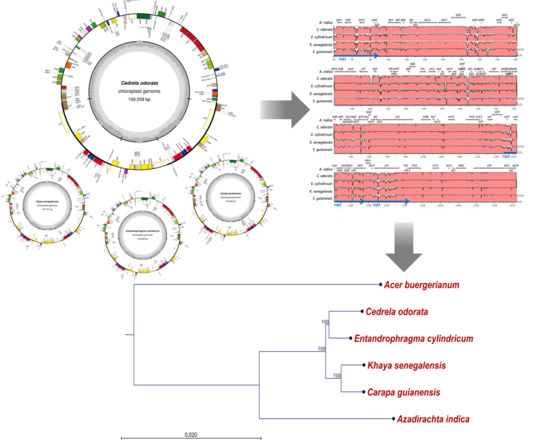
Complete Chloroplast Genome Sequences of Four Meliaceae Species and Comparative Analyses
Abstract
1. Introduction
2. Results
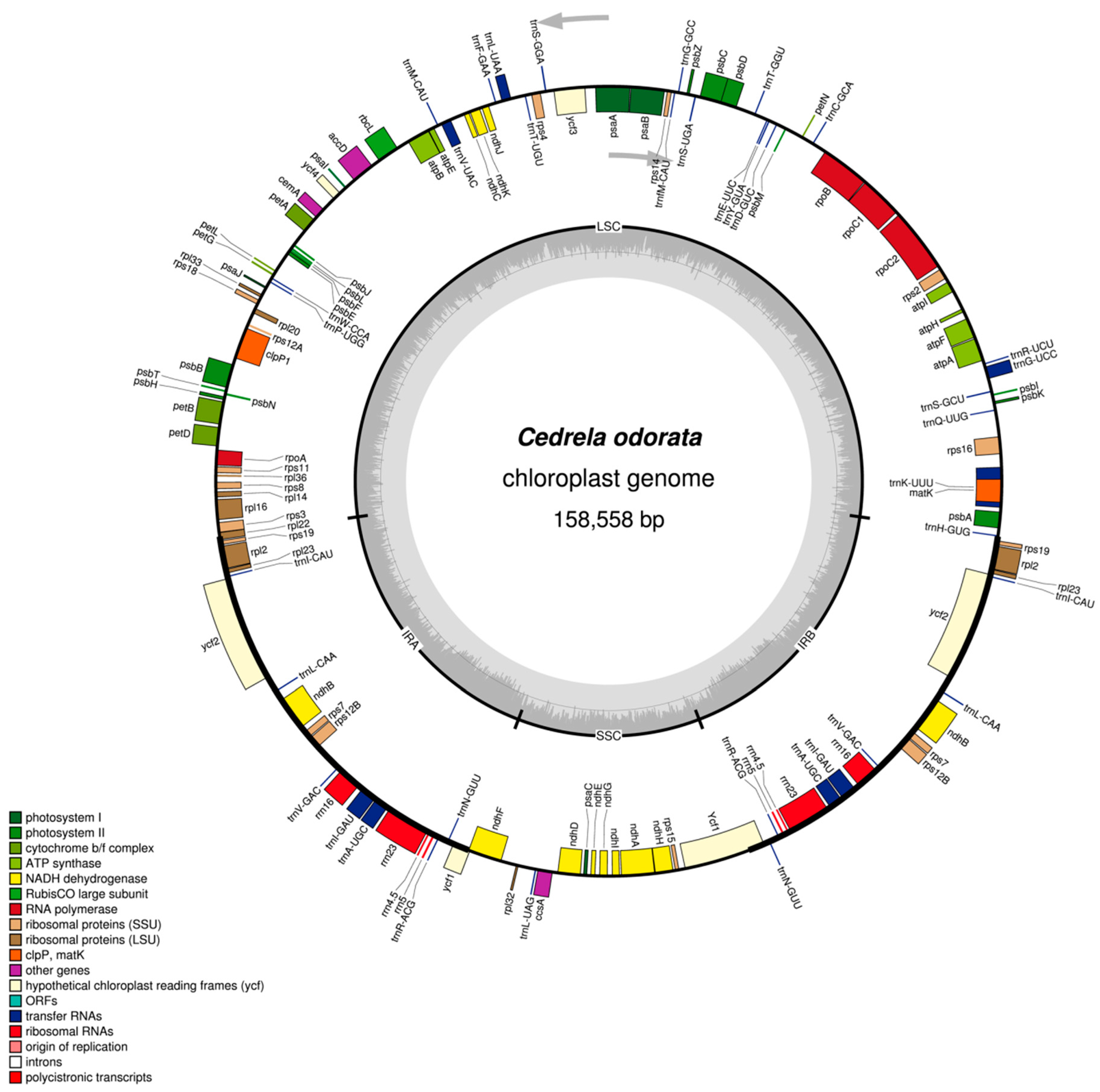
2.1. The Structure of Chloroplast Genomes from Four Meliaceae Species
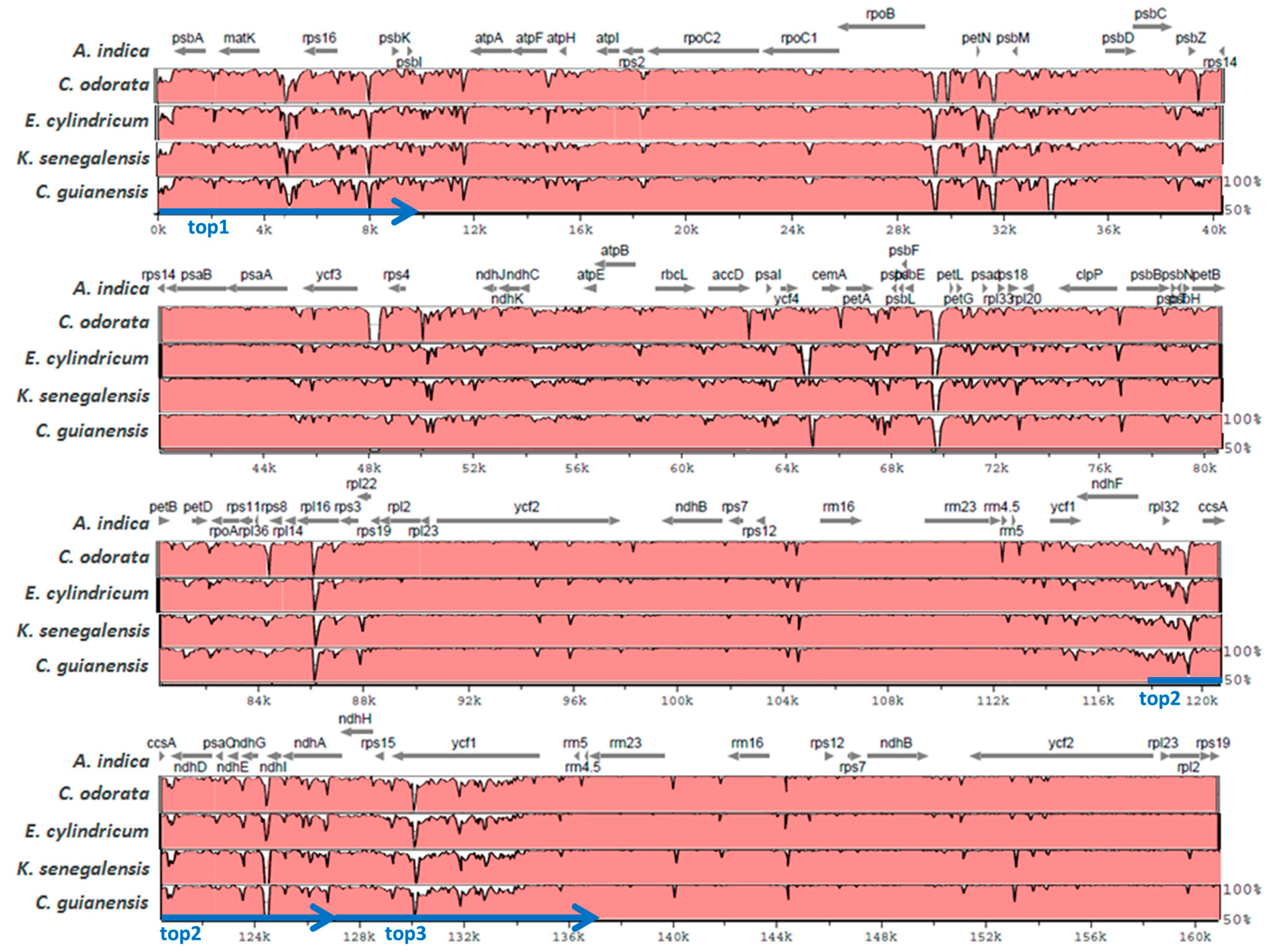
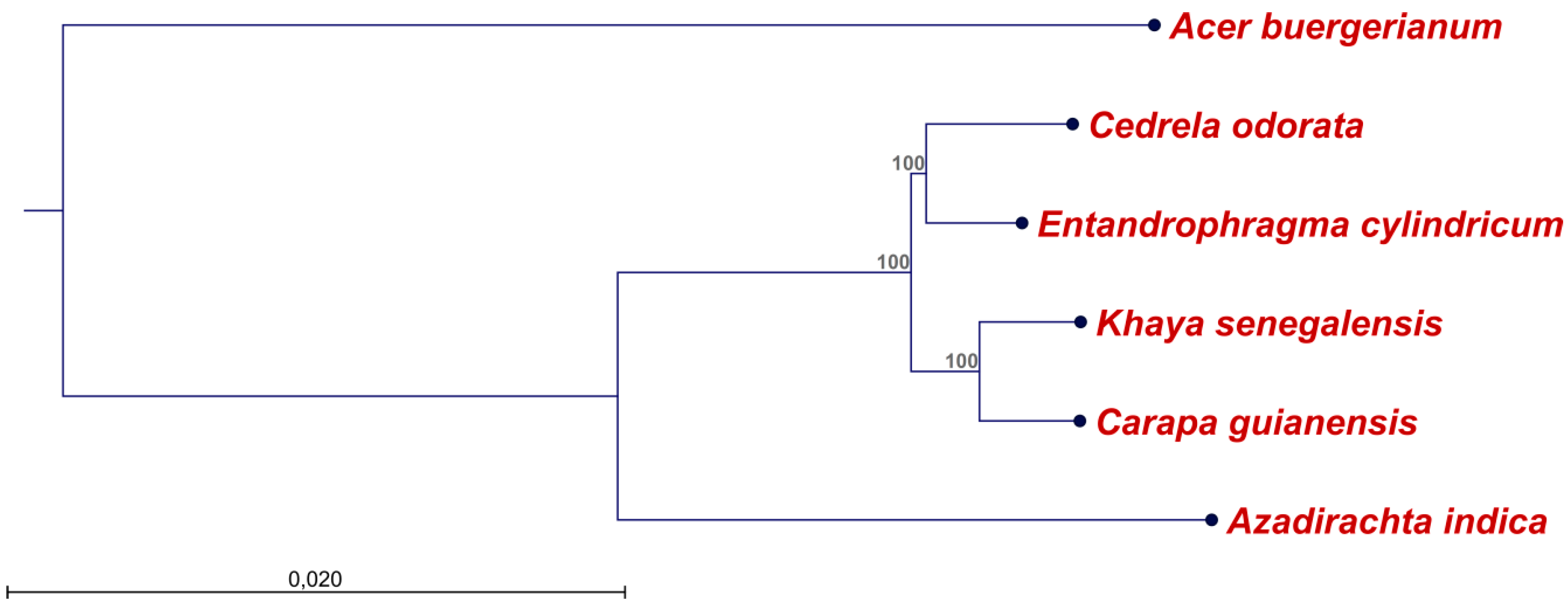
2.2. Diversity of the Meliaceae Plastome Sequences
2.3. Comparative Analyses for the Identification of Potential Meliaceae-Specific Plastome DNA Variations in One Barcoding Region
3. Discussion
4. Materials and Methods
4.1. Sampling, DNA Extraction and Sequencing
4.2. Assemblies of Chloroplast Genome Sequences and Annotation
4.2.1. Assembly of cpDNA Sequences of Khaya Senegalensis and Entandrophragma Cylindricum
4.2.2. Assembly of cpDNA Sequences of Carapa Guianensis and Cedrela Odorata
4.3. Annotation of the cpDNA Sequences and Preparation of GenBank Submission Files
4.4. Alignments and Construction of a Phylogenetic Tree
4.5. SNP and Indel Detection in Multiple Alignments Using SNiPlay
4.6. NCBI-Blast Analyses of matK and Download of matK Gene Sequences of Different Taxonomic Groups for Multiple Alignments
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| cp | chloroplast |
| cpDNA | chloroplast DNA |
| CITES | Convention on International Trade in Endangered Species of Wild Fauna and Flora |
| IRa | Inverted Repeat a |
| IRb | Inverted Repeat b |
| IUCN | International Union for Conservation of Nature |
| LSC | Large Single-Copy Region |
| NGS | Next-Generation Sequencing |
| SSC | Small Single-Copy Region |
References
- The Plant List. Available online: http://www.theplantlist.org/ (accessed on 15 January 2018).
- The IUCN Red List of Threatened Species. Available online: http://www.iucnredlist.org/ (accessed on 11 January 2018).
- CITES—Convention on International Trade in Endangered Species of Wild Fauna and Flora/the Cites Species. Available online: https://cites.org/eng/disc/species.php (accessed on 11 January 2018).
- Birky, C.W. Uniparental inheritance of mitochondrial and chloroplast genes—Mechanisms and evolution. Proc. Natl. Acad. Sci. USA 1995, 92, 11331–11338. [Google Scholar] [CrossRef] [PubMed]
- Drouin, G.; Daoud, H.; Xia, J. Relative rates of synonymous substitutions in the mitochondrial, chloroplast and nuclear genomes of seed plants. Mol. Phylogenet. Evol. 2008, 49, 827–831. [Google Scholar] [CrossRef] [PubMed]
- Barcode of Life. Available online: http://www.barcodeoflife.org/ (accessed on 1 January 2018).
- Hollingsworth, P.M.; Forrest, L.L.; Spouge, J.L.; Hajibabaei, M.; Ratnasingham, S.; van der Bank, M.; Chase, M.W.; Cowan, R.S.; Erickson, D.L.; Fazekas, A.J.; et al. A DNA barcode for land plants. Proc. Natl. Acad. Sci. USA 2009, 106, 12794–12797. [Google Scholar] [CrossRef]
- Shaw, J.; Shafer, H.L.; Leonard, O.R.; Kovach, M.J.; Schorr, M.; Morris, A.B. Chloroplast DNA sequence utility for the lowest phylogenetic and phylogeographic inferences in angiosperms: The tortoise and the hare IV. Am. J. Bot. 2014, 101, 1987–2004. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Li, H.L.; Chen, Z.D. Analysis of plastid and nuclear DNA data in plant phylogenetics-evaluation and improvement. Sci. China Life Sci. 2014, 57, 280–286. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Moore, M.J.; Soltis, P.S.; Bell, C.D.; Burleigh, J.G.; Soltis, D.E. Phylogenetic analysis of 83 plastid genes further resolves the early diversification of eudicots. Proc. Natl. Acad. Sci. USA 2010, 107, 4623–4628. [Google Scholar] [CrossRef] [PubMed]
- Carbonell-Caballero, J.; Alonso, R.; Ibanez, V.; Terol, J.; Talon, M.; Dopazo, J. A phylogenetic analysis of 34 chloroplast genomes elucidates the relationships between wild and domestic species within the genus Citrus. Mol. Biol. Evol. 2015, 32, 2015–2035. [Google Scholar] [CrossRef] [PubMed]
- Goodwin, S.; McPherson, J.D.; McCombie, W.R. Coming of age: Ten years of next-generation sequencing technologies. Nat. Rev. Genet. 2016, 17, 333–351. [Google Scholar] [CrossRef] [PubMed]
- Dodsworth, S. Genome skimming for next-generation biodiversity analysis. Trends Plant Sci. 2015, 20, 525–527. [Google Scholar] [CrossRef] [PubMed]
- Straub, S.C.K.; Parks, M.; Weitemier, K.; Fishbein, M.; Cronn, R.C.; Liston, A. Navigating the tip of the genomic iceberg: Next-generation sequencing for plant systematics. Am. J. Bot. 2012, 99, 349–364. [Google Scholar] [CrossRef] [PubMed]
- Pakull, B.; Mader, M.; Kersten, B.; Ekue, M.R.M.; Dipelet, U.G.B.; Paulini, M.; Bouda, Z.H.N.; Degen, B. Development of nuclear, chloroplast and mitochondrial SNP markers for Khaya sp. Conserv. Genet. Resour. 2016, 8, 283–297. [Google Scholar] [CrossRef]
- Schroeder, H.; Cronn, R.; Yanbaev, Y.; Jennings, T.; Mader, M.; Degen, B.; Kersten, B. Development of molecular markers for determining continental origin of wood from white oaks (Quercus l. Sect. Quercus). PLoS ONE 2016, 11, e0158221. [Google Scholar] [CrossRef] [PubMed]
- Kersten, B.; Rampant, P.F.; Mader, M.; Le Paslier, M.C.; Bounon, R.; Berard, A.; Vettori, C.; Schroeder, H.; Leple, J.C.; Fladung, M. Genome sequences of Populus tremula chloroplast and mitochondrion: Implications for holistic poplar breeding. PLoS ONE 2016, 11, e0147209. [Google Scholar] [CrossRef] [PubMed]
- Tonti-Filippini, J.; Nevill, P.G.; Dixon, K.; Small, I. What can we do with 1000 plastid genomes? Plant J. 2017, 90, 808–818. [Google Scholar] [CrossRef] [PubMed]
- Duminil, J.; Kenfack, D.; Viscosi, V.; Grumiau, L.; Hardy, O.J. Testing species delimitation in sympatric species complexes: The case of an african tropical tree, Carapa spp. (Meliaceae). Mol. Phylogenet. Evol. 2012, 62, 275–285. [Google Scholar] [CrossRef] [PubMed]
- Holtken, A.M.; Schroder, H.; Wischnewski, N.; Degen, B.; Magel, E.; Fladung, M. Development of DNA-based methods to identify cites-protected timber species: A case study in the Meliaceae family. Holzforschung 2012, 66, 97–104. [Google Scholar] [CrossRef]
- Wicke, S.; Schneeweiss, G.M.; dePamphilis, C.W.; Muller, K.F.; Quandt, D. The evolution of the plastid chromosome in land plants: Gene content, gene order, gene function. Plant Mol. Biol. 2011, 76, 273–297. [Google Scholar] [CrossRef] [PubMed]
- Lohse, M.; Drechsel, O.; Kahlau, S.; Bock, R. Organellargenomedraw-a suite of tools for generating physical maps of plastid and mitochondrial genomes and visualizing expression data sets. Nucleic Acids Res. 2013, 41, W575–W581. [Google Scholar] [CrossRef] [PubMed]
- Bausher, M.G.; Singh, N.D.; Lee, S.B.; Jansen, R.K.; Daniell, H. The complete chloroplast genome sequence of Citrus sinensis (L.) Osbeck var ‘ridge pineapple’: Organization and phylogenetic relationships to other angiosperms. BMC Plant Biol. 2006, 6, 21. [Google Scholar] [CrossRef] [PubMed]
- Organelle Resources at NCBI. Available online: http://www.ncbi.nlm.nih.gov/genome/organelle/ (accessed on 1 February 2018).
- Jansen, R.K.; Cai, Z.; Raubeson, L.A.; Daniell, H.; Depamphilis, C.W.; Leebens-Mack, J.; Muller, K.F.; Guisinger-Bellian, M.; Haberle, R.C.; Hansen, A.K.; et al. Analysis of 81 genes from 64 plastid genomes resolves relationships in angiosperms and identifies genome-scale evolutionary patterns. Proc. Natl. Acad. Sci. USA 2007, 104, 19369–19374. [Google Scholar] [CrossRef] [PubMed]
- Green, B.R. Chloroplast genomes of photosynthetic eukaryotes. Plant J. 2011, 66, 34–44. [Google Scholar] [CrossRef] [PubMed]
- Millen, R.S.; Olmstead, R.G.; Adams, K.L.; Palmer, J.D.; Lao, N.T.; Heggie, L.; Kavanagh, T.A.; Hibberd, J.M.; Giray, J.C.; Morden, C.W.; et al. Many parallel losses of infa from chloroplast DNA during angiosperm evolution with multiple independent transfers to the nucleus. Plant Cell 2001, 13, 645–658. [Google Scholar] [CrossRef] [PubMed]
- Lu, L.; Li, X.; Hao, Z.; Yang, L.; Zhang, J.; Peng, Y.; Xu, H.; Lu, Y.; Zhang, J.; Shi, J.; et al. Phylogenetic studies and comparative chloroplast genome analyses elucidate the basal position of halophyte Nitraria sibirica (Nitrariaceae) in the sapindales. Mitochondrial DNA Part A DNA Mapp. Seq. Anal. 2017, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Daniell, H.; Lin, C.S.; Yu, M.; Chang, W.J. Chloroplast genomes: Diversity, evolution, and applications in genetic engineering. Genome Biol. 2016, 17, 134. [Google Scholar] [CrossRef] [PubMed]
- Williams, A.V.; Miller, J.T.; Small, I.; Nevill, P.G.; Boykin, L.M. Integration of complete chloroplast genome sequences with small amplicon datasets improves phylogenetic resolution in Acacia. Mol. Phylogenet. Evol. 2016, 96, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Koenen, E.J.M.; Clarkson, J.J.; Pennington, T.D.; Chatrou, L.W. Recently evolved diversity and convergent radiations of rainforest mahoganies (Meliaceae) shed new light on the origins of rainforest hyperdiversity. New Phytol. 2015, 207, 327–339. [Google Scholar] [CrossRef] [PubMed]
- Pennington, T.D.; Styles, B.T. A generic monograph of the meliaceae. Blumea 1975, 22, 419–540. [Google Scholar]
- Muellner, A.N.; Samuel, R.; Johnson, S.A.; Cheek, M.; Pennington, T.D.; Chase, M.W. Molecular phylogenetics of Meliaceae (Sapindales) based on nuclear and plastid DNA sequences. Am. J. Bot. 2003, 90, 471–480. [Google Scholar] [CrossRef] [PubMed]
- Muellner, A.N.; Pennington, T.D.; Chase, M.W. Molecular phylogenetics of neotropical Cedreleae (mahogany family, Meliaceae) based on nuclear and plastid DNA sequences reveal multiple origins of “cedrela odorata”. Mol. Phylogenet. Evol. 2009, 52, 461–469. [Google Scholar] [CrossRef] [PubMed]
- Schroeder, H.; Kersten, B.; Fladung, M. Development of multiplexed marker sets to identify the most relevant poplar species for breeding. Forests 2017, 8, 492. [Google Scholar] [CrossRef]
- Soltis, D.E.; Soltis, P.S. Choosing an approach and an appropriate gene for phylogenetic analysis. In Molecular Systematics of Plants II; Soltis, D.E., Soltis, P.S., Doyle, J.J., Eds.; Kluwer Academic Publishers: Boston, MA, USA, 1998; pp. 1–42. [Google Scholar]
- Olmstead, R.G.; Palmer, J.D. Chloroplast DNA systematics—A review of methods and data-analysis. Am. J. Bot. 1994, 81, 1205–1224. [Google Scholar] [CrossRef]
- Neuhaus, H.; Link, G. The chloroplast tRNALys(UUU) gene from mustard (Sinapis alba) contains a class-II intron potentially coding for a maturase-related polypeptide. Curr. Genet. 1987, 11, 251–257. [Google Scholar] [CrossRef] [PubMed]
- Barthet, M.M.; Hilu, K.W. Evaluating evolutionary constraint on the rapidly evolving gene matK using protein composition. J. Mol. Evol. 2008, 66, 85–97. [Google Scholar] [CrossRef] [PubMed]
- Braham Bioinformatics/Fastqc. Available online: http://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed on 1 December 2017).
- Hahn, C.; Bachmann, L.; Chevreux, B. Reconstructing mitochondrial genomes directly from genomic next-generation sequencing reads-a baiting and iterative mapping approach. Nucleic Acids Res. 2013, 41. [Google Scholar] [CrossRef] [PubMed]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed]
- Camacho, C.; Coulouris, G.; Avagyan, V.; Ma, N.; Papadopoulos, J.; Bealer, K.; Madden, T.L. Blast+: Architecture and applications. BMC Bioinf. 2009, 10, 421. [Google Scholar] [CrossRef] [PubMed]
- Genbank. Available online: http://www.ncbi.nlm.nih.gov/genbank/ (accessed on 1 January 2018).
- Ncbi/Blast. Available online: https://blast.ncbi.nlm.nih.gov/Blast.cgi?PROGRAM=blastn&PAGE_TYPE=BlastSearch&LINK_LOC=blasthome (accessed on 19 January 2018).
- Liu, C.; Shi, L.C.; Zhu, Y.J.; Chen, H.M.; Zhang, J.H.; Lin, X.H.; Guan, X.J. Cpgavas, an integrated web server for the annotation, visualization, analysis, and genbank submission of completely sequenced chloroplast genome sequences. BMC Genom. 2012, 13, 715. [Google Scholar] [CrossRef] [PubMed]
- CpGAVAS: Chloroplast Genome Annotation, Visualization, Analysis and Genbank. Available online: http://www.herbalgenomics.org/0506/cpgavas (accessed on 1 February 2018).
- Chlorobox/genbank2sequin. Available online: https://chlorobox.mpimp-golm.mpg.de/GenBank2Sequin.html (accessed on 1 February 2018).
- NCBI/sequin. Available online: https://www.ncbi.nlm.nih.gov/Sequin/ (accessed on 1 February 2018).
- OrganellarGenomeDRAW. Available online: http://ogdraw.mpimp-golm.mpg.de/ (accessed on 1 February 2018).
- VISTA. Available online: http://genome.lbl.gov/vista/index.shtml (accessed on 1 February 2018).
- Mayor, C.; Brudno, M.; Schwartz, J.R.; Poliakov, A.; Rubin, E.M.; Frazer, K.A.; Pachter, L.S.; Dubchak, I. VISTA: Visualizing global DNA sequence alignments of arbitrary length. Bioinformatics 2000, 16, 1046–1047. [Google Scholar] [CrossRef] [PubMed]
- SNiPlay Pipeline v2. Available online: http://sniplay.cirad.fr/cgi-bin/analysis.cgi (accessed on 1 February 2018).
- Dereeper, A.; Homa, F.; Andres, G.; Sempere, G.; Sarah, G.; Hueber, Y.; Dufayard, J.F.; Ruiz, M. SNiPlay3: A web-based application for exploration and large scale analyses of genomic variations. Nucleic Acids Res. 2015, 43, W295–W300. [Google Scholar] [CrossRef] [PubMed]
| Species | Individual (Thuenen-ID) | Origin/Location | Longitude | Latitude | Trimmed Reads | Coverage * |
|---|---|---|---|---|---|---|
| Cedrela odorata | CEODO_205_2 | Cuba, population Guisa | −76.68 | 20.16 | 254214 | 101× |
| Entandrophragma cylindricum | c-5-ENTC-46 | Cameroon, FBR, Parc National de Lobeke | 15.6442 | 2.26286 | 2206300 | 165× |
| Khaya senegalensis | KS | Unknown/Green house, Thünen Institute Hamburg-Lohbrügge | 2783117 | 346× | ||
| Carapa guianensis | CAGUI_332_1 | French Guiana, region Rorota | −52.262392 | 4.87761 | 422759 | 135× |
| Cedrela Odorata (MG724915) | Entandrophragma Cylindricum (KY923074.1) | Khaya Senegalensis (KX364458.1) | Carapa Guianensis (MF401522.1) | Azadirachta Indica * (KF986530.1) | |
|---|---|---|---|---|---|
| Genome size (bp) | 158,558 | 159,609 | 159,787 | 159,483 | 160,737 |
| LSC length (bp) | 86,390 | 87,117 | 87,404 | 87,054 | 88,137 |
| SSC length (bp) | 18,380 | 18,532 | 18,311 | 18,277 | 18,636 |
| IR length (bp) | 26,894 | 26,980 | 27,036 | 27,076 | 26,982 |
| Number of genes | 130 | 130 | 130 | 130 | 131 |
| Function | Genes |
| RNAs, ribosomal | rrn23, rrn16, rrn5, rrn4.5 |
| RNAs, transfer | trnA-UGC *, trnC-GCA, trnD-GUC, trnE-UUC, trnF-GAA, trnG-GCC, trnG-UCC *, trnH-GUG, trnI-CAU, trnI-GAU *, trnK-UUU *, trnL-CAA, trnL-UAA *, trnL-UAG, trnM-CAU, trnfM-CAU, trnN-GUU, trnP-UGG, trnQ-UUG, trnR-ACG, trnR-UCU, trnS-GCU, trnS-GGA, trnS-UGA, trnT-GGU, trnT-UGU, trnV-GAC, trnV-UAC *, trnW-CCA, trnY-GUA |
| Transcription and splicing | rpoA, rpoB, rpoC1 *, rpoC2, matK |
| Translation, ribosomal proteins | |
| Small subunit | rps2, rps3, rps4, rps7, rps8, rps11, rps12 **,T, rps14, rps15, rps16 *, rps18, rps19 |
| Large subunit | rpl2 *, rpl14, rpl16 *, rpl20, rpl22, rpl23, rpl32, rpl33, rpl36 |
| Photosynthesis | |
| ATP synthase | atpA, atpB, atpE, atpF *, atpH, atpI |
| Photosystem I | psaA, psaB, psaC, psaI, psaJ, ycf3 **, ycf4 |
| Photosystem II | psbA, psbB, psbC, psbD, psbE, psbF, psbH, psbI, psbJ, psbK, psbL, psbM, psbN, psbT, psbZ |
| Calvin cycle | rbcL |
| Cytochrome complex | petA, petB *, petD *, petG, petL, petN |
| NADH dehydrogenase | NdhA *, ndhB *, ndhC, ndhD, ndhE, ndhF, ndhG, ndhH, ndhI, ndhJ, ndhK |
| Others | clpP1 **, accD, cemA, ccsA, ycf1, ycf2 |
| Position (Consensus) | Position (Cedrela odorata) | Meliaceae | Sapindales without Meliaceae | Rosids without Sapindales | All without Rosids |
|---|---|---|---|---|---|
| 208 | 208 | T | A or C or T | C or A | C or A |
| 280 | 262 | T or C 1 | |||
| 346 | 328 | G | C or T | C or A | C or A |
| 402 | 378 | A | C or T or A or G | A or C | A or C |
| 574 | 550 | C or T 3 | |||
| 618 | 588 | C or A 1 | |||
| 639 | 609 | G | T or G or A or C | T or G or A or C | G or A or T |
| 861 | 819 | T | C | C or T or G | C |
| 995 | 953 | C or T 4 | |||
| 1194 | 1146 | T | C or T | C or T | C |
| 1237 | 1189 | G or A 2 or C 1 | |||
| 1239 | 1191 | G or A 3 | |||
| 1318 | 1270 | G | A | A | A |
| 1389 | 1341 | C or T 1 | |||
| 1478 | 1430 | C | G or A | G or A or T | A or G |
| 1494 | 1446 | T | A or G | G or A | G or C |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mader, M.; Pakull, B.; Blanc-Jolivet, C.; Paulini-Drewes, M.; Bouda, Z.H.-N.; Degen, B.; Small, I.; Kersten, B. Complete Chloroplast Genome Sequences of Four Meliaceae Species and Comparative Analyses. Int. J. Mol. Sci. 2018, 19, 701. https://doi.org/10.3390/ijms19030701
Mader M, Pakull B, Blanc-Jolivet C, Paulini-Drewes M, Bouda ZH-N, Degen B, Small I, Kersten B. Complete Chloroplast Genome Sequences of Four Meliaceae Species and Comparative Analyses. International Journal of Molecular Sciences. 2018; 19(3):701. https://doi.org/10.3390/ijms19030701
Chicago/Turabian StyleMader, Malte, Birte Pakull, Céline Blanc-Jolivet, Maike Paulini-Drewes, Zoéwindé Henri-Noël Bouda, Bernd Degen, Ian Small, and Birgit Kersten. 2018. "Complete Chloroplast Genome Sequences of Four Meliaceae Species and Comparative Analyses" International Journal of Molecular Sciences 19, no. 3: 701. https://doi.org/10.3390/ijms19030701
APA StyleMader, M., Pakull, B., Blanc-Jolivet, C., Paulini-Drewes, M., Bouda, Z. H.-N., Degen, B., Small, I., & Kersten, B. (2018). Complete Chloroplast Genome Sequences of Four Meliaceae Species and Comparative Analyses. International Journal of Molecular Sciences, 19(3), 701. https://doi.org/10.3390/ijms19030701