Sequencing, Characterization, and Comparative Analyses of the Plastome of Caragana rosea var. rosea
Abstract
1. Introduction
2. Results
2.1. General Features of the Plastome
2.2. Gene Loss Analysis
2.3. Repeat Analysis
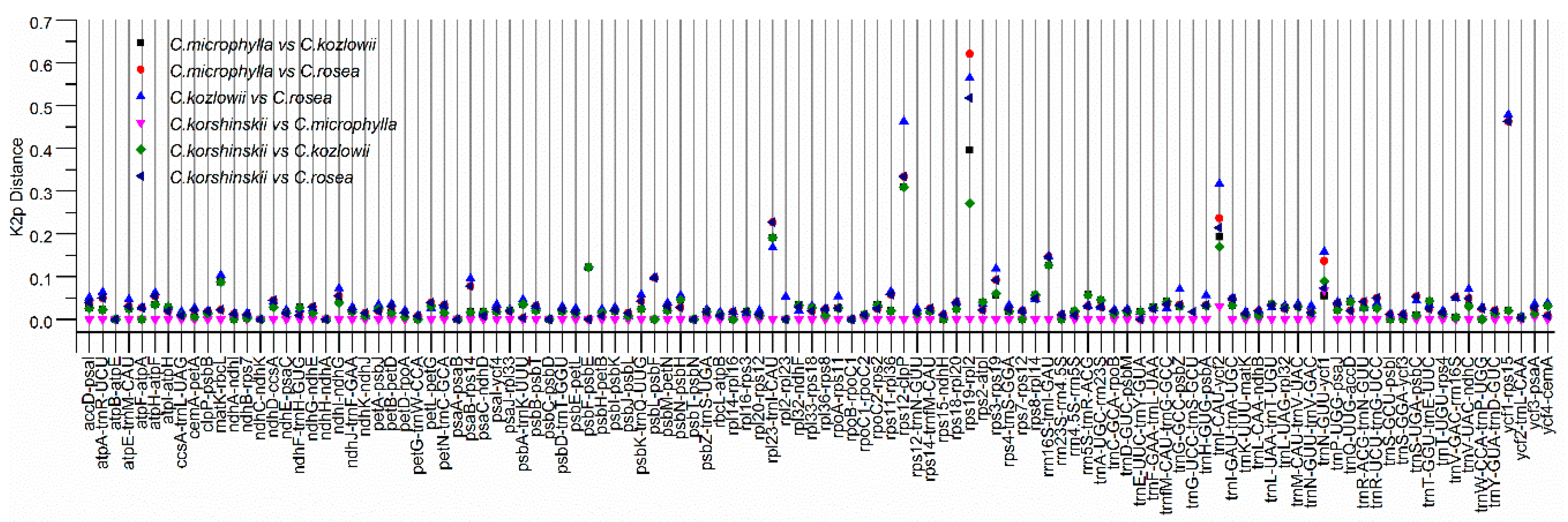
2.4. Sequence Divergence Analysis among Caragana Species
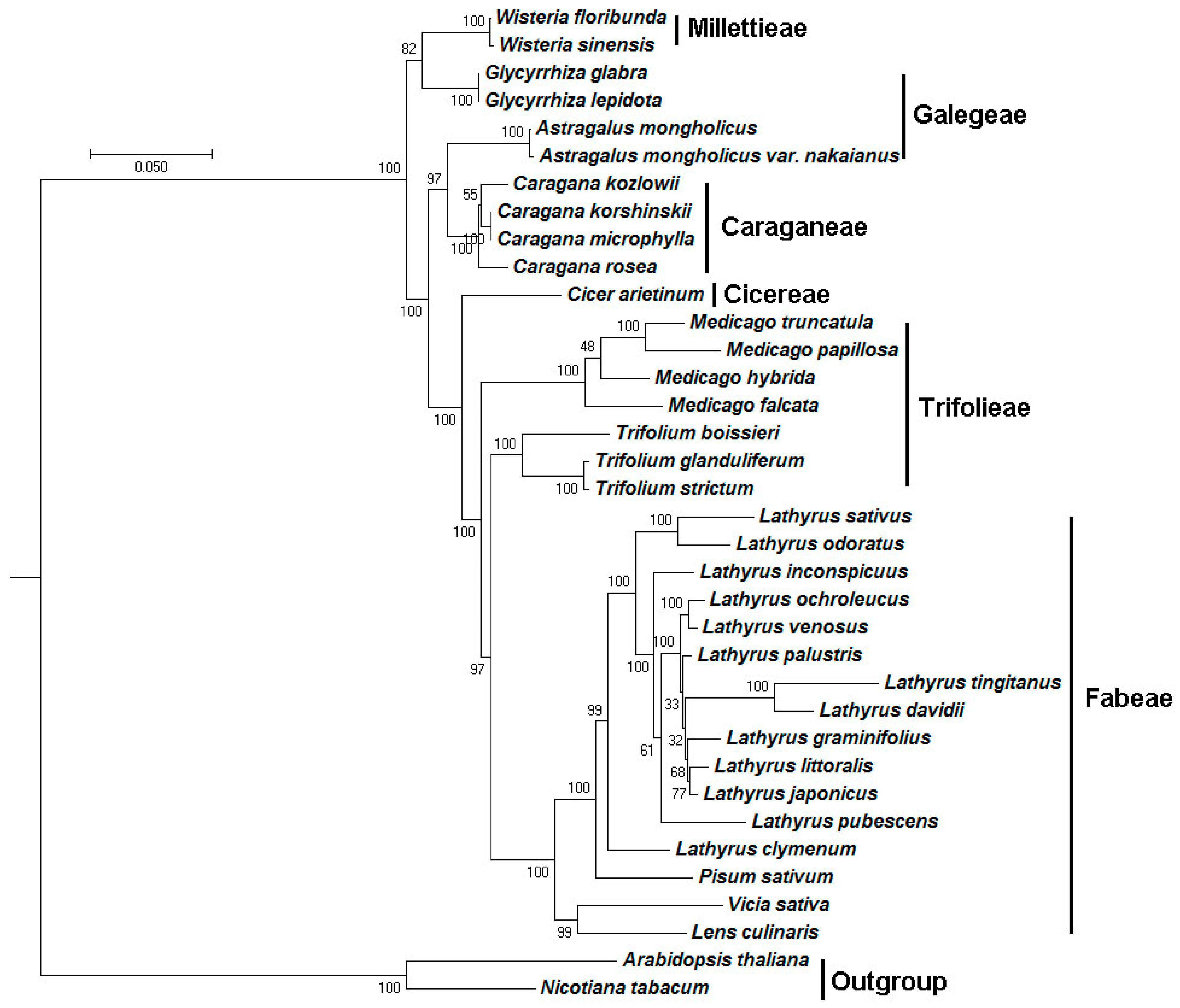
2.5. Phylogenetic Analysis
2.6. Selective Pressure Analysis
3. Discussion
4. Materials and Methods
4.1. Plant Material, DNA Extraction, and Sequencing
4.2. Genome Assembly and Gap Filling
4.3. Genome Annotation and Characteristics Analysis
4.4. Repeat and SSR Analysis
4.5. Comparative Genomic Analysis
4.6. Phylogenetic Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Meng, Q.; Niu, Y.; Niu, X.; Roubin, R.H.; Hanrahan, J.R. Ethnobotany, phytochemistry and pharmacology of the genus Caragana used in traditional chinese medicine. J. Ethnopharmacol. 2009, 124, 350–368. [Google Scholar] [CrossRef] [PubMed]
- Delectis Flora Reipublicae Popularis Sinicae Agendae Academiae Sinicae Edita. Flora Reipublicae Popularis Sinicae; Science Press: Beijing, China, 1993; Volume 42, p. 18. [Google Scholar]
- Ma, F.; Na, X.; Xu, T. Drought responses of three closely related Caragana species: Implication for their vicarious distribution. Ecol. Evol. 2016, 6, 2763–2773. [Google Scholar] [CrossRef] [PubMed]
- Delectis Flora Reipublicae Popularis Sinicae Agendae Academiae Sinicae Edita. Flora Reipublicae Popularis Sinicae; Science Press: Beijing, China, 1993; Volume 42, p. 31. [Google Scholar]
- Delectis Flora Reipublicae Popularis Sinicae Agendae Academiae Sinicae Edita. Flora Reipublicae Popularis Sinicae; Science Press: Beijing, China, 1993; Volume 42, p. 60. [Google Scholar]
- Yang, G.X.; Qi, J.B.; Cheng, K.J.; Hu, C.Q. Anti-HIV chemical constituents of aerial parts of Caragana rosea. Yao Xue Xue Bao (Acta Pharm. Sin.) 2007, 42, 179–182. [Google Scholar]
- Zhu, J.K. Abiotic stress signaling and responses in plants. Cell 2016, 167, 313–324. [Google Scholar] [CrossRef] [PubMed]
- Hu, S.; Sablok, G.; Wang, B.; Qu, D.; Barbaro, E.; Viola, R.; Li, M.; Varotto, C. Plastome organization and evolution of chloroplast genes in Cardamine species adapted to contrasting habitats. BMC Genom. 2015, 16, 306. [Google Scholar] [CrossRef] [PubMed]
- Ometto, L.; Li, M.; Bresadola, L.; Varotto, C. Rates of evolution in stress-related genes are associated with habitat preference in two Cardamine lineages. BMC Evol. Biol. 2012, 12, 7. [Google Scholar] [CrossRef] [PubMed]
- Gong, C.; Bai, J.; Wang, J.; Zhou, Y.; Kang, T.; Wang, J.; Hu, C.; Guo, H.; Chen, P.; Xie, P.; et al. Carbon storage patterns of Caragana korshinskii in areas of reduced environmental moisture on the loess plateau, China. Sci. Rep. 2016, 6, 28883. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Liu, D.; Liu, Y. Evaluation on drought-resistant characteristics of ten Caragana species based on leaf micromorphological structure. J. Desert Res. 2016, 3, 708–717. [Google Scholar] [CrossRef]
- Yamamoto, Y.; Aminaka, R.; Yoshioka, M.; Khatoon, M.; Komayama, K.; Takenaka, D.; Yamashita, A.; Nijo, N.; Inagawa, K.; Morita, N.; et al. Quality control of photosystem II: Impact of light and heat stresses. Photosynth. Res. 2008, 98, 589–608. [Google Scholar] [CrossRef] [PubMed]
- Mulo, P.; Sakurai, I.; Aro, E.M. Strategies for psba gene expression in cyanobacteria, green algae and higher plants: From transcription to psii repair. Biochim. Biophys. Acta 2012, 1817, 247–257. [Google Scholar] [CrossRef] [PubMed]
- Wright, I.J.; Reich, P.B.; Westoby, M.; Ackerly, D.D.; Baruch, Z.; Bongers, F.; Cavender-Bares, J.; Chapin, T.; Cornelissen, J.H.; Diemer, M.; et al. The worldwide leaf economics spectrum. Nature 2004, 428, 821–827. [Google Scholar] [CrossRef] [PubMed]
- Neufeld, H.S. Plant physiological ecology. Photosynthetica 1999, 80, 1785–1787. [Google Scholar] [CrossRef]
- Sabir, J.; Schwarz, E.; Ellison, N.; Zhang, J.; Baeshen, N.A.; Mutwakil, M.; Jansen, R.; Ruhlman, T. Evolutionary and biotechnology implications of plastid genome variation in the inverted-repeat-lacking clade of legumes. Plant Biotechnol. J. 2014, 12, 743–754. [Google Scholar] [CrossRef] [PubMed]
- Cardoso, D.; de Queiroz, L.P.; Pennington, R.T.; de Lima, H.C.; Fonty, E.; Wojciechowski, M.F.; Lavin, M. Revisiting the phylogeny of papilionoid legumes: New insights from comprehensively sampled early-branching lineages. Am. J. Bot. 2012, 99, 1991–2013. [Google Scholar] [CrossRef] [PubMed]
- Dugas, D.V.; Hernandez, D.; Koenen, E.J.; Schwarz, E.; Straub, S.; Hughes, C.E.; Jansen, R.K.; Nageswara-Rao, M.; Staats, M.; Trujillo, J.T.; et al. Mimosoid legume plastome evolution: Ir expansion, tandem repeat expansions, and accelerated rate of evolution in clpP. Sci. Rep. 2015, 5, 16958. [Google Scholar] [CrossRef] [PubMed]
- Jansen, R.K.; Wojciechowski, M.F.; Sanniyasi, E.; Lee, S.B.; Daniell, H. Complete plastid genome sequence of the chickpea (Cicer arietinum) and the phylogenetic distribution of rps12 and clpP intron losses among legumes (leguminosae). Mol. Phylogenet. Evol. 2008, 48, 1204–1217. [Google Scholar] [CrossRef] [PubMed]
- Qian, J.; Song, J.; Gao, H.; Zhu, Y.; Xu, J.; Pang, X.; Yao, H.; Sun, C.; Li, X.; Li, C.; et al. The complete chloroplast genome sequence of the medicinal plant Salvia miltiorrhiza. PLoS ONE 2013, 8, e57607. [Google Scholar] [CrossRef] [PubMed]
- Shen, X.; Wu, M.; Liao, B.; Liu, Z.; Bai, R.; Xiao, S.; Li, X.; Zhang, B.; Xu, J.; Chen, S. Complete chloroplast genome sequence and phylogenetic analysis of the medicinal plant Artemisia annua. Molecules 2017, 22, 1330. [Google Scholar] [CrossRef] [PubMed]
- He, L.; Qian, J.; Li, X.; Sun, Z.; Xu, X.; Chen, S. Complete chloroplast genome of medicinal plant Lonicera japonica: Genome rearrangement, intron gain and loss, and implications for phylogenetic studies. Molecules 2017, 22, 249. [Google Scholar] [CrossRef] [PubMed]
- Daniell, H.; Lin, C.S.; Yu, M.; Chang, W.J. Chloroplast genomes: Diversity, evolution, and applications in genetic engineering. Genome Biol. 2016, 17, 134. [Google Scholar] [CrossRef] [PubMed]
- Keller, J.; Rousseau-Gueutin, M.; Martin, G.E.; Morice, J.; Boutte, J.; Coissac, E.; Ourari, M.; Ainouche, M.; Salmon, A.; Cabello-Hurtado, F.; et al. The evolutionary fate of the chloroplast and nuclear rps16 genes as revealed through the sequencing and comparative analyses of four novel legume chloroplast genomes from Lupinus. DNA Res. Int. J. Rapid Publ. Rep. Genes Genomes 2017, 24, 343–358. [Google Scholar] [CrossRef] [PubMed]
- Jansen, R.K.; Saski, C.; Lee, S.B.; Hansen, A.K.; Daniell, H. Complete plastid genome sequences of three rosids (Castanea, Prunus, Theobroma): Evidence for at least two independent transfers of rpl22 to the nucleus. Mol. Biol. Evol. 2011, 28, 835–847. [Google Scholar] [CrossRef] [PubMed]
- Gantt, J.S.; Baldauf, S.L.; Calie, P.J.; Weeden, N.F.; Palmer, J.D. Transfer of rpl22 to the nucleus greatly preceded its loss from the chloroplast and involved the gain of an intron. EMBO J. 1991, 10, 3073–3078. [Google Scholar] [PubMed]
- Steane, D.A. Complete nucleotide sequence of the chloroplast genome from the tasmanian blue gum, Eucalyptus globulus (Myrtaceae). DNA Res. Int. J. Rapid Publ. Rep. Genes Genomes 2005, 12, 215–220. [Google Scholar] [CrossRef] [PubMed]
- Jansen, R.K.; Cai, Z.; Raubeson, L.A.; Daniell, H.; Depamphilis, C.W.; Leebens-Mack, J.; Muller, K.F.; Guisinger-Bellian, M.; Haberle, R.C.; Hansen, A.K.; et al. Analysis of 81 genes from 64 plastid genomes resolves relationships in angiosperms and identifies genome-scale evolutionary patterns. Proc. Nat. Acad. Sci. USA 2007, 104, 19369–19374. [Google Scholar] [CrossRef] [PubMed]
- Magee, A.M.; Aspinall, S.; Rice, D.W.; Cusack, B.P.; Semon, M.; Perry, A.S.; Stefanovic, S.; Milbourne, D.; Barth, S.; Palmer, J.D.; et al. Localized hypermutation and associated gene losses in legume chloroplast genomes. Genome Res. 2010, 20, 1700–1710. [Google Scholar] [CrossRef] [PubMed]
- Park, S.; Jansen, R.K.; Park, S. Complete plastome sequence of Thalictrum coreanum (ranunculaceae) and transfer of the rpl32 gene to the nucleus in the ancestor of the subfamily thalictroideae. BMC Plant Biol. 2015, 15, 40. [Google Scholar] [CrossRef] [PubMed]
- Ueda, M.; Fujimoto, M.; Arimura, S.; Murata, J.; Tsutsumi, N.; Kadowaki, K. Loss of the rpl32 gene from the chloroplast genome and subsequent acquisition of a preexisting transit peptide within the nuclear gene in Populus. Gene 2007, 402, 51–56. [Google Scholar] [CrossRef] [PubMed]
- Jo, Y.D.; Park, J.; Kim, J.; Song, W.; Hur, C.G.; Lee, Y.H.; Kang, B.C. Complete sequencing and comparative analyses of the pepper (Capsicum annuum L.) plastome revealed high frequency of tandem repeats and large insertion/deletions on pepper plastome. Plant Cell Rep. 2011, 30, 217–229. [Google Scholar] [CrossRef] [PubMed]
- Sloan, D.B.; Triant, D.A.; Forrester, N.J.; Bergner, L.M.; Wu, M.; Taylor, D.R. A recurring syndrome of accelerated plastid genome evolution in the angiosperm tribe Sileneae (caryophyllaceae). Mol. Phylogenet. Evol. 2014, 72, 82–89. [Google Scholar] [CrossRef] [PubMed]
- Martin, G.E.; Rousseau-Gueutin, M.; Cordonnier, S.; Lima, O.; Michon-Coudouel, S.; Naquin, D.; de Carvalho, J.F.; Ainouche, M.; Salmon, A.; Ainouche, A. The first complete chloroplast genome of the genistoid legume Lupinus luteus: Evidence for a novel major lineage-specific rearrangement and new insights regarding plastome evolution in the legume family. Annu. Bot. 2014, 113, 1197–1210. [Google Scholar] [CrossRef] [PubMed]
- Haberle, R.C.; Fourcade, H.M.; Boore, J.L.; Jansen, R.K. Extensive rearrangements in the chloroplast genome of Trachelium caeruleum are associated with repeats and tRNA genes. J. Mol. Evol. 2008, 66, 350–361. [Google Scholar] [CrossRef] [PubMed]
- Provan, J.; Corbett, G.; McNicol, J.W.; Powell, W. Chloroplast DNA variability in wild and cultivated rice (Oryza spp.) revealed by polymorphic chloroplast simple sequence repeats. Genome 1997, 40, 104–110. [Google Scholar] [CrossRef] [PubMed]
- Xue, J.; Wang, S.; Zhou, S.L. Polymorphic chloroplast microsatellite loci in Nelumbo (nelumbonaceae). Am. J. Bot. 2012, 99, e240–e244. [Google Scholar] [CrossRef] [PubMed]
- Pauwels, M.; Vekemans, X.; Gode, C.; Frerot, H.; Castric, V.; Saumitou-Laprade, P. Nuclear and chloroplast DNA phylogeography reveals vicariance among european populations of the model species for the study of metal tolerance, Arabidopsis halleri (Brassicaceae). New Phytol. 2012, 193, 916–928. [Google Scholar] [CrossRef] [PubMed]
- Kuang, D.Y.; Wu, H.; Wang, Y.L.; Gao, L.M.; Zhang, S.Z.; Lu, L. Complete chloroplast genome sequence of Magnolia kwangsiensis (Magnoliaceae): Implication for DNA barcoding and population genetics. Genome 2011, 54, 663–673. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.K. Salt and drought stress signal transduction in plants. Annu. Rev. Plant Biol. 2002, 53, 247–273. [Google Scholar] [CrossRef] [PubMed]
- Mignolet-Spruyt, L.; Xu, E.; Idanheimo, N.; Hoeberichts, F.A.; Muhlenbock, P.; Brosche, M.; van Breusegem, F.; Kangasjarvi, J. Spreading the news: Subcellular and organellar reactive oxygen species production and signalling. J. Exp. Bot. 2016, 67, 3831–3844. [Google Scholar] [CrossRef] [PubMed]
- Ichimura, K.; Mizoguchi, T.; Yoshida, R.; Yuasa, T.; Shinozaki, K. Various abiotic stresses rapidly activate Arabidopsis MAP kinases ATMPK4 and ATMPK6. Plant J. Cell Mol. Biol. 2000, 24, 655–665. [Google Scholar] [CrossRef]
- Iuchi, S.; Kobayashi, M.; Taji, T.; Naramoto, M.; Seki, M.; Kato, T.; Tabata, S.; Kakubari, Y.; Yamaguchi-Shinozaki, K.; Shinozaki, K. Regulation of drought tolerance by gene manipulation of 9-cis-epoxycarotenoid dioxygenase, a key enzyme in abscisic acid biosynthesis in Arabidopsis. Plant J. Cell Mol. Biol. 2001, 27, 325–333. [Google Scholar] [CrossRef]
- Ishitani, M.; Liu, J.; Halfter, U.; Kim, C.S.; Shi, W.; Zhu, J.K. Sos3 function in plant salt tolerance requires n-myristoylation and calcium binding. Plant Cell 2000, 12, 1667–1678. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.H.; Li, P.H.; Brenner, M.L. Involvement of abscisic acid in potato cold acclimation. Plant Physiol. 1983, 71, 362–365. [Google Scholar] [CrossRef] [PubMed]
- Choi, H.; Hong, J.; Ha, J.; Kang, J.; Kim, S.Y. ABFs, a family of ABA-responsive element binding factors. J. Biol. Chem. 2000, 275, 1723–1730. [Google Scholar] [CrossRef] [PubMed]
- Gustin, M.C.; Albertyn, J.; Alexander, M.; Davenport, K. MAP kinase pathways in the yeast Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. (MMBR) 1998, 62, 1264–1300. [Google Scholar] [PubMed]
- Jacob, T.; Ritchie, S.; Assmann, S.M.; Gilroy, S. Abscisic acid signal transduction in guard cells is mediated by phospholipase D activity. Proc. Nat. Acad. Sci. USA 1999, 96, 12192–12197. [Google Scholar] [CrossRef] [PubMed]
- Ingram, J.; Bartels, D. The molecular basis of dehydration tolerance in plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1996, 47, 377–403. [Google Scholar] [CrossRef] [PubMed]
- Jaglo-Ottosen, K.R.; Gilmour, S.J.; Zarka, D.G.; Schabenberger, O.; Thomashow, M.F. Arabidopsis CBF1 overexpression induces COR genes and enhances freezing tolerance. Science 1998, 280, 104–106. [Google Scholar] [CrossRef] [PubMed]
- Jonak, C.; Kiegerl, S.; Ligterink, W.; Barker, P.J.; Huskisson, N.S.; Hirt, H. Stress signaling in plants: A mitogen-activated protein kinase pathway is activated by cold and drought. Proc. Nat. Acad. Sci. USA 1996, 93, 11274–11279. [Google Scholar] [CrossRef] [PubMed]
- Knight, H.; Trewavas, A.J.; Knight, M.R. Calcium signalling in Arabidopsis thaliana responding to drought and salinity. Plant J. Cell Mol. Biol. 1997, 12, 1067–1078. [Google Scholar] [CrossRef]
- Katagiri, T.; Takahashi, S.; Shinozaki, K. Involvement of a novel Arabidopsis phospholipase D, AtPLDδ, in dehydration-inducible accumulation of phosphatidic acid in stress signalling. Plant J. Cell Mol. Biol. 2001, 26, 595–605. [Google Scholar] [CrossRef]
- Kovtun, Y.; Chiu, W.L.; Tena, G.; Sheen, J. Functional analysis of oxidative stress-activated mitogen-activated protein kinase cascade in plants. Proc. Nat. Acad. Sci. USA 2000, 97, 2940–2945. [Google Scholar] [CrossRef] [PubMed]
- Camacho, C.; Coulouris, G.; Avagyan, V.; Ma, N.; Papadopoulos, J.; Bealer, K.; Madden, T.L. Blast+: Architecture and applications. BMC Bioinform. 2009, 10, 421. [Google Scholar] [CrossRef] [PubMed]
- Simpson, J.T.; Wong, K.; Jackman, S.D.; Schein, J.E.; Jones, S.J.; Birol, I. ABySS: A parallel assembler for short read sequence data. Genome Res. 2009, 19, 1117–1123. [Google Scholar] [CrossRef] [PubMed]
- Langmead, B.; Trapnell, C.; Pop, M.; Salzberg, S.L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009, 10, R25. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Shi, L.; Zhu, Y.; Chen, H.; Zhang, J.; Lin, X.; Guan, X. CpGAVAS, an integrated web server for the annotation, visualization, analysis, and genbank submission of completely sequenced chloroplast genome sequences. BMC Genom. 2012, 13, 715. [Google Scholar] [CrossRef] [PubMed]
- Laslett, D.; Canback, B. ARAGORN, a program to detect tRNA genes and tmRNA genes in nucleotide sequences. Nucleic Acids Res. 2004, 32, 11–16. [Google Scholar] [CrossRef] [PubMed]
- Schattner, P.; Brooks, A.N.; Lowe, T.M. The tRNAscan-SE, snoscan and snoGPS web servers for the detection of tRNAs and snoRNAs. Nucleic Acids Res. 2005, 33, W686–W689. [Google Scholar] [CrossRef] [PubMed]
- Misra, S.; Harris, N. Using apollo to browse and edit genome annotations. Curr. Protocol. Bioinform. 2006. Chapter 9, Unit 9 5. [Google Scholar] [CrossRef]
- Lohse, M.; Drechsel, O.; Bock, R. Organellargenomedraw (OGDRAW): A tool for the easy generation of high-quality custom graphical maps of plastid and mitochondrial genomes. Curr. Genet. 2007, 52, 267–274. [Google Scholar] [CrossRef] [PubMed]
- Rice, P.; Longden, I.; Bleasby, A. Emboss: The European molecular biology open software suite. Trends Genet. (TIG) 2000, 16, 276–277. [Google Scholar] [CrossRef]
- Kurtz, S.; Choudhuri, J.V.; Ohlebusch, E.; Schleiermacher, C.; Stoye, J.; Giegerich, R. REPuter: The manifold applications of repeat analysis on a genomic scale. Nucleic Acids Res. 2001, 29, 4633–4642. [Google Scholar] [CrossRef] [PubMed]
- Tangphatsornruang, S.; Sangsrakru, D.; Chanprasert, J.; Uthaipaisanwong, P.; Yoocha, T.; Jomchai, N.; Tragoonrung, S. The chloroplast genome sequence of mungbean (Vigna radiata) determined by high-throughput pyrosequencing: Structural organization and phylogenetic relationships. DNA Res. Int. J. Rapid Publ. Rep. Genes Genomes 2010, 17, 11–22. [Google Scholar] [CrossRef] [PubMed]
- Benson, G. Tandem repeats finder: A program to analyze DNA sequences. Nucleic Acids Res. 1999, 27, 573–580. [Google Scholar] [CrossRef] [PubMed]
- Lei, W.; Ni, D.; Wang, Y.; Shao, J.; Wang, X.; Yang, D.; Wang, J.; Chen, H.; Liu, C.; Lei, W. Intraspecific and heteroplasmic variations, gene losses and inversions in the chloroplast genome of Astragalus membranaceus. Sci. Rep. 2016, 6, 21669. [Google Scholar] [CrossRef] [PubMed]
- Frazer, K.A.; Pachter, L.; Poliakov, A.; Rubin, E.M.; Dubchak, I. Vista: Computational tools for comparative genomics. Nucleic Acids Res. 2004, 32, W273–W279. [Google Scholar] [CrossRef] [PubMed]
- Stamatakis, A. Raxml version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef] [PubMed]
- Kimura, M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 1980, 16, 111–120. [Google Scholar] [CrossRef] [PubMed]
- Wernersson, R.; Pedersen, A.G. RevTrans: Multiple alignment of coding DNA from aligned amino acid sequences. Nucleic Acids Res. 2003, 31, 3537–3539. [Google Scholar] [CrossRef] [PubMed]
- Smith, M.D.; Wertheim, J.O.; Weaver, S.; Murrell, B.; Scheffler, K.; Kosakovsky Pond, S.L. Less is more: An adaptive branch-site random effects model for efficient detection of episodic diversifying selection. Mol. Biol. Evol. 2015, 32, 1342–1353. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Nielsen, R. Codon-substitution models for detecting molecular adaptation at individual sites along specific lineages. Mol. Biol. Evol. 2002, 19, 908–917. [Google Scholar] [CrossRef] [PubMed]
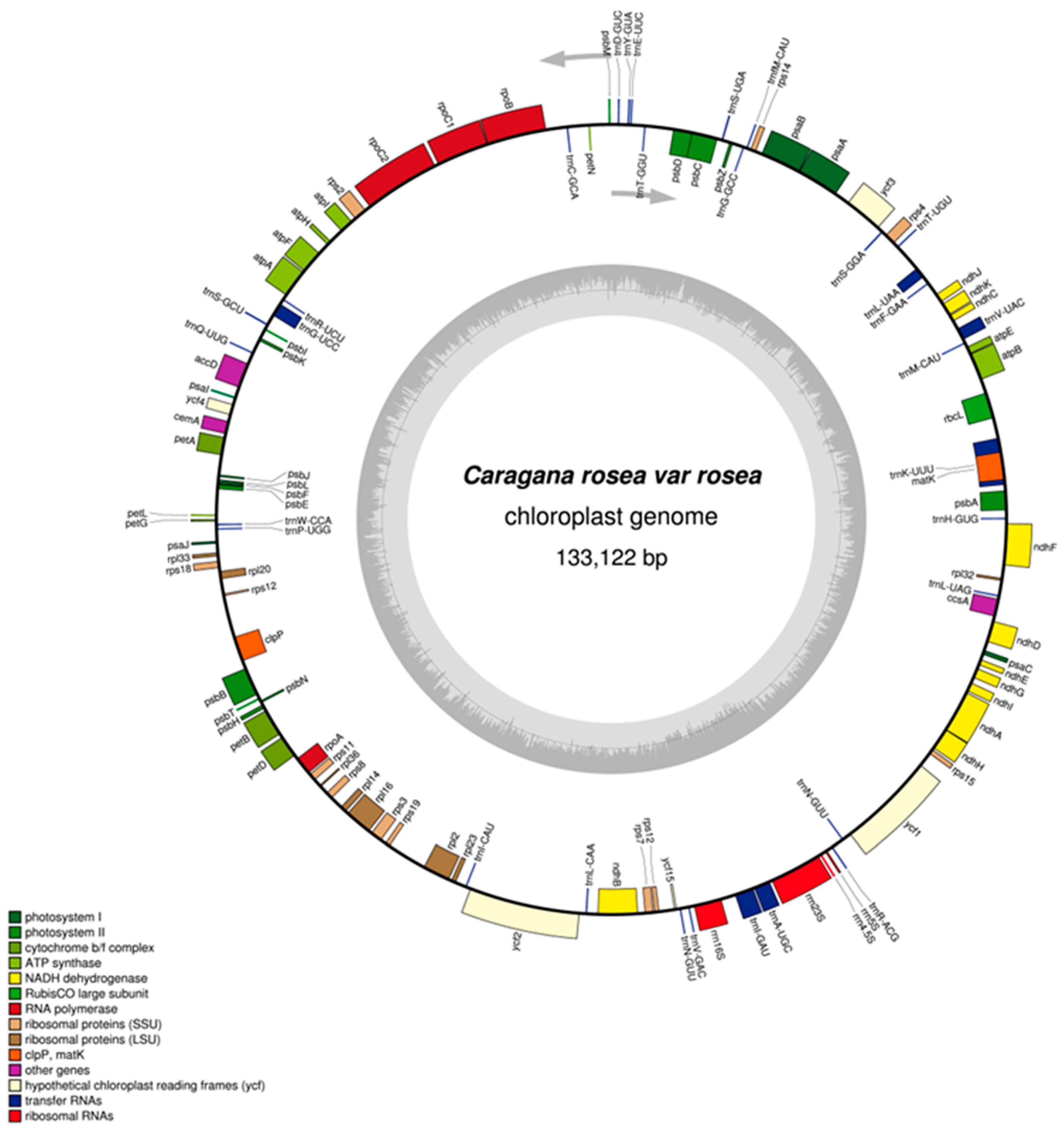
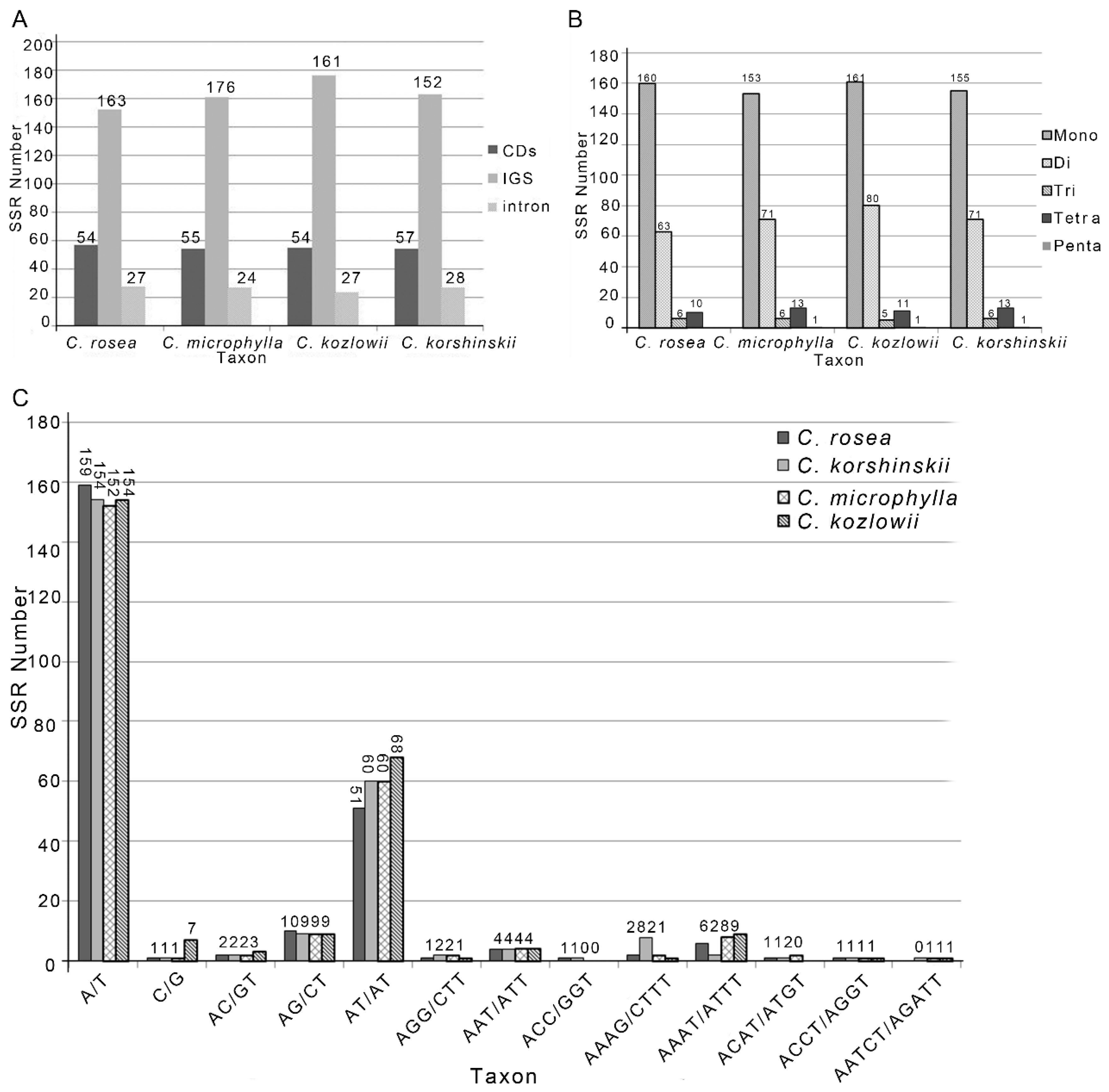
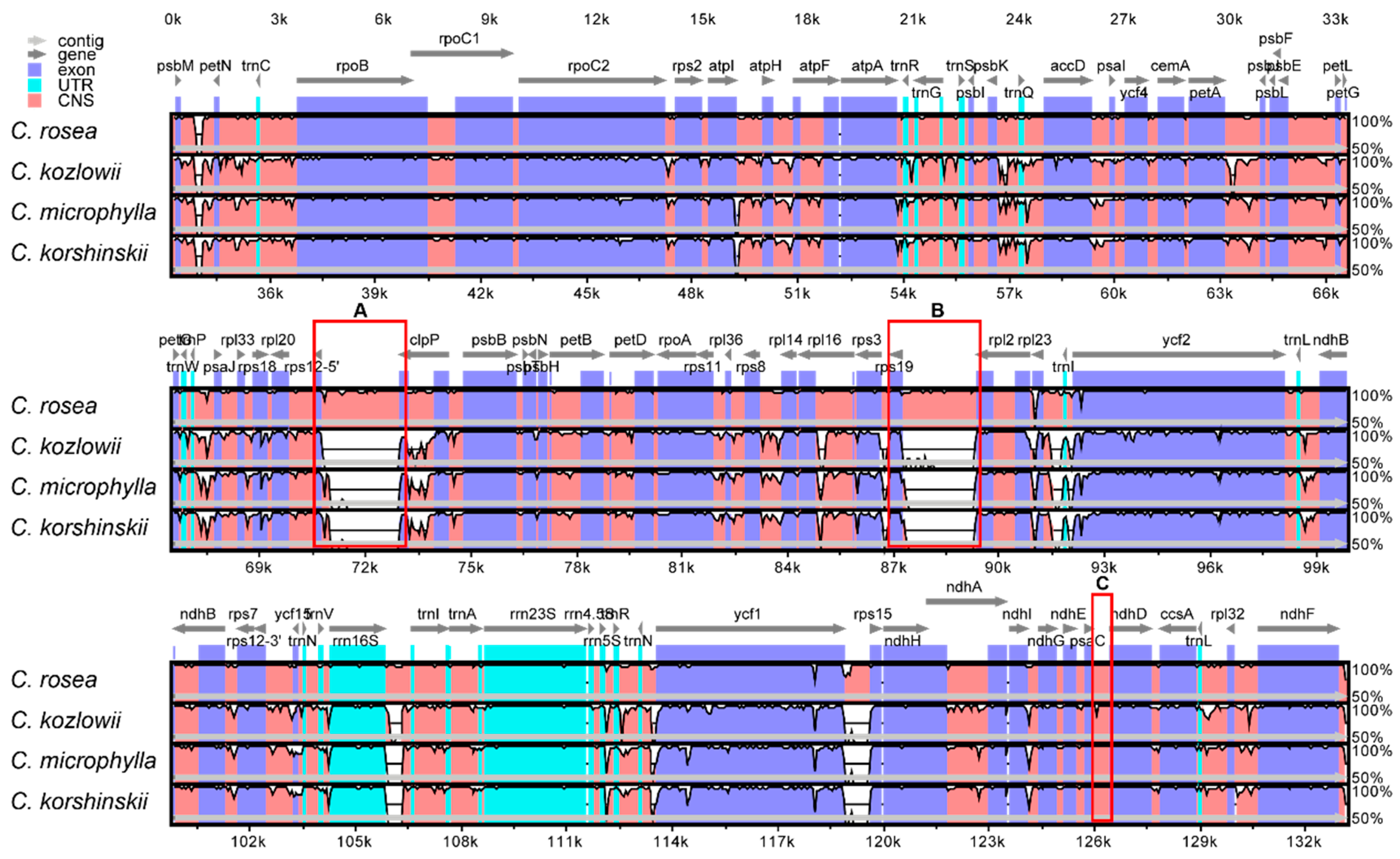
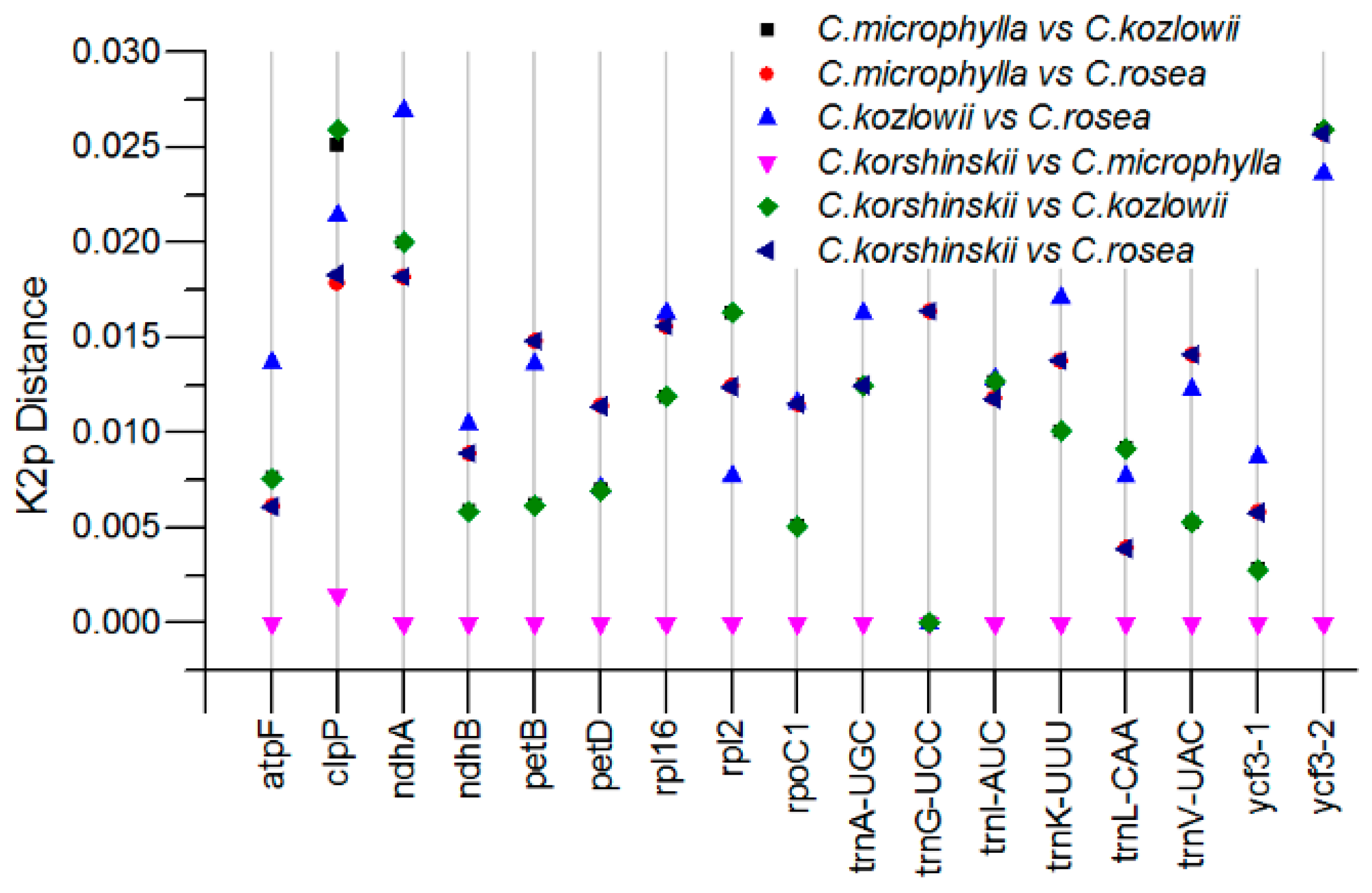
| Plastome Characteristics | C. rosea | C. microphylla | C. kozlowii | C. korshinskii |
|---|---|---|---|---|
| complete genome length | 133,122 bp | 130,029 bp | 131,274 bp | 129,331 bp |
| No.of unique genes | 110 | 110 | 110 | 110 |
| No.of unique protein-coding genes | 76 | 76 | 76 | 76 |
| No.of unique tRNA genes | 30 | 30 | 30 | 30 |
| No.of unique rRNA genes | 4 | 4 | 4 | 4 |
| Size of protein-coding genes | 66,243 bp (49.76%) | 66,231 bp (50.94%) | 66,234 bp (50.45%) | 66,231 bp (51.21%) |
| Size of tRNA genes | 2359 bp (1.77%) | 2370 bp (1.82%) | 2285 bp (1.74%) | 2370 bp (1.83) |
| Size of rRNA genes | 4537 bp (3.4%) | 4520 bp (3.48%) | 4521 bp (3.44%) | 4520 bp (3.49) |
| Overall GC contents | 34.84% | 34.26% | 34.50% | 34.36% |
| GC contents of protein-coding genes | Coding GC 37.13% | Coding GC 36.88% | Coding GC 37.03% | Coding GC 36.88% |
| #1st position 45.36% | #1st position 44.98% | #1st position 45.30% | #1st position 44.96% | |
| #2nd position 37.62% | #2nd position 37.67% | #2nd position 37.58% | #2nd position 37.67% | |
| #3rd position 28.41% | #3rd position 27.99% | #3rd position 28.21% | #3rd position 27.99% | |
| GC contents of tRNA genes | 52.73% | 53.14% | 53.15% | 53.05% |
| GC contents of rRNA genes | 54.77% | 54.82% | 54.75% | 54.82% |
| Category | Name of Species | rps16 | rpl22 | ycf15 | ycf4 | psaI | ycf1 | rpl23 | rps18 | ndhB |
|---|---|---|---|---|---|---|---|---|---|---|
| Millettieae | W. floribunda | − | − | + | + | + | − | + | + | + |
| W. sinensis | − | − | + | + | + | − | + | + | − | |
| Galegeae | G. glabra | − | − | − | + | + | + | + | + | + |
| G. lepidota | − | − | − | + | + | + | + | + | + | |
| A. mongholicus | − | − | − | + | + | + | + | + | + | |
| A. mongholicus var. nakaianus | − | − | − | + | + | + | + | + | + | |
| Caraganeae | C. kozlowii | − | − | − | + | + | + | + | + | + |
| C. korshinskii | − | − | − | + | + | + | + | + | + | |
| C. microphylla | − | − | − | + | + | + | + | + | + | |
| C. rosea var. rosea | − | − | − | + | + | + | + | + | + | |
| Cicereae | C. arietinum | − | − | − | − | + | + | + | + | + |
| Trifolieae | M. truncatula | − | − | − | − | + | + | + | + | + |
| M. papillosa | − | − | − | − | + | + | + | + | + | |
| M. hybrida | − | − | − | − | + | + | + | + | + | |
| M. falcata | − | − | − | + | + | − | + | + | − | |
| T. boissieri | − | − | − | − | + | + | + | + | + | |
| T. glanduliferum | − | − | − | − | + | + | + | + | + | |
| Fabeae | T. strictum | − | − | − | − | + | + | + | + | + |
| L. sativus | − | − | − | + | − | + | − | + | + | |
| L. odoratus | − | − | − | − | − | + | + | − | + | |
| L. inconspicuus | − | − | − | − | − | + | + | + | + | |
| L. ochroleucus | − | − | − | − | + | + | + | + | + | |
| L. venosus | − | − | − | − | + | + | + | + | + | |
| L. palustris | − | − | − | − | + | + | + | + | + | |
| L. tingitanus | − | − | − | − | + | + | + | − | + | |
| L. davidii | − | − | − | − | − | + | + | + | + | |
| L. graminifolius | − | − | − | − | + | + | + | + | + | |
| L. littoralis | − | − | − | − | + | + | + | + | + | |
| L. japonicus | − | − | − | − | + | + | + | + | + | |
| L. pubescens | − | − | − | − | − | + | + | + | + | |
| L. clymenum | − | − | − | − | + | + | + | + | + | |
| P. sativum | − | − | − | − | + | + | − | + | + | |
| V. sativa | − | − | − | − | + | + | − | + | + | |
| L. culinaris | − | − | − | − | + | − | + | − | + | |
| Number a | 34 | 34 | 32 | 22 | 5 | 4 | 3 | 3 | 2 |
| Gene | B | LRT | Test p-Value | Uncorrected p-Value | ω Distribution over Sites |
|---|---|---|---|---|---|
| rpl16 | 0.0128 | 17.4234 | 0.0037 | 0.0001 | ω1 = 0.121 (98%) |
| ω2 = 781 (1.9%) | |||||
| rpl20 | 0.0128 | 31.0401 | 0 | 0 | ω1 = 0.147 (99%) |
| ω2 = 290 (0.92%) | |||||
| rps11 | 0.0128 | 12.0303 | 0.0452 | 0.0008 | ω1 = 0.226 (100%) |
| ω2 = 0.824 (0.20%) | |||||
| ω3 = 10000 (0.14%) | |||||
| rps7 | 0.0128 | 24.3289 | 0.0001 | 0 | ω1 = 0.00 (100%) |
| ω2 = 286 (0.43%) | |||||
| ycf1 | 0.0128 | 34.4372 | 0 | 0 | ω1 = 0.384 (99%) |
| ω2 = 91.3 (0.74%) |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jiang, M.; Chen, H.; He, S.; Wang, L.; Chen, A.J.; Liu, C. Sequencing, Characterization, and Comparative Analyses of the Plastome of Caragana rosea var. rosea. Int. J. Mol. Sci. 2018, 19, 1419. https://doi.org/10.3390/ijms19051419
Jiang M, Chen H, He S, Wang L, Chen AJ, Liu C. Sequencing, Characterization, and Comparative Analyses of the Plastome of Caragana rosea var. rosea. International Journal of Molecular Sciences. 2018; 19(5):1419. https://doi.org/10.3390/ijms19051419
Chicago/Turabian StyleJiang, Mei, Haimei Chen, Shuaibing He, Liqiang Wang, Amanda Juan Chen, and Chang Liu. 2018. "Sequencing, Characterization, and Comparative Analyses of the Plastome of Caragana rosea var. rosea" International Journal of Molecular Sciences 19, no. 5: 1419. https://doi.org/10.3390/ijms19051419
APA StyleJiang, M., Chen, H., He, S., Wang, L., Chen, A. J., & Liu, C. (2018). Sequencing, Characterization, and Comparative Analyses of the Plastome of Caragana rosea var. rosea. International Journal of Molecular Sciences, 19(5), 1419. https://doi.org/10.3390/ijms19051419




