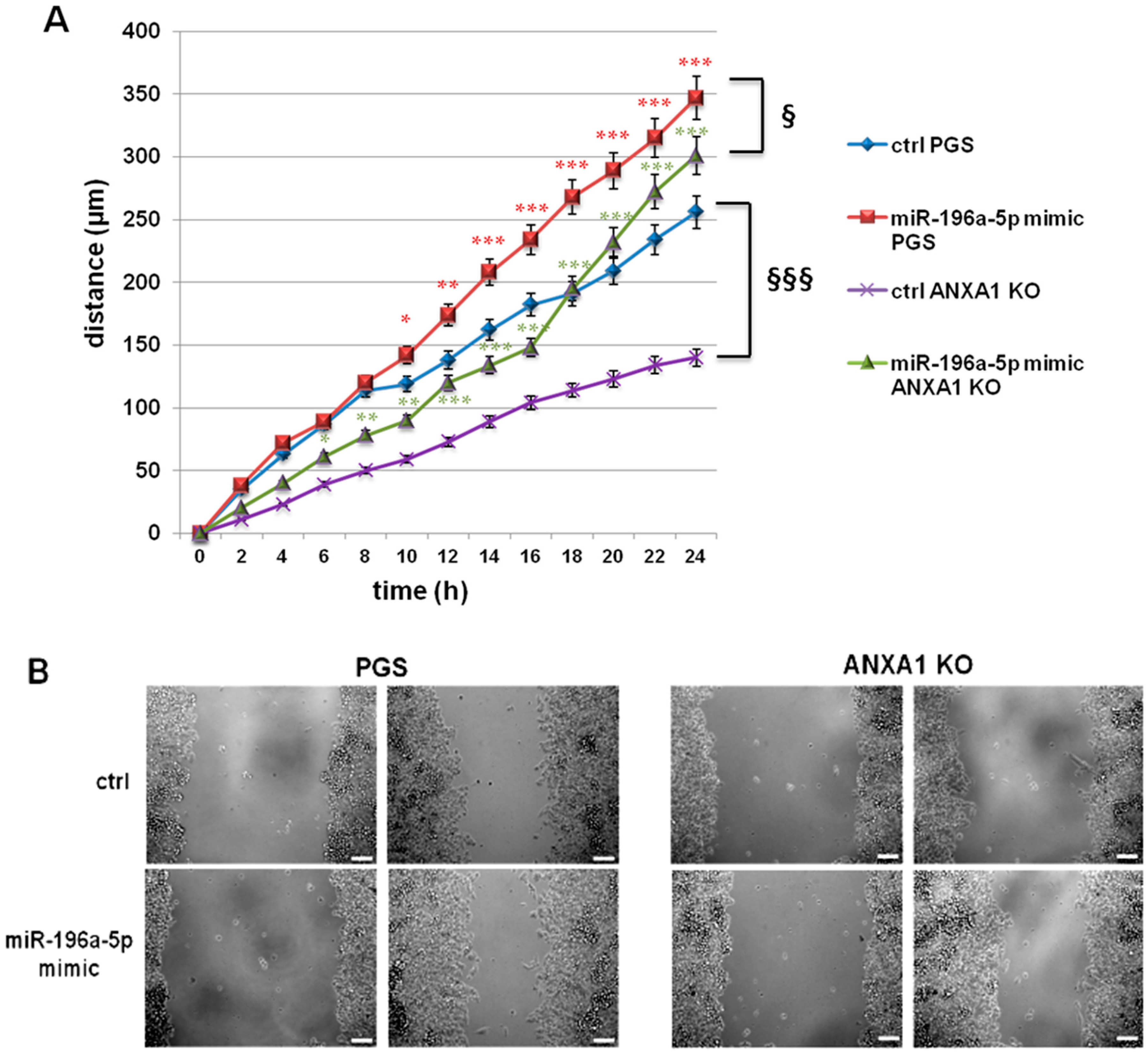
2.2. The miR-196a-5p Mimic Increased the Migration of PGS and ANXA1 KO MIA PaCa-2 Cells
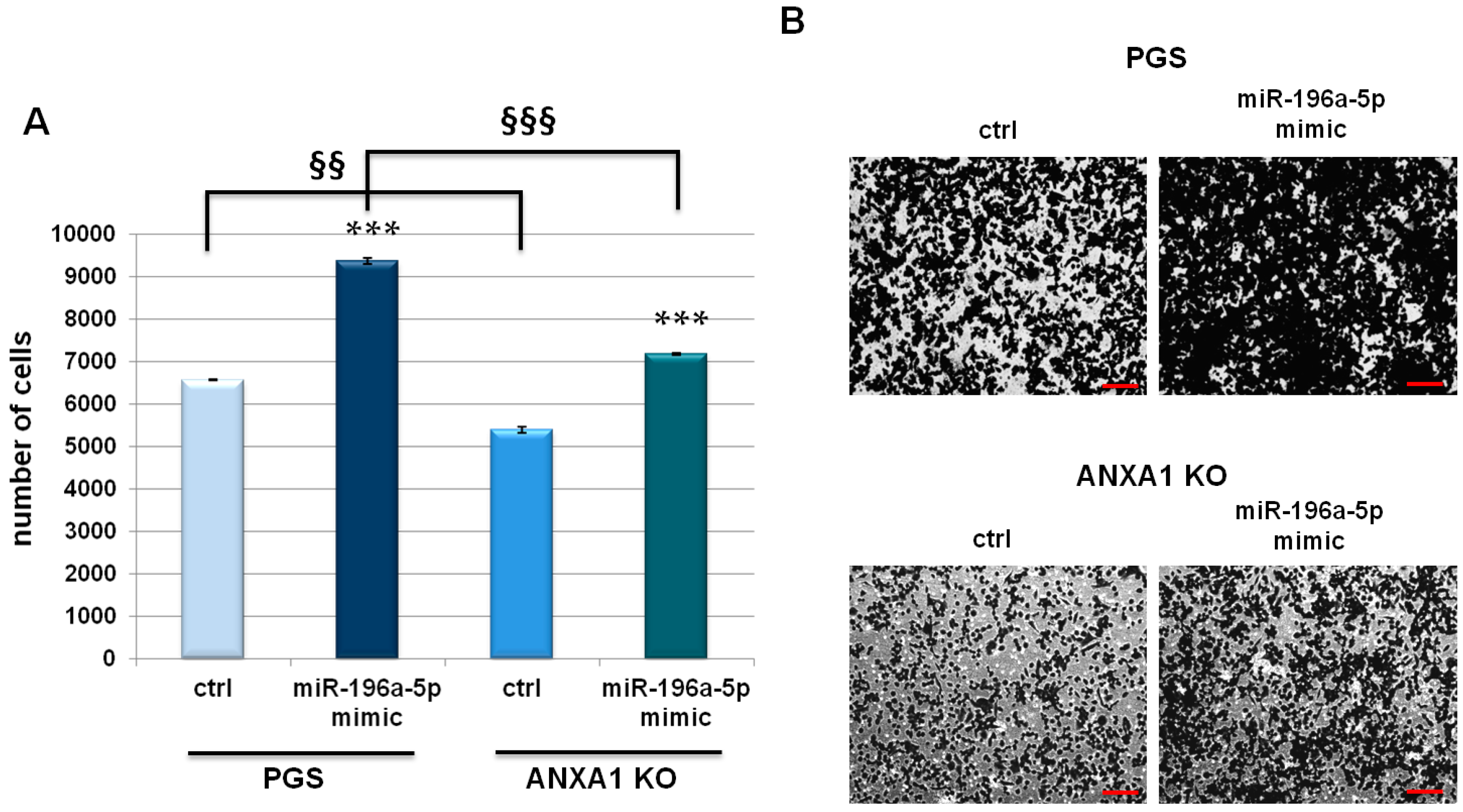
2.3. miR-196a-5p Affected PGS and ANXA1 KO MIA PaCa-2 Invasive Behaviour
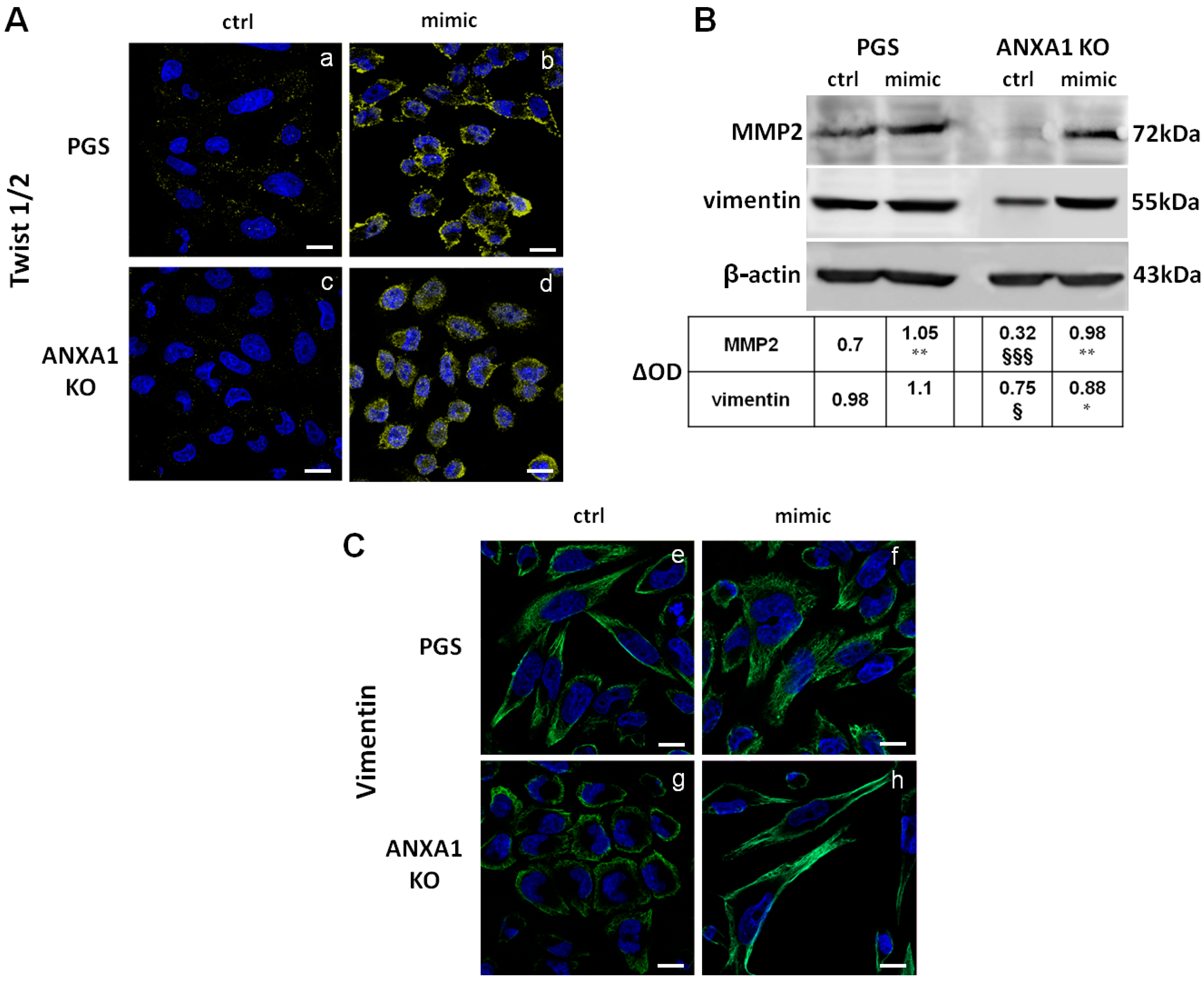
2.4. The miR-196a-5p Mimic Induced the Increase of Some EMT Markers in PGS and ANXA1 KO MIA PaCa-2 Cells
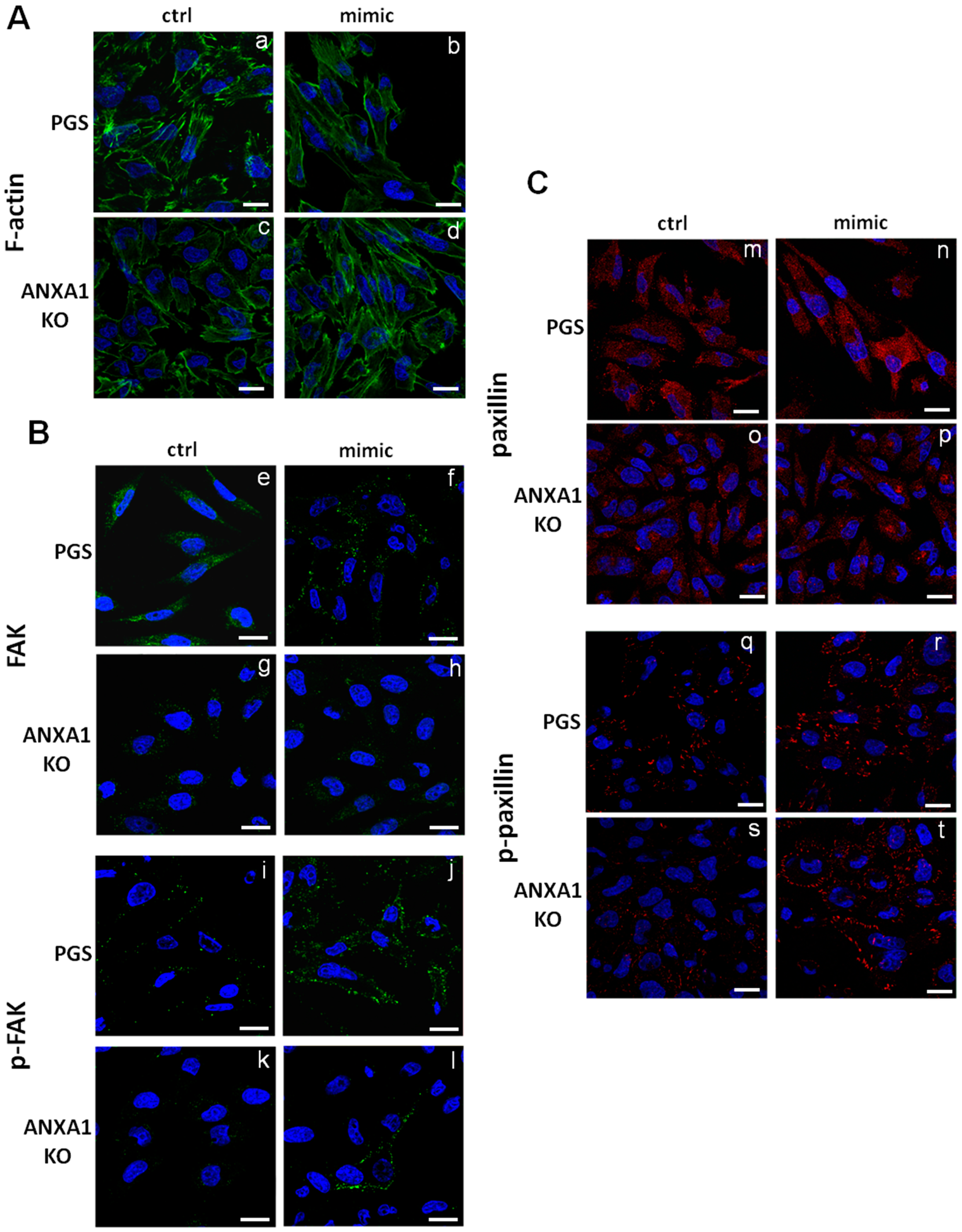
2.5. The miR-196a-5p Mimic Led to Cytoskeletal Organization Particularly on ANXA1 KO MIA PaCa-2 Cells
3. Discussion
4. Material and Methods
4.1. Cell Cultures
4.2. RNA Isolation and Quality Controls
4.3. Small RNA Sequencing and Data Analysis
4.4. Mimic Transfection
4.5. In Vitro Wound-Healing
4.6. Invasion Assay
4.7. Confocal Microscopy
4.8. Western Blotting
4.9. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Perretti, M.; Di Filippo, C.; D’Amico, M.; Dalli, J. Characterizing the anti-inflammatory and tissue protective actions of a novel Annexin A1 peptide. PLoS ONE 2017, 12, e0175786. [Google Scholar] [CrossRef] [PubMed]
- Perucci, L.O.; Sugimoto, M.A.; Gomes, K.; Dusse, L.M.; Teixeira, M.M.; Sousa, L.P. Annexin A1 and specialized proresolving lipid mediators: Promoting resolution as a therapeutic strategy in human inflammatory diseases. Expert Opin Ther. Targets 2017, 21, 879–896. [Google Scholar] [CrossRef] [PubMed]
- D’Acunto, C.W.; Gbelcova, H.; Festa, M.; Ruml, T. The complex understanding of Annexin A1 phosphorylation. Cell Signal 2014, 26, 173–178. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yi, M.; Schnitzer, J.E. Impaired tumor growth, metastasis, angiogenesis and wound healing in annexin A1-null mice. Proc. Natl. Acad. Sci. USA 2009, 106, 17886–17891. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rondepierre, F.; Bouchon, B.; Papon, J.; Bonnet-Duquennoy, M.; Kintossou, R.; Moins, N.; Maublant, J.; Madelmont, J.C.; D’Incan, M.; Degoul, F. Proteomic studies of B16 lines: Involvement of annexin A1 in melanoma dissemination. Biochim. Biophys. Acta 2009, 1794, 61–69. [Google Scholar] [CrossRef] [PubMed]
- Bhardwaj, A.; Ganesan, N.; Tachibana, K.; Rajapakshe, K.; Albarracin, C.T.; Gunaratne, P.H.; Coarfa, C.; Bedrosian, I. Annexin A1 Preferentially Predicts Poor Prognosis of Basal-Like Breast Cancer Patients by Activating mTOR-S6 Signaling. PLoS ONE 2015, 10, e0127678. [Google Scholar] [CrossRef] [PubMed]
- Bizzarro, V.; Belvedere, R.; Migliaro, V.; Romano, E.; Parente, L.; Petrella, A. Hypoxia regulates ANXA1 expression to support prostate cancer cell invasion and aggressiveness. Cell Adhes. Migr. 2017, 11, 247–260. [Google Scholar] [CrossRef] [PubMed]
- Boudhraa, Z.; Bouchon, B.; Viallard, C.; D’Incan, M.; Degoul, F. Annexin A1 localization and its relevance to cancer. Clin. Sci. 2016, 130, 205–220. [Google Scholar] [CrossRef] [PubMed]
- Rong, B.; Cai, X.; Yang, S. Annexin A1 in malignant tumors: Current opinions and controversies. Int. J. Biol. Markers 2014, 29, e8–e20. [Google Scholar]
- Chen, C.Y.; Shen, J.Q.; Wang, F.; Wan, R.; Wang, X.P. Prognostic significance of annexin A1 expression in pancreatic ductal adenocarcinoma. Asian Pac. J. Cancer Prev. 2012, 13, 4707–4712. [Google Scholar] [CrossRef] [PubMed]
- Bai, X.F.; Ni, X.G.; Zhao, P.; Liu, S.M.; Wang, H.X.; Guo, B.; Zhou, L.P.; Liu, F.; Zhang, J.S.; Wang, K.; et al. Overexpression of annexin 1 in pancreatic cancer and its clinical significance. World J. Gastroenterol. 2004, 10, 1466–1470. [Google Scholar] [CrossRef] [PubMed]
- Belvedere, R.; Bizzarro, V.; Popolo, A.; Dal Piaz, F.; Vasaturo, M.; Picardi, P.; Parente, L.; Petrella, A. Role of intracellular and extracellular annexin A1 in migration and invasion of human pancreatic carcinoma cells. BMC Cancer 2014, 14, 961. [Google Scholar] [CrossRef] [PubMed]
- Belvedere, R.; Bizzarro, V.; Forte, G.; Dal Piaz, F.; Parente, L.; Petrella, A. Annexin A1 contributes to pancreatic cancer cell phenotype, behaviour and metastatic potential independently of Formyl Peptide Receptor pathway. Sci. Rep. 2016, 6, 29660. [Google Scholar] [CrossRef] [PubMed]
- Reddy, K.B. MicroRNA (miRNA) in cancer. Cancer Cell Int. 2015, 15, 38. [Google Scholar] [CrossRef] [PubMed]
- Ha, M.; Kim, V.N. Regulation of microRNA biogenesis. Nat. Rev. Mol. Cell Biol. 2014, 158, 509–524. [Google Scholar] [CrossRef] [PubMed]
- Bartel, D.P. MicroRNAs: Target recognition and regulatory functions. Cell 2009, 136, 215–233. [Google Scholar] [CrossRef] [PubMed]
- Lewis, B.P.; Burge, C.B.; Bartel, D.P. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 2005, 1201, 15–20. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Zhang, Y.; Zhang, L.; Weakley, S.M.; Yao, Q. MicroRNA-196: Critical roles and clinical applications in development and cancer. J. Cell Mol. Med. 2011, 15, 14–23. [Google Scholar] [CrossRef] [PubMed]
- Calin, G.A.; Sevignani, C.; Dumitru, C.D.; Hyslop, T.; Noch, E.; Yendamuri, S.; Shimizu, M.; Rattan, S.; Bullrich, F.; Negrini, M.; et al. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc. Natl. Acad. Sci. USA 2004, 1019, 2999–3004. [Google Scholar] [CrossRef] [PubMed]
- Jafri, M.A.; Al-Qahtani, M.H.; Shay, J.W. Role of miRNAs in human cancer metastasis: Implications for therapeutic intervention. Semin. Cancer Biol. 2017, 44, 117–131. [Google Scholar] [CrossRef] [PubMed]
- Slater, E.P.; Strauch, K.; Rospleszcz, S.; Ramaswamy, A.; Esposito, I.; Kloppel, G.; Matthai, E.; Heeger, K.; Fendrich, V.; Langer, P.; et al. MicroRNA-196a and -196b as Potential Biomarkers for the Early Detection of Familial Pancreatic Cancer. Transl. Oncol. 2014, 74, 464–471. [Google Scholar] [CrossRef] [PubMed]
- Kong, X.; Du, Y.; Wang, G.; Gao, J.; Gong, Y.; Li, L.; Zhang, Z.; Zhu, J.; Jing, Q.; Qin, Y.; et al. Detection of differentially expressed microRNAs in serum of pancreatic ductal adenocarcinoma patients: MiR-196a could be a potential marker for poor prognosis. Digest. Dis. Sci. 2011, 562, 602–609. [Google Scholar] [CrossRef] [PubMed]
- Debernardi, S.; Skoulakis, S.; Molloy, G.; Chaplin, T.; Dixon-McIver, A.; Young, B.D. MicroRNA miR-181a correlates with morphological sub-class of acute myeloid leukaemia and the expression of its target genes in global genome-wide analysis. Leukemia 2007, 21, 912–916. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Braig, S.; Mueller, D.W.; Rothhammer, T.; Bosserhoff, A.K. MicroRNA miR-196a is a central regulator of HOX-B7 and BMP4 expression in malignant melanoma. Cell Mol. Life Sci. 2010, 67, 3535–3548. [Google Scholar] [CrossRef] [PubMed]
- Mueller, D.W.; Bosserhoff, A.K. MicroRNA miR-196a controls melanoma-associated genes by regulating HOX-C8 expression. Int. J. Cancer 2011, 129, 1064–1074. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, J.; Chen, J.; Chang, P.; LeBlanc, A.; Li, D.; Abbruzzesse, J.L.; Frazier, M.L.; Killary, A.M.; Sen, S. MicroRNAs in plasma of pancreatic ductal adenocarcinoma patients as novel blood-based biomarkers of disease. Cancer Prev. Res. 2009, 2, 807–813. [Google Scholar] [CrossRef] [PubMed]
- Szafranska, A.E.; Davison, T.S.; John, J.; Cannon, T.; Sipos, B.; Maghnouj, A.; Labourier, E.; Hahn, S.A. MicroRNA expression alterations are linked to tumorigenesis and non-neoplastic processes in pancreatic ductal adenocarcinoma. Oncogene 2007, 26, 4442–4452. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gaianigo, N.; Melisi, D.; Carbone, C. EMT and Treatment Resistance in Pancreatic Cancer. Cancers 2017, 9, 122. [Google Scholar] [CrossRef] [PubMed]
- Huang, F.; Tang, J.; Zhuang, X.; Zhuang, Y.; Cheng, W.; Chen, W.; Yao, H.; Zhang, S. MiR-196a Promotes Pancreatic Cancer Progression by Targeting Nuclear Factor Kappa-B-Inhibitor Alpha. PLoS ONE 2014, 9, e87897. [Google Scholar] [CrossRef] [PubMed]
- Luthra, R.; Singh, R.R.; Luthra, M.G.; Li, Y.X.; Hannah, C.; Romans, A.M.; Barkoh, B.A.; Chen, S.S.; Ensor, J.; Maru, D.M.; et al. MicroRNA-196a targets annexin A1: A microRNA-mediated mechanism of annexin A1 downregulation in cancers. Oncogene 2008, 27, 6667–6678. [Google Scholar] [CrossRef] [PubMed]
- Yuan, Y.; Anbalagan, D.; Lee, L.H.; Samy, R.P.; Shanmugam, M.K.; Kumar, A.P.; Sethi, G.; Lobie, P.E.; Lim, L.H. ANXA1 inhibits miRNA-196a in a negative feedback loop through NF-kB and c-Myc to reduce breast cancer proliferation. Oncotarget 2016, 7, 27007–27020. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Panero, R.; Rinaldi, A.; Memoli, D.; Nassa, G.; Ravo, M.; Rizzo, F.; Tarallo, R.; Milanesi, L.; Weisz, A.; Giurato, G. iSmaRT: A toolkit for a comprehensive analysis of small RNA-Seq data. Bioinformatics 2017, 33, 938–940. [Google Scholar] [CrossRef] [PubMed]
- Yonemori, K.; Kurahara, H.; Maemura, K.; Natsugoe, S. MicroRNA in pancreatic cancer. J. Hum. Genet. 2017, 62, 33–40. [Google Scholar] [CrossRef] [PubMed]
- Lamouille, S.; Subramanyam, D.; Blelloch, R.; Derynck, R. Regulation of epithelial-mesenchymal and mesenchymal-epithelial transitions by microRNAs. Curr. Opin. Cell Biol. 2013, 25, 200–207. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Wang, Y.; Chen, Z.; Sternlicht, M.D.; Hidalgo, M.; Steffensen, B. Matrix metalloproteinase-2 contributes to cancer cell migration on collagen. Cancer Res. 2005, 65, 130–136. [Google Scholar] [PubMed]
- Zhao, X.; Guan, J.L. Focal adhesion kinase and its signaling pathways in cell migration and angiogenesis. Adv. Drug Deliv. Rev. 2011, 63, 610–615. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Turner, C.E. Paxillin and focal adhesion signalling. Nat. Cell Biol. 2000, 2, E231–E236. [Google Scholar] [CrossRef] [PubMed]
- Jin, C.; Liang, R. miR-205 promotes epithelial-mesenchymal transition by targeting AKT signaling in endometrial cancer cells. J. Obstet. Gynaecol. Res. 2015, 41, 1653–1660. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Li, L.; Li, Z.; Gong, G.; Chen, P.; Liu, H.; Wang, J.; Liu, Y.; Wu, X. The role of miR-205 in the VEGF-mediated promotion of human ovarian cancer cell invasion. Gynecol. Oncol. 2015, 137, 125–133. [Google Scholar] [CrossRef] [PubMed]
- Yan, Y.; Luo, Y.C.; Wan, H.Y.; Wang, J.; Zhang, P.P.; Liu, M.; Li, X.; Li, S.; Tang, H. MicroRNA-10a is involved in the metastatic process by regulating Eph tyrosine kinase receptor A4-mediated epithelial-mesenchymal transition and adhesion in hepatoma cells. Hepatology 2013, 57, 667–677. [Google Scholar] [CrossRef] [PubMed]
- Yan, Y.; Wang, Q.; Yan, X.L.; Zhang, Y.; Li, W.; Tang, F.; Li, X.; Yang, P. miR-10a controls glioma migration and invasion through regulating epithelial-mesenchymal transition via EphA8. FEBS Lett. 2015, 589, 756–765. [Google Scholar] [CrossRef] [PubMed]
- Ma, L.; Teruya-Feldstein, J.; Weinberg, R.A. Tumour invasion and metastasis initiated by microRNA-10b in breast cancer. Nature 2007, 449, 682–688. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Zhu, J.; Cao, H.; Ren, H.; Fang, X. miR-10b promotes cell invasion through RhoC-AKT signaling pathway by targeting HOXD10 in gastric cancer. Int. J. Oncol. 2012, 40, 1553–1560. [Google Scholar] [PubMed]
- Zhang, P.; Hong, H.; Sun, X.; Jiang, H.; Ma, S.; Zhao, S.; Zhang, M.; Wang, Z.; Jiang, C.; Liu, H. MicroRNA-10b regulates epithelial-mesenchymal transition by modulating KLF4/Notch1/E-cadherin in cisplatin-resistant nasopharyngeal carcinoma cells. Am. J. Cancer Res. 2016, 6, 141–156. [Google Scholar] [PubMed]
- Nakata, K.; Ohuchida, K.; Mizumoto, K.; Kayashima, T.; Ikenaga, N.; Sakai, H.; Lin, C.; Fujita, H.; Otsuka, T.; Aishima, S.; et al. MicroRNA-10b is overexpressed in pancreatic cancer, promotes its invasiveness, and correlates with a poor prognosis. Surgery 2011, 150, 916–922. [Google Scholar] [CrossRef] [PubMed]
- Hagman, Z.; Haflidadottir, B.S.; Ansari, M.; Persson, M.; Bjartell, A.; Edsjö, A.; Ceder, Y. The tumour suppressor miR-34c targets MET in prostate cancer cells. Br. J. Cancer 2013, 109, 1271–1278. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cannell, I.G.; Kong, Y.W.; Johnston, S.J.; Chen, M.L.; Collins, H.M.; Dobbyn, H.C.; Elia, A.; Kress, T.R.; Dickens, M.; Clemens, M.J.; et al. p38 MAPK/MK2-mediated induction of miR-34c following DNA damage prevents Myc-dependent DNA replication. Proc. Natl. Acad. Sci. USA 2010, 107, 5375–5380. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Y.J.; Ping, C.; Tang, J.; Zhang, W. MicroRNA-455 suppresses non-small cell lung cancer through targeting ZEB1. Cell Biol. Int. 2016, 40, 621–628. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Z.; Lv, B.; Zhang, L.; Zhao, N.; Lv, Y. miR-202 functions as a tumor suppressor in non-small cell lung cancer by targeting STAT3. Mol. Med. Rep. 2017, 16, 2281–2289. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Li, C.; Wang, M.; Su, L.; Qu, Y.; Li, J.; Yu, B.; Yan, M.; Yu, Y.; Liu, B.; Zhu, Z. Decrease of miR-202-3p expression, a novel tumor suppressor, in gastric cancer. PLoS ONE 2013, 8, e69756. [Google Scholar] [CrossRef] [PubMed]
- Xiao, J.; Peng, F.; Yu, C.; Wang, M.; Li, X.; Li, Z.; Jiang, J.; Sun, C. microRNA-137 modulates pancreatic cancer cells tumor growth, invasion and sensitivity to chemotherapy. Int. J. Clin. Exp. Pathol. 2014, 7, 7442–7450. [Google Scholar] [PubMed]
- Neault, M.; Mallette, F.A.; Richard, S. miR-137 Modulates a Tumor Suppressor Network-Inducing Senescence in Pancreatic Cancer Cells. Cell Rep. 2016, 14, 1966–1978. [Google Scholar] [CrossRef] [PubMed]
- Guerriero, I.; D’Angelo, D.; Pallante, P.; Santos, M.; Scrima, M.; Malanga, D.; De Marco, C.; Ravo, M.; Weisz, A.; Laudanna, C.; et al. Analysis of miRNA profiles identified miR-196a as a crucial mediator of aberrant PI3K/AKT signaling in lung cancer cells. Oncotarget 2017, 8, 19172–19191. [Google Scholar] [CrossRef] [PubMed]
- Hoffman, A.E.; Zheng, T.; Yi, C.; Leaderer, D.; Weidhaas, J.; Slack, F.; Zhang, Y.; Paranjape, T.; Zhu, Y. microRNA miR-196a-2 and breast cancer: A genetic and epigenetic association study and functional analysis. Cancer Res. 2009, 69, 5970–5977. [Google Scholar] [CrossRef] [PubMed]
- Yang, B.; Li, S.Z.; Ma, L.; Liu, H.L.; Liu, J.; Shao, J.J. Expression and mechanism of action of miR-196a in epithelial ovarian cancer. Asian Pac. J. Trop. Med. 2016, 9, 1105–1110. [Google Scholar] [CrossRef] [PubMed]
- Saito, K.; Inagaki, K.; Kamimoto, T.; Ito, Y.; Sugita, T.; Nakajo, S.; Hirasawa, A.; Iwamaru, A.; Ishikura, T.; Hanaoka, H.; et al. MicroRNA-196a is a putative diagnostic biomarker and therapeutic target for laryngeal cancer. PLoS ONE. 2013, 8, e71480. [Google Scholar] [CrossRef] [PubMed]
- Tsai, K.W.; Liao, Y.L.; Wu, C.W.; Hu, L.Y.; Li, S.C.; Chan, W.C.; Ho, M.R.; Lai, C.H.; Kao, H.W.; Fang, W.L.; et al. Aberrant expression of miR-196a in gastric cancers and correlation with recurrence. Genes Chromosomes Cancer 2012, 51, 394–401. [Google Scholar] [CrossRef] [PubMed]
- Suh, Y.E.; Raulf, N.; Gäken, J.; Lawler, K.; Urbano, T.G.; Bullenkamp, J.; Gobeil, S.; Huot, J.; Odell, E.; Tavassoli, M. microRNA-196a promotes an oncogenic effect in head and neck cancer cells by suppressing annexin A1 and enhancing radioresistance. Int. J. Cancer 2014, 137, 1021–1034. [Google Scholar] [CrossRef] [PubMed]
- Pin, A.L.; Houle, F.; Fournier, P.; Guillonneau, M.; Paquet, É.R.; Simard, M.J.; Royal, I.; Huot, J. Annexin-1-mediated endothelial cell migration and angiogenesis are regulated by vascular endothelial growth factor (VEGF)-induced inhibition of miR-196a expression. J. Biol. Chem. 2012, 287, 30541–30551. [Google Scholar] [CrossRef] [PubMed]
- Álvarez-Teijeiro, S.; Menéndez, S.T.; Villaronga, M.Á.; Pena-Alonso, E.; Rodrigo, J.P.; Morgan, R.O.; Granda-Díaz, R.; Salom, C.; Fernandez, M.P.; García-Pedrero, J.M. Annexin A1 down-regulation in head and neck squamous cell carcinoma is mediated via transcriptional control with direct involvement of miR-196a/b. Sci. Rep. 2017, 7, 6790. [Google Scholar] [CrossRef] [PubMed]
- Bist, P.; Phua, Q.H.; Shu, S.; Yi, Y.; Anbalagan, D.; Lee, L.H.; Sethi, G.; Low, B.C.; Lim, L.H. Annexin-A1 controls an ERK-RhoA-NFκB activation loop in breast cancer cells. Biochem. Biophys. Res. Commun. 2015, 461, 47–53. [Google Scholar] [CrossRef] [PubMed]
- Bizzarro, V.; Belvedere, R.; Milone, M.R.; Pucci, B.; Lombardi, R.; Bruzzese, F.; Popolo, A.; Parente, L.; Budillon, A.; Petrella, A. Annexin A1 is involved in the acquisition and maintenance of a stem cell-like/aggressive phenotype in prostate cancer cells with acquired resistance to zoledronic acid. Oncotarget 2015, 6, 25076–25092. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Girard, P.R.; Nerem, R.M. Shear stress modulates endothelial cell morphology and F-actin organization through the regulation of focal adhesion-associated proteins. J. Cell Physiol. 1995, 163, 179–193. [Google Scholar] [CrossRef] [PubMed]
- Parsons, J.T.; Martin, K.H.; Slack, J.K.; Taylor, J.M.; Weed, S.A. Focal adhesion kinase: A regulator of focal adhesion dynamics and cell movement. Oncogene 2000, 19, 5606–5613. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]
- Saeed, A.I.; Sharov, V.; White, J.; Li, J.; Liang, W.; Bhagabati, N.; Braisted, J.; Klapa, M.; Currier, T.; Thiagarajan, M.; et al. TM4: A free, open-source system for microarray data management and analysis. Biotechniques 2003, 34, 374–378. [Google Scholar] [PubMed]
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Belvedere, R.; Saggese, P.; Pessolano, E.; Memoli, D.; Bizzarro, V.; Rizzo, F.; Parente, L.; Weisz, A.; Petrella, A. miR-196a Is Able to Restore the Aggressive Phenotype of Annexin A1 Knock-Out in Pancreatic Cancer Cells by CRISPR/Cas9 Genome Editing. Int. J. Mol. Sci. 2018, 19, 1967. https://doi.org/10.3390/ijms19071967
Belvedere R, Saggese P, Pessolano E, Memoli D, Bizzarro V, Rizzo F, Parente L, Weisz A, Petrella A. miR-196a Is Able to Restore the Aggressive Phenotype of Annexin A1 Knock-Out in Pancreatic Cancer Cells by CRISPR/Cas9 Genome Editing. International Journal of Molecular Sciences. 2018; 19(7):1967. https://doi.org/10.3390/ijms19071967
Chicago/Turabian StyleBelvedere, Raffaella, Pasquale Saggese, Emanuela Pessolano, Domenico Memoli, Valentina Bizzarro, Francesca Rizzo, Luca Parente, Alessandro Weisz, and Antonello Petrella. 2018. "miR-196a Is Able to Restore the Aggressive Phenotype of Annexin A1 Knock-Out in Pancreatic Cancer Cells by CRISPR/Cas9 Genome Editing" International Journal of Molecular Sciences 19, no. 7: 1967. https://doi.org/10.3390/ijms19071967





