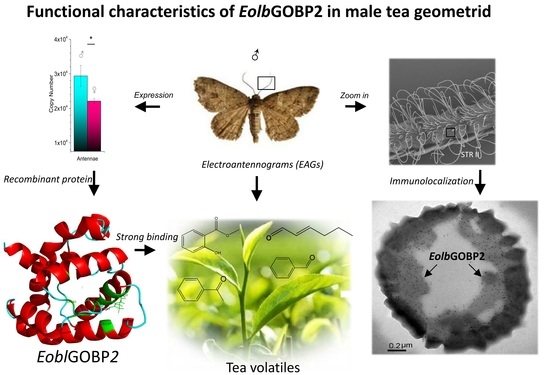
Functional Characteristics, Electrophysiological and Antennal Immunolocalization of General Odorant-Binding Protein 2 in Tea Geometrid, Ectropis obliqua
Abstract
1. Introduction
2. Results
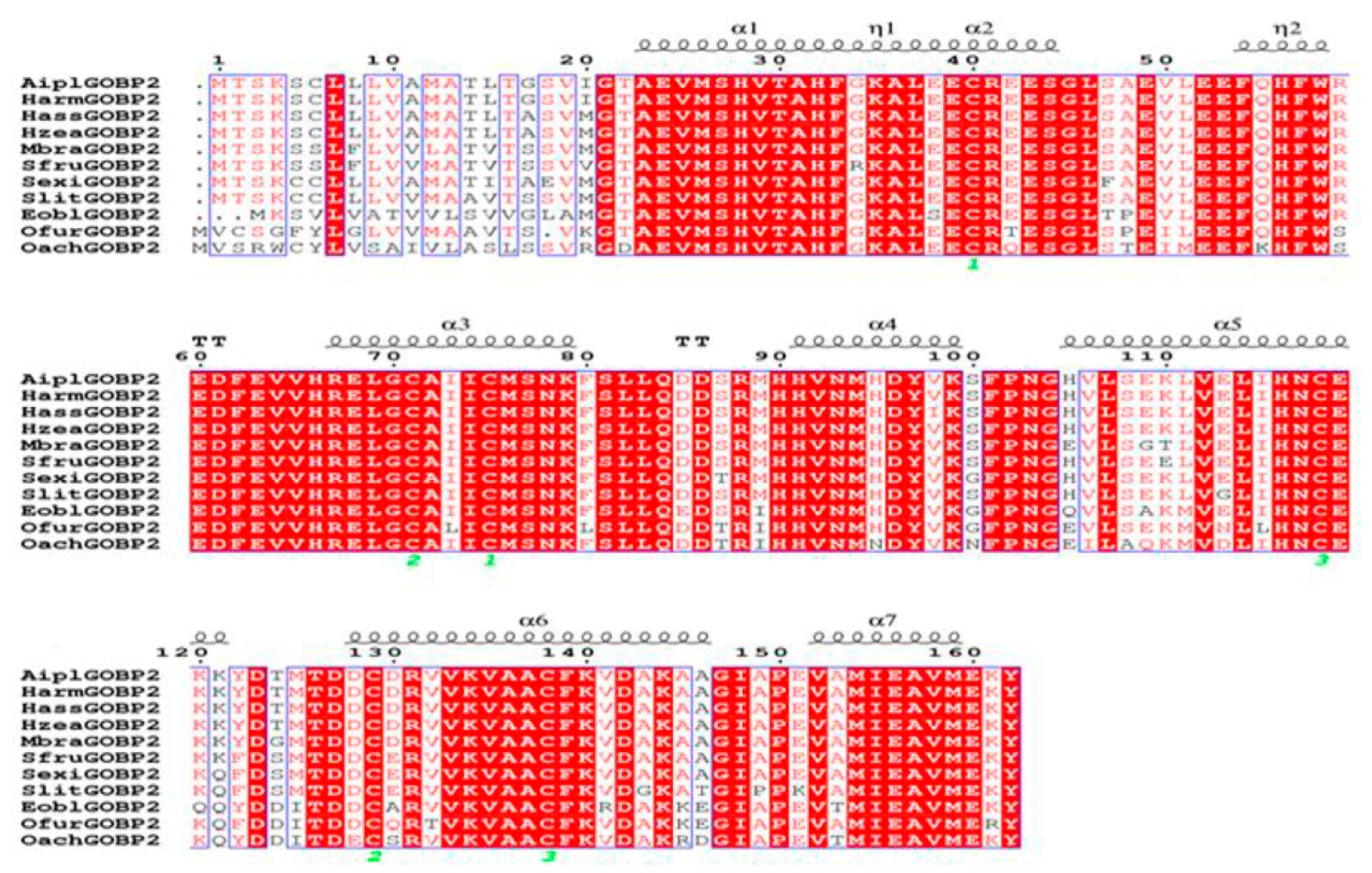
2.1. Identification and Sequence Analysis of EoblGOBP2
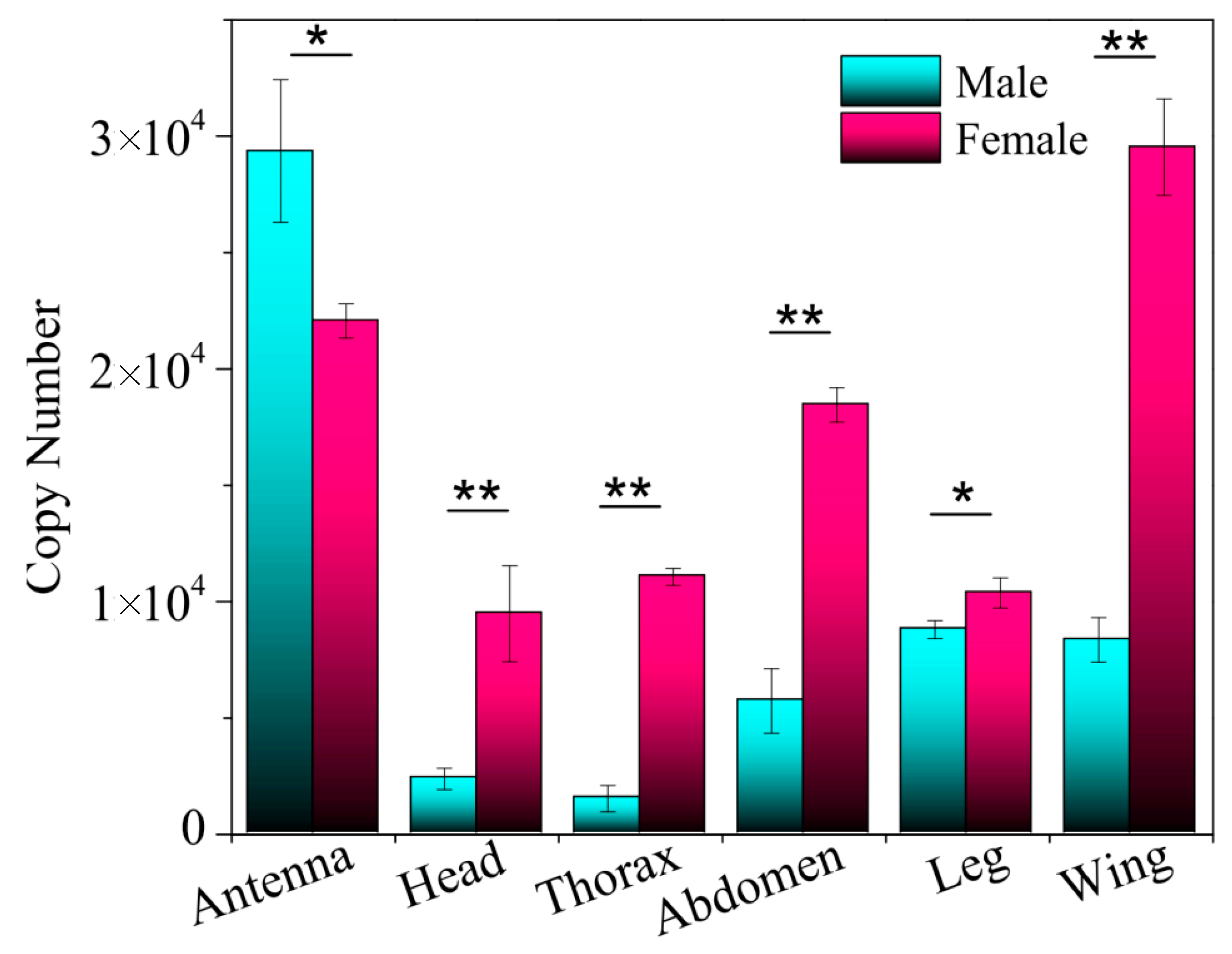
2.2. Tissue Expression Profile of EoblGOBP2
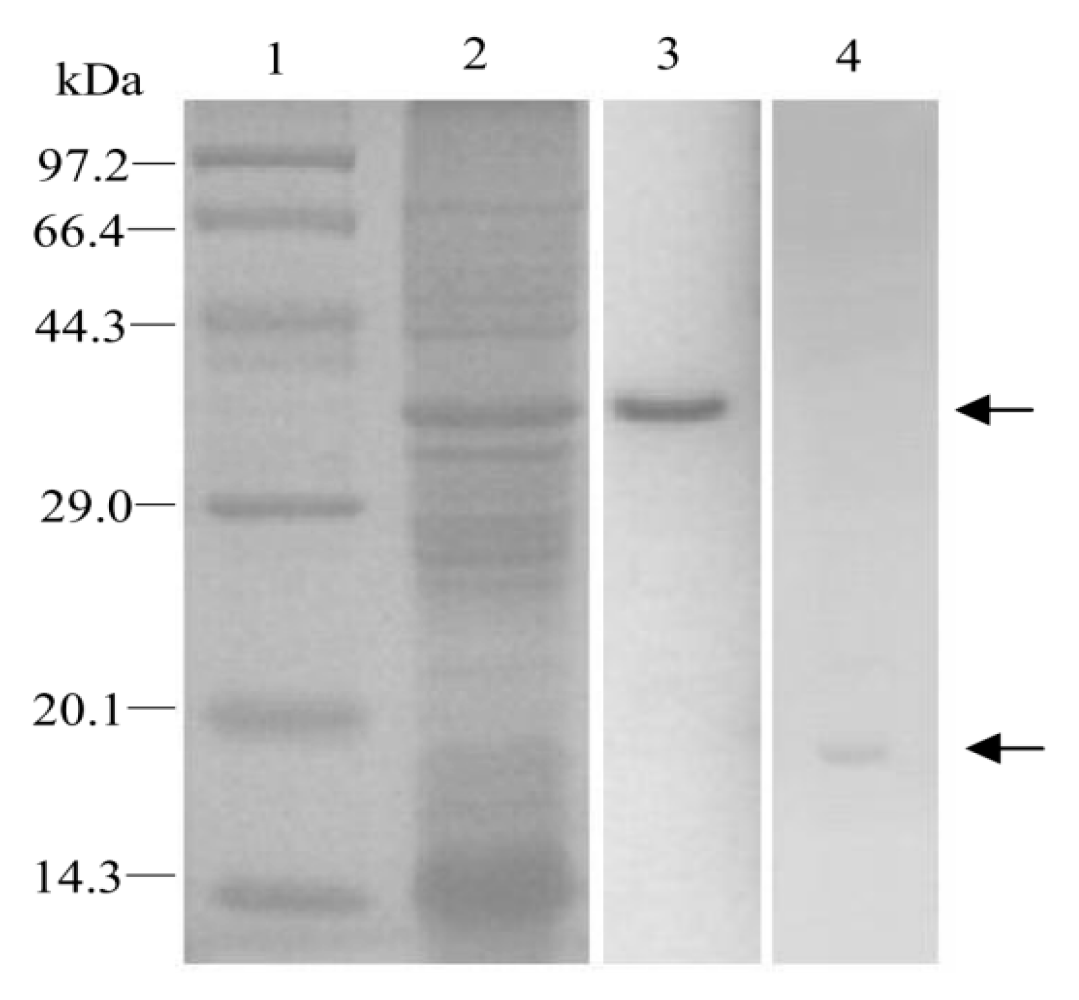
2.3. Preparation and Purification of Recombinant EoblGOBP2 Protein
2.4. Competitive Fluorescence Ligand-Binding Assay
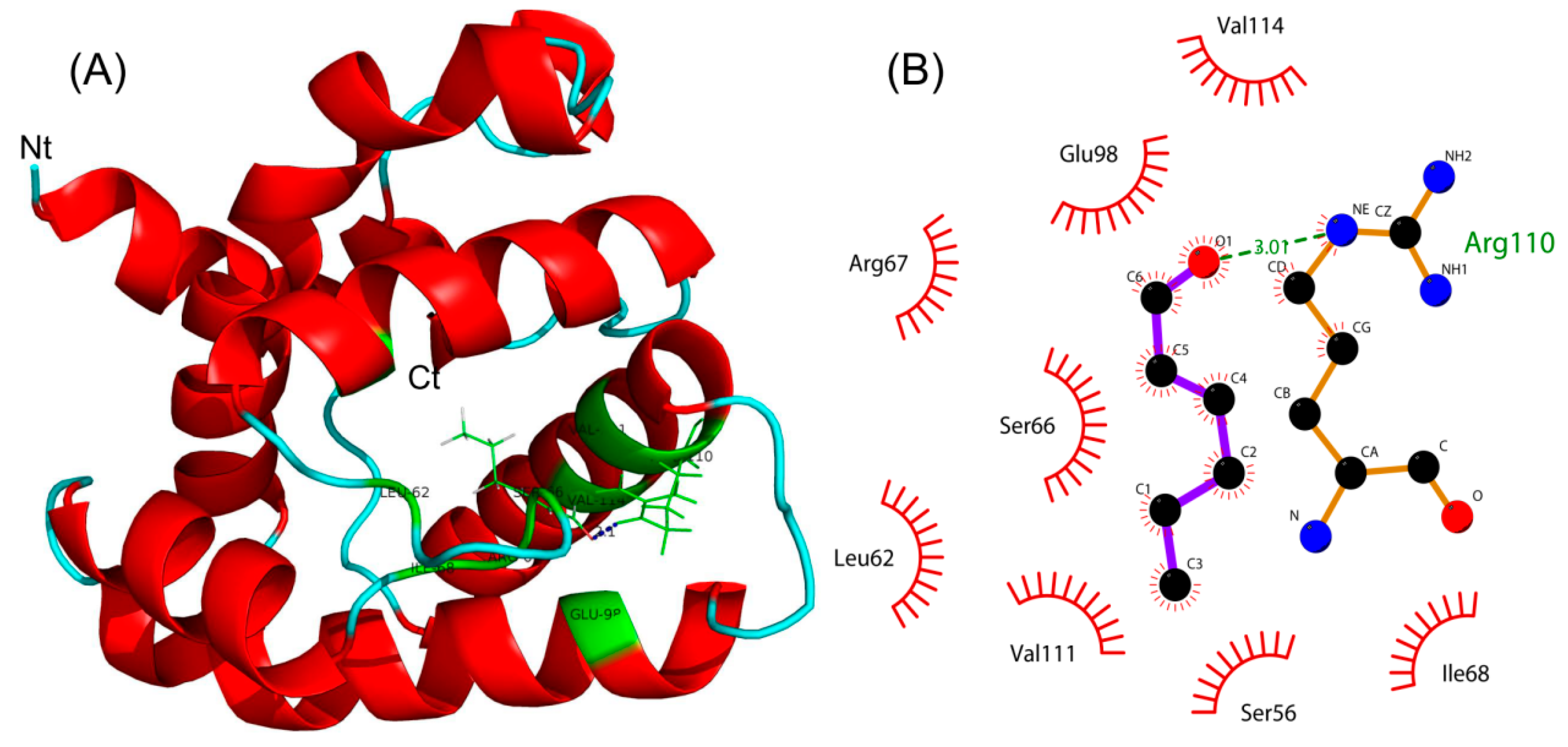
2.5. Molecular Docking and Interaction Analysis
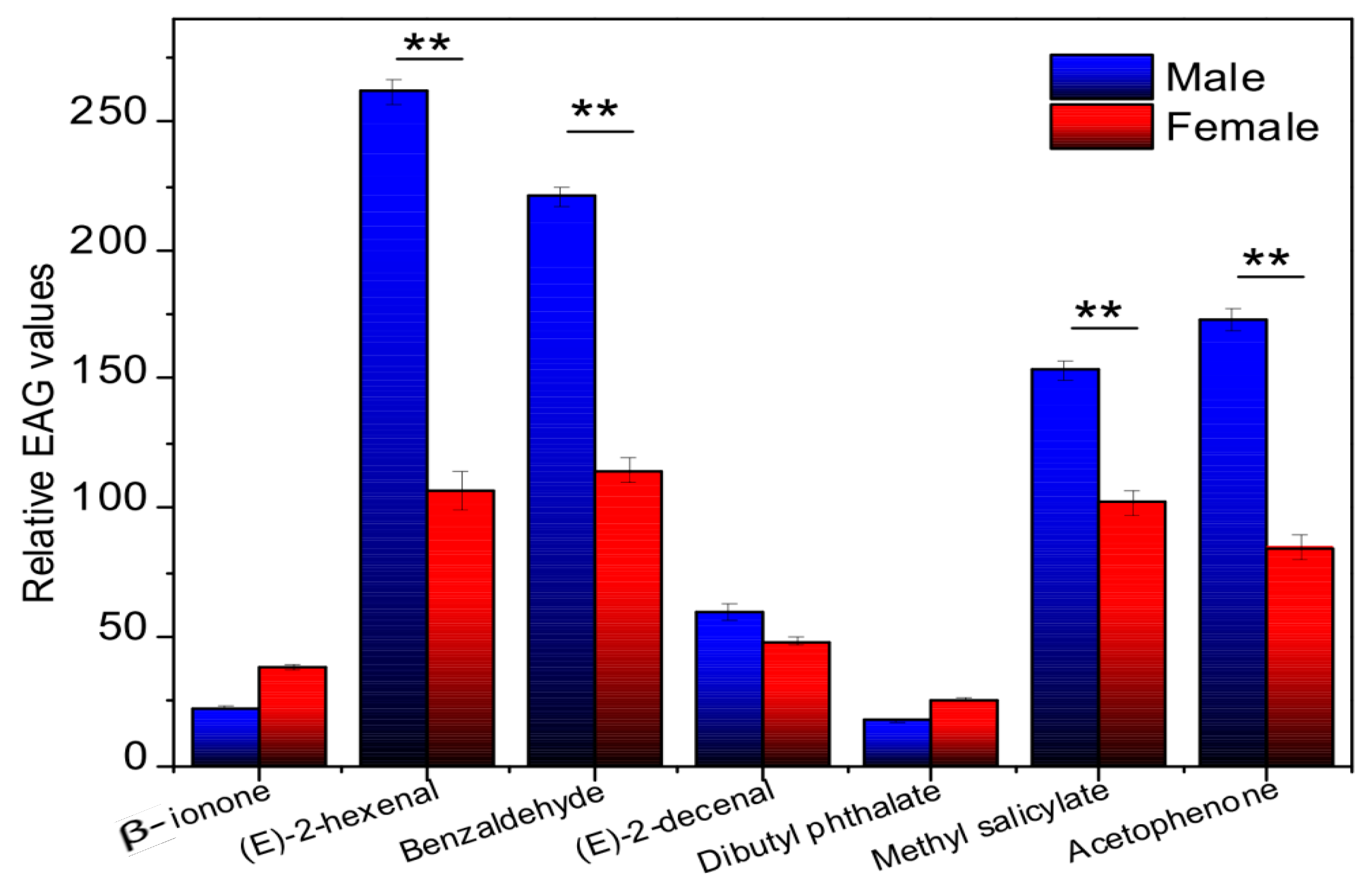
2.6. Electroantennograms
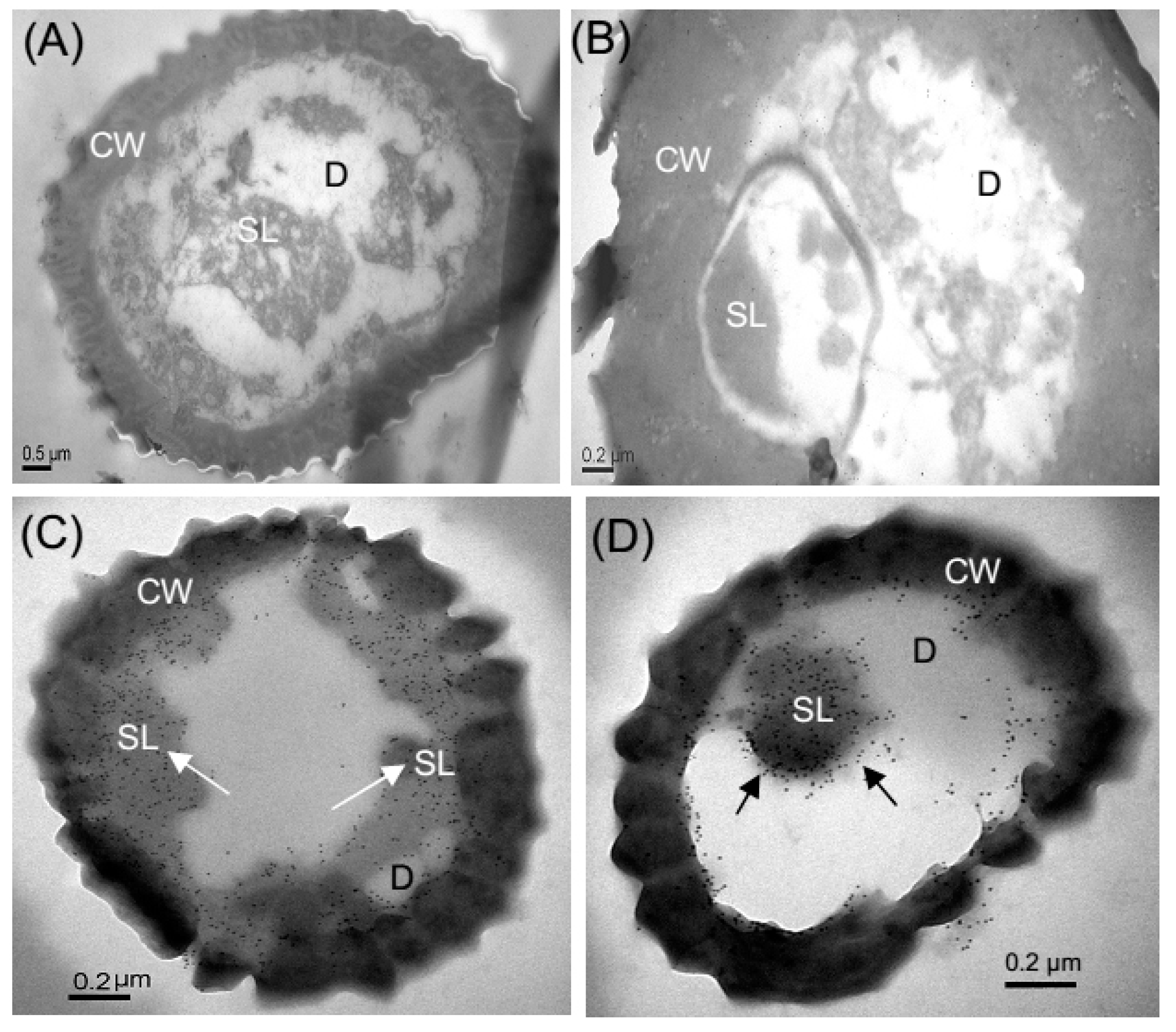
2.7. Immunocytochemical Localization
3. Discussion
4. Materials and Methods
4.1. Insects and Tissue Collection
4.2. Chemical Ligands
4.3. RNA Extraction and Cloning of EoblGOBP2
4.4. Expression Profiles of EoblGOBP2 in Various Tissues
4.5. Expression and Purification of Recombinant EoblGOBP2 Protein
4.6. Competitive Ligand-Binding Assay
4.7. Molecular Docking
4.8. Electroantennograms (EAGs)
4.9. Scanning Electron Microscopy (SEM)
4.10. Immunocytochemical Localization
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Logan, J.G.; Birkett, M.A. Semiochemicals for biting fly control: Their identification and exploitation. Pest Manag. Sci. 2007, 63, 647–657. [Google Scholar] [CrossRef] [PubMed]
- Vosshall, L.B.; Stocker, R.F. Molecular architecture of smell and taste in Drosophila. Annu. Rev. Neurosci. 2007, 30, 505–533. [Google Scholar] [CrossRef] [PubMed]
- Leal, W.S. Odorant reception in insects: Roles of receptors, binding proteins, and degrading enzymes. Annu. Rev. Entomol. 2013, 58, 373–391. [Google Scholar] [CrossRef] [PubMed]
- Vogt, R.G.; Prestwich, G.D.; Lerner, M.R. Odorant-binding-protein subfamilies associate with distinct classes of olfactory receptor neurons in insects. J. Neurobiol. 1991, 22, 74–84. [Google Scholar] [CrossRef] [PubMed]
- Pelosi, P.; Iovinella, I.; Felicioli, A.; Dani, F.R. Soluble proteins of chemical communication: An overview across arthropods. Front. Physiol. 2014, 5, 320. [Google Scholar] [CrossRef] [PubMed]
- Vogt, R.G.; Riddiford, L.M. Pheromone binding and inactivation by moth antennae. Nature 1981, 293, 161–163. [Google Scholar] [CrossRef] [PubMed]
- Pelosi, P.; Zhou, J.J.; Ban, L.P.; Calvello, M. Soluble proteins in insect chemical communication. Cell. Mol. Life Sci. 2006, 63, 1658–1676. [Google Scholar] [CrossRef] [PubMed]
- Vogt, R.G.; Grosse-Wilde, E.; Zhou, J.J. The Lepidoptera odorant binding protein gene family: Gene gain and loss within the GOBP/PBP complex of moths and butterflies. Insect Biochem. Mol. Biol. 2015, 62, 142–153. [Google Scholar] [CrossRef] [PubMed]
- Scaloni, A.; Monti, M.; Angeli, S.; Pelosi, P. Structural analysis and disulfide-bridge pairing of two odorant-binding proteins from Bombyx mori. Biochem. Biophys. Res. Commun. 1999, 266, 386–391. [Google Scholar] [CrossRef] [PubMed]
- Leal, W.S.; Nikonova, L.; Peng, G. Disulfide structure of the pheromone binding protein from the silkworm moth, Bombyx mori. FEBS Lett. 1999, 464, 85–90. [Google Scholar] [CrossRef]
- Zhou, J.J. Odorant-binding proteins in insects. Vitam. Horm. 2010, 83, 241–272. [Google Scholar] [PubMed]
- Leal, W.S. Pheromone reception. In The Chemistry of Pheromones and Other Semiochemicals II; Springer: Berlin, Germany, 2005; pp. 1–36. [Google Scholar]
- Zhang, S.; Zhang, Z.; Wang, H.; Kong, X. Molecular characterization, expression pattern, and ligand-binding property of three odorant binding protein genes from Dendrolimus tabulaeformis. J. Chem. Ecol. 2014, 40, 396–406. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Yin, J.; Feng, H.; Sun, H.; Xi, J.; Cao, Y.; Li, K. Functional analysis of general odorant binding protein 2 from the meadow moth, Loxostege sticticalis L. (Lepidoptera: Pyralidae). PLoS ONE 2012, 7, e33589. [Google Scholar] [CrossRef] [PubMed]
- He, X.; Tzotzos, G.; Woodcock, C.; Pickett, J.A.; Hooper, T.; Field, L.M.; Zhou, J.J. Binding of the general odorant binding protein of Bombyx mori BmorGOBP2 to the moth sex pheromone components. J. Chem. Ecol. 2010, 36, 1293–1305. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.J.; Robertson, G.; He, X.; Dufour, S.; Hooper, A.M.; Pickett, J.A.; Keep, N.H.; Field, L.M. Characterisation of Bombyx mori odorant-binding proteins reveals that a general odorant-binding protein discriminates between sex pheromone components. J. Mol. Biol. 2009, 389, 529–545. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.; Ban, L.P.; Song, L.M.; Liu, Y.; Pelosi, P.; Wang, G.R. General odorant-binding proteins and sex pheromone guide larvae of Plutella xylostella to better food. Insect Biochem. Mol. Biol. 2016, 72, 10–19. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.H.; Tan, J.C. Non-Pollution Control of Tea Pests in China; Anhui Science and Technology Press: Hefei, China, 2004. [Google Scholar]
- Ma, X.C.; Xu, H.J.; Tang, M.J.; Xiao, Q.; Hong, J.; Zhang, C.X. Morphological, phylogenetic and biological characteristics of Ectropis obliqua single-nucleocapsid nucleopolyhedrovirus. J. Microbiol. 2006, 44, 77–82. [Google Scholar] [PubMed]
- Lu, J.; Zhang, J.; Wang, X.; Jiang, H.; Liu, C.; Hu, Y. In vitro and in vivo identification of structural and sequence elements in the 5′ untranslated region of Ectropis obliqua picorna-like virus required for internal initiation. J. Gen. Virol. 2006, 87, 3667–3677. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.J.; Field, L.M.; He, X.L. Insect odorant-binding proteins: Do they offer an alternative pest control strategy? Outlooks Pest Manag. 2010, 21, 31–34. [Google Scholar] [CrossRef]
- Sun, X.L.; Li, X.W.; Xin, Z.J.; Han, J.J.; Ran, W.; Lei, S. Development of synthetic volatile attractant for male Ectropis obliqua moths. J. Integr. Agric. 2016, 15, 1532–1539. [Google Scholar] [CrossRef]
- Watanabe, H.; Tabunoki, H.; Miura, N.; Sato, R.; Ando, T. Analysis of odorant-binding proteins in antennae of a geometrid species, Ascotis selenaria cretacea, which produces lepidopteran Type II sex pheromone components. Invertebr. Neurosci. 2007, 7, 109–118. [Google Scholar] [CrossRef] [PubMed]
- Ma, L.; Li, Z.-Q.; Bian, L.; Cai, X.-M.; Luo, Z.-X.; Zhang, Y.-J.; Chen, Z.-M. Identification and comparative study of chemosensory genes related to host selection by legs transcriptome analysis in the tea geometrid Ectropis obliqua. PLoS ONE 2016, 11, e0149591. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; Mao, T.F.; Zhang, Y.X.; Wu, J.J.; Bai, J.H.; Zhang, Y.N.; Jiang, X.C.; Yin, K.S.; Guo, Y.Y.; Zhang, Y.J. Characterization of candidate odorant-binding proteins and chemosensory proteins in the tea geometrid Ectropis obliqua Prout (Lepidoptera: Geometridae). Arch. Insect Biochem. Physiol. 2017, 94, e21383. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.-Q.; Luo, Z.-X.; Cai, X.-M.; Bian, L.; Xin, Z.-J.; Liu, Y.; Chu, B.; Chen, Z.-M. Chemosensory gene families in Ectropis grisescens and candidates for detection of Type-II sex pheromones. Front. Physiol. 2017, 8, 953. [Google Scholar] [CrossRef] [PubMed]
- Saitou, N.; Nei, M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987, 4, 406. [Google Scholar] [PubMed]
- Arnold, K.; Bordoli, L.; Kopp, J.; Schwede, T. The SWISS-MODEL workspace: A web-based environment for protein structure homology modelling. Bioinformatics 2006, 22, 195–201. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.J.; Liu, N.Y.; He, P.; Li, Z.Q.; Dong, S.L.; Mu, L.F. Molecular characterization, expression patterns, and ligand-binding properties of two odorant-binding protein genes from Orthaga achatina (Butler) (Lepidoptera: Pyralidae). Arch. Insect Biochem. Physiol. 2012, 80, 123–139. [Google Scholar] [CrossRef] [PubMed]
- Hong, J.; Ye, G.Y.; Hu, C. Scanning electron microscopy of adult antennal sensilla of the tea geometrid, Ectropis obliqua hypulina Wehrli. J. Zhejiang Agric. Univ. 1993, 19, 53–56. [Google Scholar]
- Ma, L.; Bian, L.; Li, Z.Q.; Cai, X.M.; Luo, Z.X.; Chen, Z.M. Ultrastructure of chemosensilla on antennae and tarsi of Ectropis obliqua (Lepidoptera: Geometridae). Ann. Entomol. Soc. Am. 2016, 109, 574–584. [Google Scholar] [CrossRef]
- Zhu, J.Y.; Zhang, L.F.; Ze, S.Z.; Wang, D.W.; Yang, B. Identification and tissue distribution of odorant binding protein genes in the beet armyworm, Spodoptera exigua. J. Insect Physiol. 2013, 59, 722–728. [Google Scholar] [CrossRef] [PubMed]
- Gong, Z.J.; Zhou, W.W.; Yu, H.Z.; Mao, C.G.; Zhang, C.X.; Cheng, J.A.; Zhu, Z.R. Cloning, expression and functional analysis of a general odorant-binding protein 2 gene of the rice striped stem borer, Chilo suppressalis (Walker) (Lepidoptera: Pyralidae). Insect Mol. Biol. 2009, 18, 405–417. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.L.; Wang, G.C.; Gao, Y.; Zhang, X.Z.; Xin, Z.J.; Chen, Z.M. Volatiles emitted from tea plants infested by Ectropis obliqua larvae are attractive to conspecific moths. J. Chem. Ecol. 2014, 40, 1080–1089. [Google Scholar] [CrossRef] [PubMed]
- Feng, L.; Prestwich, G.D. Expression and characterization of a lepidopteran general odorant binding protein. Insect Biochem. Mol. Biol. 1997, 27, 405–412. [Google Scholar] [CrossRef]
- Wang, P.; Lyman, R.F.; Shabalina, S.A.; Mackay, T.F.; Anholt, R.R. Association of polymorphisms in odorant-binding protein genes with variation in olfactory response to benzaldehyde in Drosophila. Genetics 2007, 177, 1655–1665. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.J.; Zhang, G.A.; Huang, W.; Birkett, M.A.; Field, L.M.; Pickett, J.A.; Pelosi, P. Revisiting the odorant-binding protein LUSH of Drosophila melanogaster: Evidence for odour recognition and discrimination. FEBS Lett. 2004, 558, 23–26. [Google Scholar] [CrossRef]
- Sun, L.; Gu, S.H.; Xiao, H.J.; Zhou, J.J.; Guo, Y.Y.; Liu, Z.W.; Zhang, Y.J. The preferential binding of a sensory organ specific odorant binding protein of the alfalfa plant bug Adelphocoris lineolatus AlinOBP10 to biologically active host plant volatiles. J. Chem. Ecol. 2013, 39, 1221–1231. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Bian, L.; Sun, X.; Luo, Z.; Xin, Z.; Luo, F.; Chen, Z. Electrophysiological and behavioural responses of the tea geometrid Ectropis obliqua (Lepidoptera: Geometridae) to volatiles from a non-host plant, rosemary, Rosmarinus officinalis (Lamiaceae). Pest Manag. Sci. 2014, 71, 96. [Google Scholar] [CrossRef] [PubMed]
- Deng, S.; Yin, J.; Zhong, T.; Cao, Y.; Li, K. Function and immunocytochemical localization of two novel odorant-binding proteins in olfactory sensilla of the scarab beetle Holotrichia oblita Faldermann (Coleoptera: scarabaeidae). Chem. Senses 2012, 37, 141–150. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Maida, R.; Steinbrecht, R.A. Immunolocalization of odorant-binding proteins in noctuid moths (Insecta, Lepidoptera). Chem. Senses 2001, 26, 885–896. [Google Scholar] [CrossRef] [PubMed]
- Laue, M.; Steinbrecht, R.A.; Ziegelberger, G. Immunocytochemical localization of general odorant-binding protein in olfactory sensilla of the silkmoth Antheraea polyphemus. Naturwissenschaften 1994, 81, 178–180. [Google Scholar] [CrossRef]
- Wang, G.R.; Wu, K.M.; Guo, Y.Y. Cloning, expression and immunocytochemical localization of a general odorant-binding protein gene from Helicoverpa armigera (Hubner). Insect Biochem. Mol. Biol. 2003, 33, 115–124. [Google Scholar] [CrossRef]
- Steinbrecht, R.A. Structure and function of insect olfactory sensilla. In Ciba Foundation Symposium 200—Olfaction in Mosquito-Host Interactions; John Wiley & Sons: Hoboken, NJ, USA, 1996; pp. 158–174. [Google Scholar]
- Maeda, Y.; Ueda, T.; Imoto, T. Effective renaturation of denatured and reduced immunoglobulin G in vitro without assistance of chaperone. Protein Eng. Des. Sel. 1996, 9, 95–100. [Google Scholar] [CrossRef][Green Version]
- Sideris, E.E.; Valsami, G.N.; Koupparis, M.A.; Macheras, P.E. Determination of association constants in cyclodextrin/drug complexation using the Scatchard plot: Application to β-cyclodextrin-anilinonaphthalenesulfonates. Pharm. Res. 1992, 9, 1568–1574. [Google Scholar] [CrossRef] [PubMed]
- Ban, L.; Scaloni, A.; D’ambrosio, C.; Zhang, L.; Yan, Y.; Pelosi, P. Biochemical characterization and bacterial expression of an odorant-binding protein from Locusta migratoria. Cell. Mol. Life Sci. 2003, 60, 390–400. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Thiessen, P.A.; Bolton, E.E.; Chen, J.; Fu, G.; Gindulyte, A.; Han, L.; He, J.; He, S.; Shoemaker, B.A. PubChem substance and compound databases. Nucleic Acids Res. 2015, 44, D1202–D1213. [Google Scholar] [CrossRef] [PubMed]
- Thomsen, R.; Christensen, M.H. MolDock: A new technique for high-accuracy molecular docking. J. Med. Chem. 2006, 49, 3315–3321. [Google Scholar] [CrossRef] [PubMed]
- Laskowski, R.A.; Swindells, M.B. LigPlot+: Multiple ligand–protein interaction diagrams for drug discovery. J. Chem. Inf. Model. 2011, 51, 2778–2786. [Google Scholar] [CrossRef] [PubMed]
- DeLano, W.L. The PyMOL Molecular Graphics System; DeLano Scientific: San Carlos, CA, USA, 2002. [Google Scholar]
- Fu, X.W.; Guo, X.R.; Luo, M.H.; Yuan, G.H.; Li, W.Z.; Wu, S.Y. Electrophysiological and behavioral responses of Helicoverpa assulta (Guenée) and H. armigera (Hübner) (Lepidoptera:Noctuidae) to tobacco volatile compounds of high concentration. Acta Entomol. Sin. 2008, 51, 902–909. [Google Scholar]

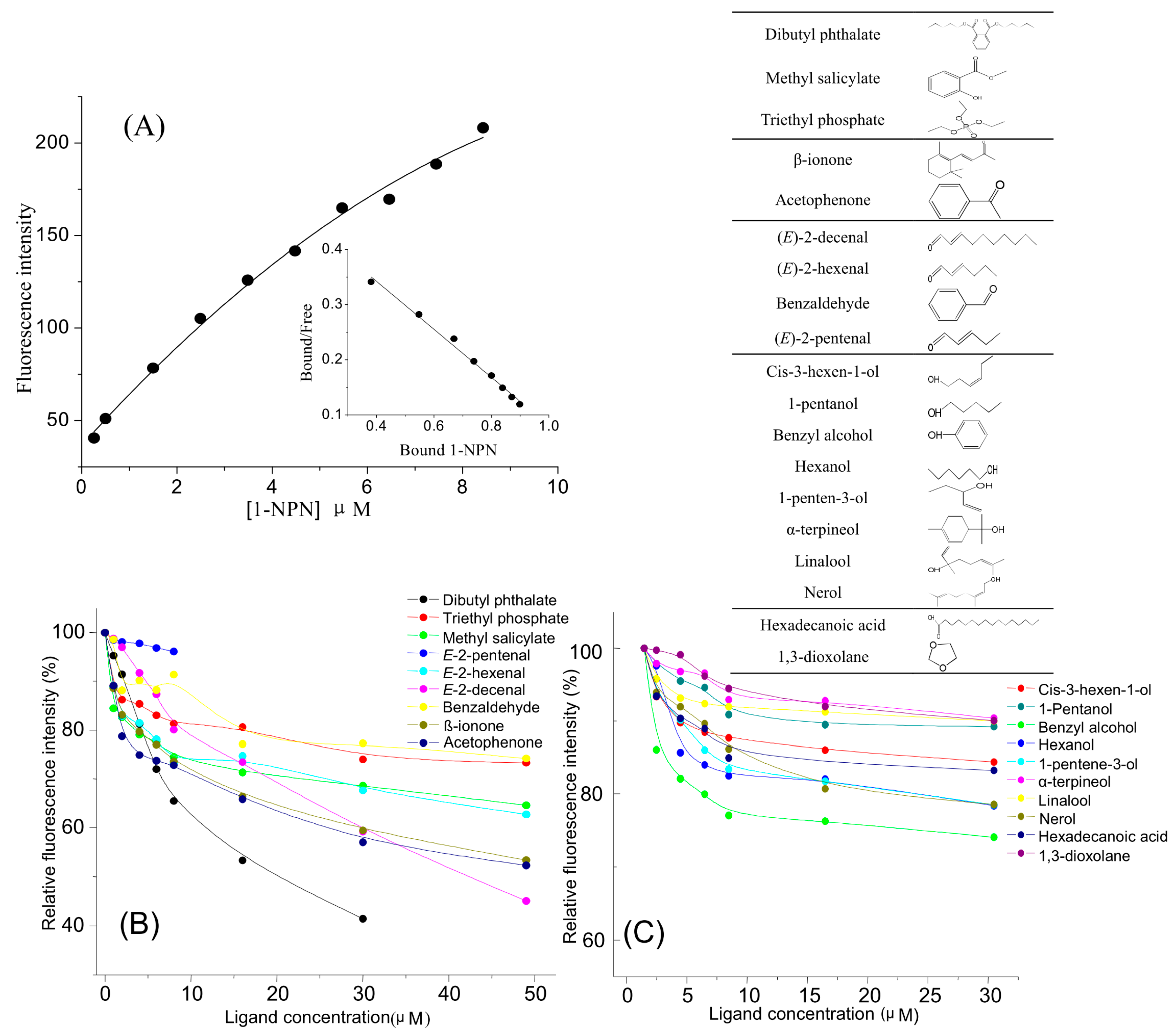

| Chemical Category | Tea Volatiles | [IC50] (μM) | KD (μM) Dissociation Constant | Percentage in the Total Tea Leaves Volatiles (%) |
|---|---|---|---|---|
| Esters | Dibutyl phthalate | 19.42 | 4.35 | 0.08 |
| Methyl salicylate | 194.05 | 43.51 | 2.37 | |
| Triethyl phosphate | ― | ― | 0.53 | |
| Ketones | β-ionone | 56.13 | 12.59 | 0.11 |
| Acetophenone | 57.86 | 12.97 | 0.31 | |
| Aldehydes | (E)-2-decenal | 92.41 | 20.72 | 0.23 |
| (E)-2-hexenal | 143.08 | 32.24 | 0.46 | |
| Benzaldehyde | 197.20 | 44.21 | 0.13 | |
| (E)-2-pentenal | ― | ― | 0 | |
| Alcohols | (Z)-3-hexenol | ― | ― | 0 |
| n-pentanol | ― | ― | 0.45 | |
| Benzyl alcohol | ― | ― | 0 | |
| n-hexanol | ― | ― | 0.06 | |
| 1-penten-3-ol | ― | ― | 0.09 | |
| α-terpineol | ― | ― | 0.31 | |
| Linalool | ― | ― | 0 | |
| Nerol | ― | ― | 0.200 | |
| Others | Hexadecanoic acid | ― | ― | 0.251 |
| 1,3-dioxolane | ― | ― | 0.435 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, Y.-L.; Fu, X.-B.; Cui, H.-C.; Zhao, L.; Yu, J.-Z.; Li, H.-L. Functional Characteristics, Electrophysiological and Antennal Immunolocalization of General Odorant-Binding Protein 2 in Tea Geometrid, Ectropis obliqua. Int. J. Mol. Sci. 2018, 19, 875. https://doi.org/10.3390/ijms19030875
Zhang Y-L, Fu X-B, Cui H-C, Zhao L, Yu J-Z, Li H-L. Functional Characteristics, Electrophysiological and Antennal Immunolocalization of General Odorant-Binding Protein 2 in Tea Geometrid, Ectropis obliqua. International Journal of Molecular Sciences. 2018; 19(3):875. https://doi.org/10.3390/ijms19030875
Chicago/Turabian StyleZhang, Ya-Li, Xiao-Bin Fu, Hong-Chun Cui, Lei Zhao, Ji-Zhong Yu, and Hong-Liang Li. 2018. "Functional Characteristics, Electrophysiological and Antennal Immunolocalization of General Odorant-Binding Protein 2 in Tea Geometrid, Ectropis obliqua" International Journal of Molecular Sciences 19, no. 3: 875. https://doi.org/10.3390/ijms19030875
APA StyleZhang, Y.-L., Fu, X.-B., Cui, H.-C., Zhao, L., Yu, J.-Z., & Li, H.-L. (2018). Functional Characteristics, Electrophysiological and Antennal Immunolocalization of General Odorant-Binding Protein 2 in Tea Geometrid, Ectropis obliqua. International Journal of Molecular Sciences, 19(3), 875. https://doi.org/10.3390/ijms19030875