1. Introduction
Citrus fruits have been consumed by humans for thousands of years, and are increasingly consumed worldwide for their nutritional and medicinal value [
1]. Functional compounds, such as β-cryptoxanthin, β-carotene, folate, vitamin C, and quercetin are some recognized bioactive ingredients of citrus that defend against cancers [
2,
3,
4,
5]. For instance, some studies have reported that intake of citrus fruits results in reduced risk of breast and stomach cancers [
6,
7]. Other studies have focused on evaluating the antioxidant capacity of citrus extracts, their protective effect against oxidative damage, and other defensive responses against certain diseases such as lowering high blood-pressure risk or preventing kidney-stone formation [
8,
9]. However, despite the increased demand for citrus, studies related to the potential toxicity of citrus fruits are limited and restricted. Therefore, the focal point of our study was to investigate and identify the toxicity of citrus fruits to ensure their safety for various applications.
The citron (
Citrus medica) is one of the three original citrus species and has already developed extensive applications for medicinal purposes, such as to treat sea sickness, pulmonary troubles, and intestinal ailments. With the improvement of processing technology, various citron fruit applications that maximize its functional advantages have been developed, but are still limited because of the botanical characteristics of the citron. Unlike other citrus fruits, its thick white rind is the main content of the citron and its pulp is dry with little juice [
10]. Hence, citron has had only restricted use in the food industry, mainly as candied peel. Meanwhile, few studies have provided a biological examination of citron, such as of its bioactivity, pharmacology, toxicology, or bioavailability. Therefore, experimental studies to improve use of the citron and to determine its biological properties are essential.
Nanotechnology, as a cutting-edge scientific field, has many prospects for applications in various fields, such as medicine delivery, computer electronics, materials science, and for the food industry [
11]. The application of nanotechnology in food science not only significantly increases the economic benefits, but also remarkably reforms food quality and safety [
12,
13,
14]. Given the special nature of nanoparticles, the changes in the properties of food products are apparent, such as the improvement of food bioavailability, solubility, stability, biological activity, and safety [
14]. To broaden the application of the citron and to further elaborate its biological benefits, the citron was prepared in nanometer scale by means of nanotechnology, and experiments were performed to measure its cell viability and genotoxicity by employing diverse in vitro models. By conducting comparative studies, we evaluated the different effects of citron extracts on different cell lines at the cellular and genomic levels.
3. Discussion
Functional foods, as one area of the food industry, have grown a lot over the years [
18]. Benefiting from the improvement of food technology, especially with the emergence of nanotechnology, it is expected that the functional foods will further advance, with promising prospects [
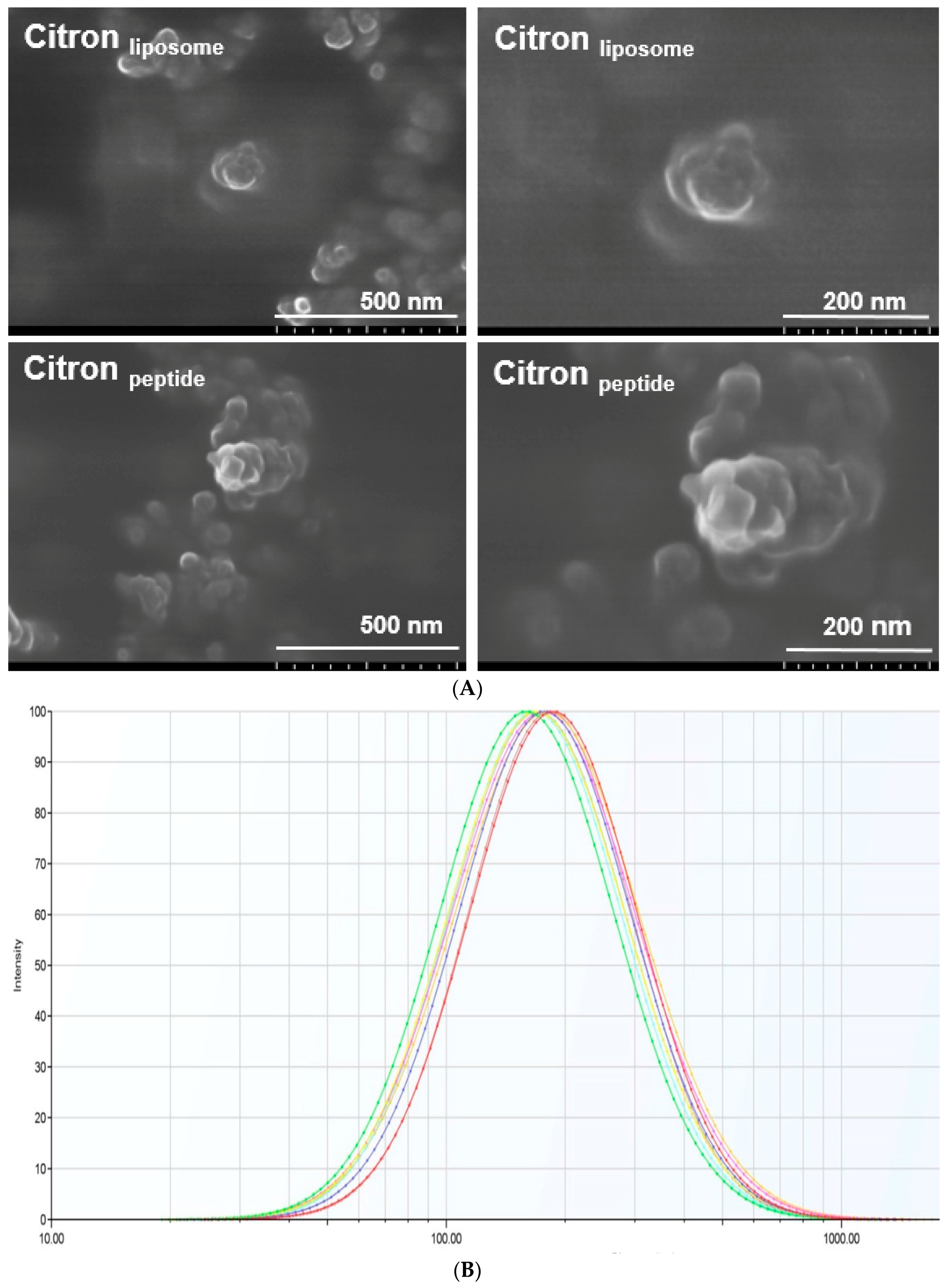
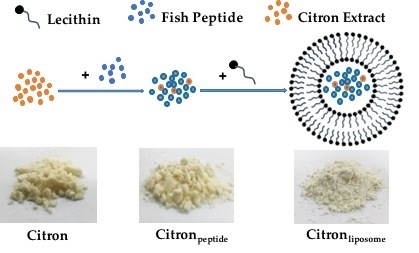
19]. In this study, by using nanotechnology, we prepared citron extract at a nanometer scale by incorporating functional ingredients, liposome, and peptide. We employed fish oligopeptide and water-soluble liposome to formulate citron nanoparticles. With predominant properties in viscosity and foaming, oligopeptide has been widely used in food processing. Meanwhile, because of its high yield, low cost, and the beneficial effects in human body, we used fish collagen peptide to process citron
peptide into nanoparticle, as shown in
Figure 1A. The typical size of citron
peptide nanoparticles was around 100 nm. However, because of the biological limitation of oligopeptide, such as chemical degradation, low stability, and bioavailability, physicochemical instability, and low permeability, its development in the food industry is restricted. To avoid the deficiencies of peptide, we used both liposome and oligopeptide to formulate citron nanoparticles, labeled as citron
liposome in
Figure 1A. The content ratio of three types of citron samples (Citron, Citron
peptide, and Citron
liposome) are depicted in
supplementary data Table S1. Furthermore, the description of the citron extract compositions such as general composition (
Table S2), amino acid (
Table S3), fatty acid (
Table S4), minerals (
Table S5), and vitamins (
Table S6) are provided in
supplementary data file as well (obtained from Rural Development Administration, Korea).
Next, regarding the determination of citron nanoparticle, the morphology of citron
peptide and citron
liposome clearly demonstrated the formation of citron extracts nanoparticles. On the other hand, concerns remain with the significantly reduced size of particles when utilizing nanotechnology. For example, it has been reported that particles with a diameter of less than 100 nm may induce toxic effects [
20]. To assess the toxicity of citron nanoparticles and provide quantitative references for various applications, we confirmed the biological toxicity, as shown in the results section.
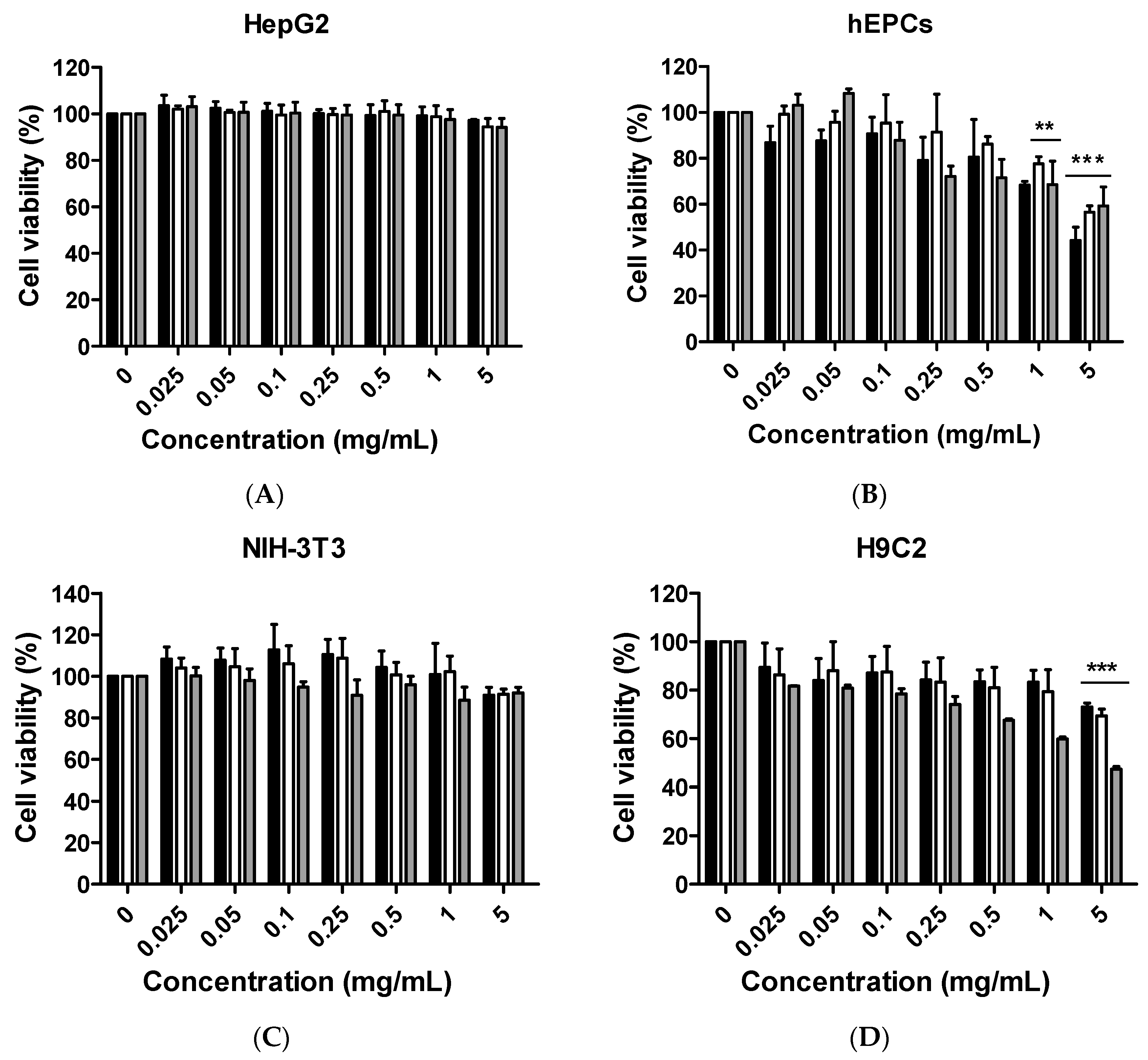
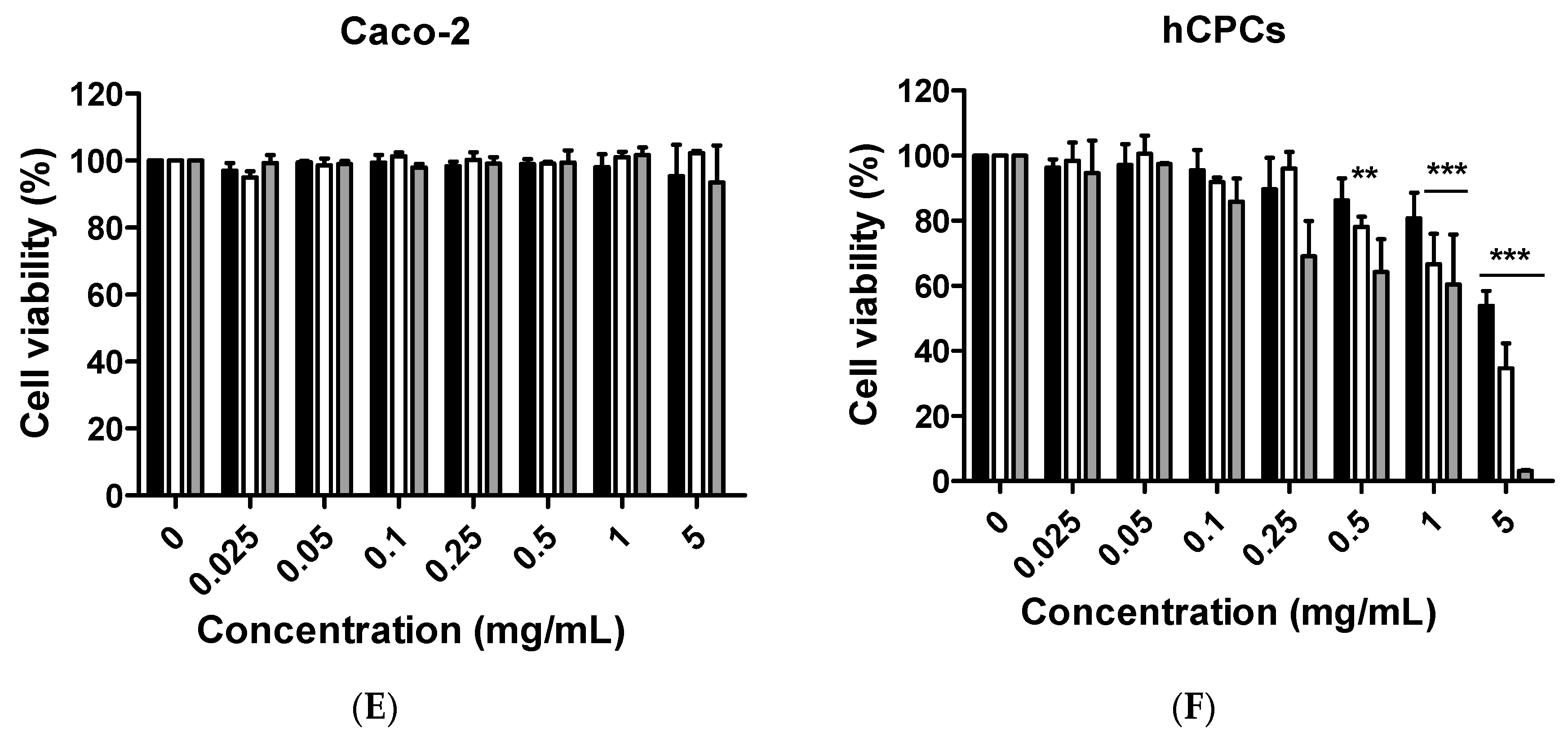
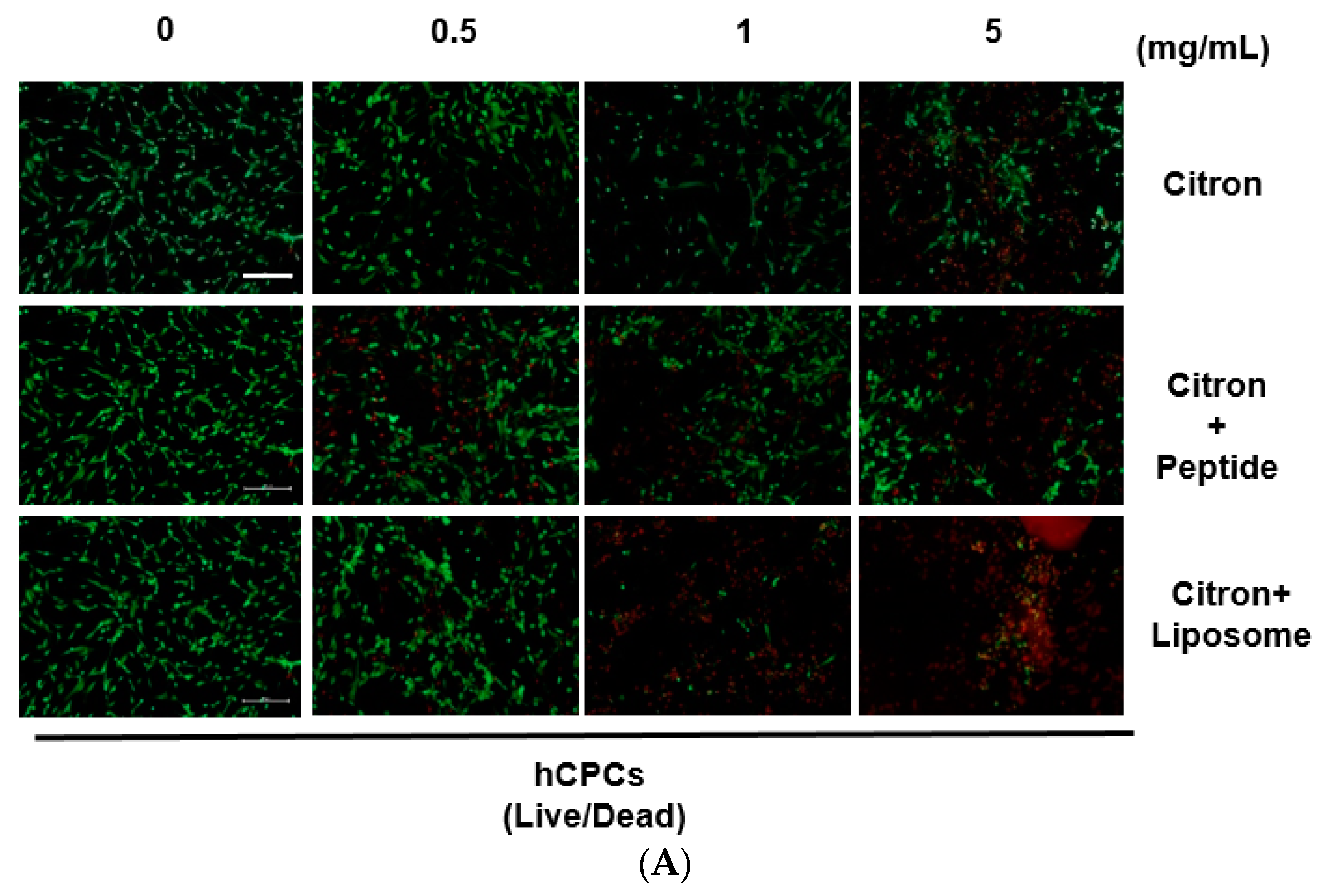
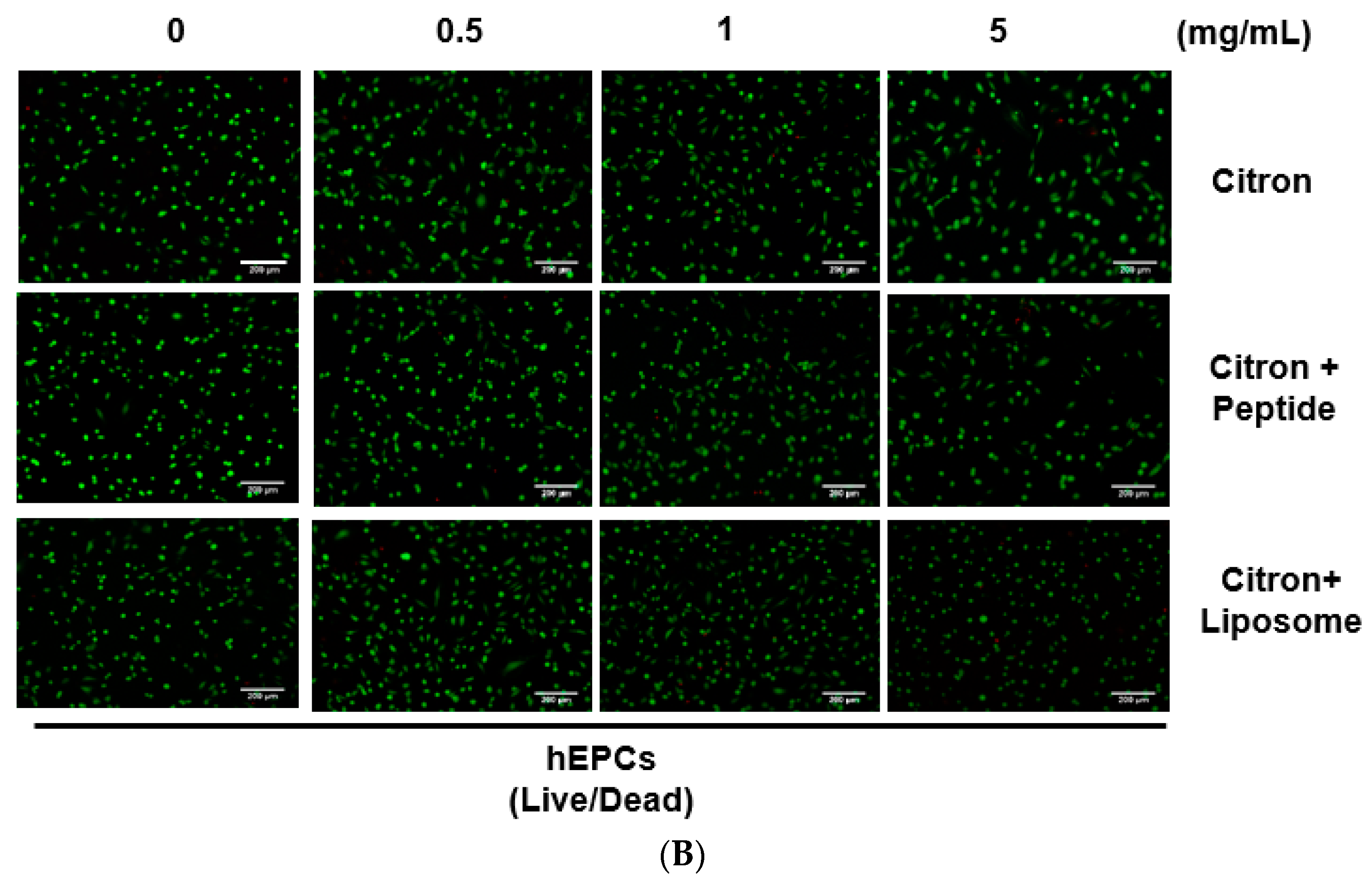
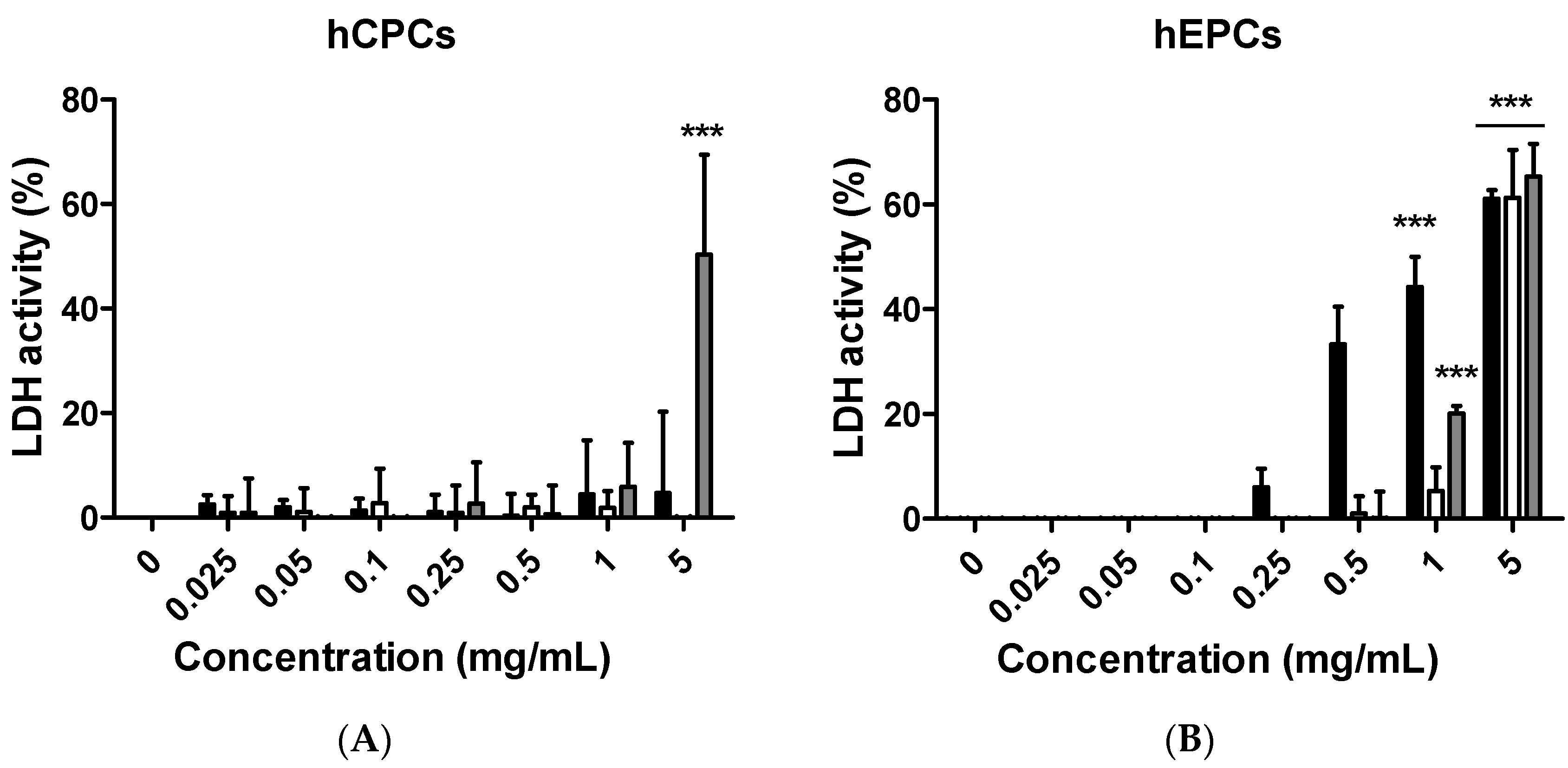
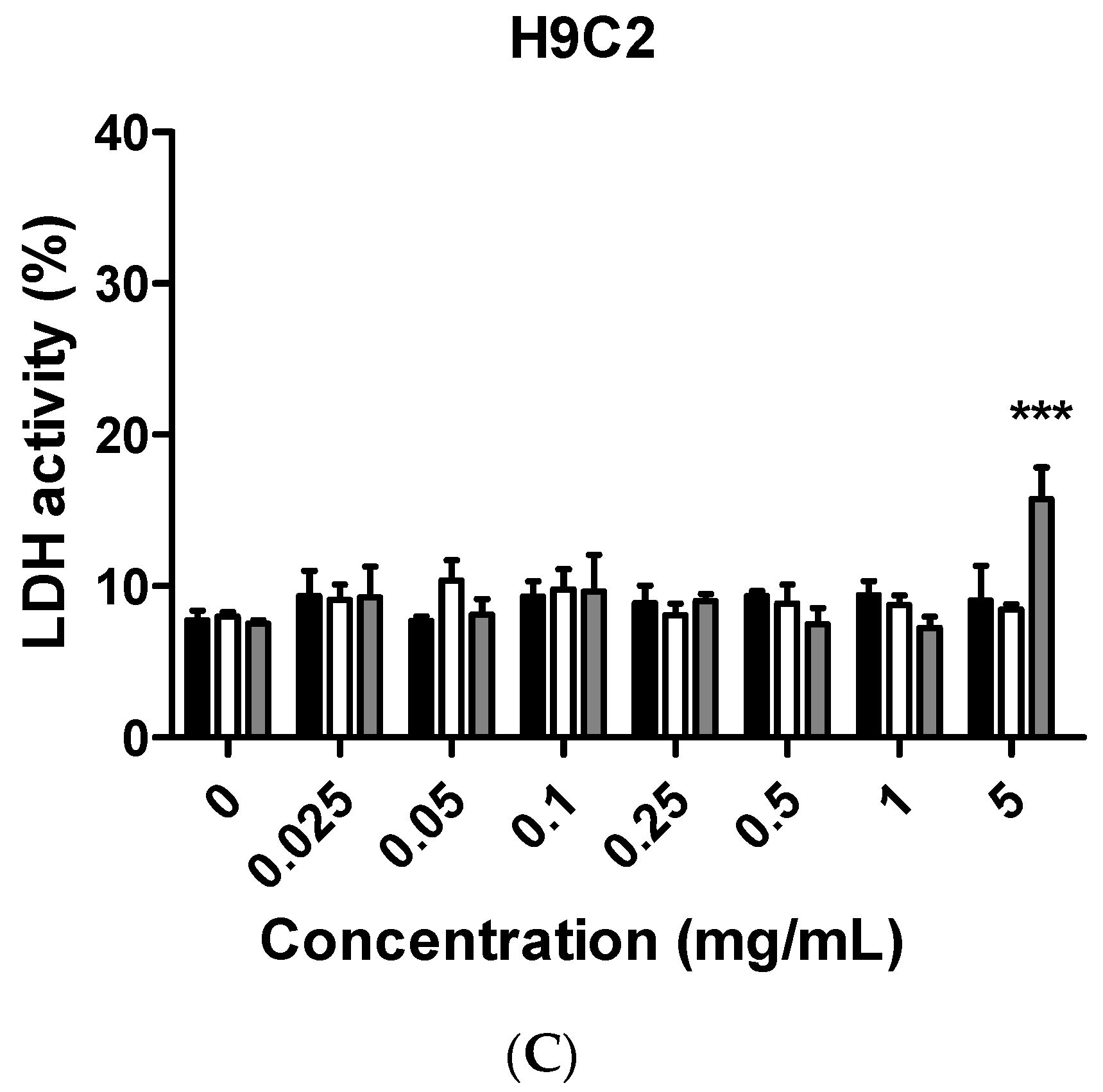
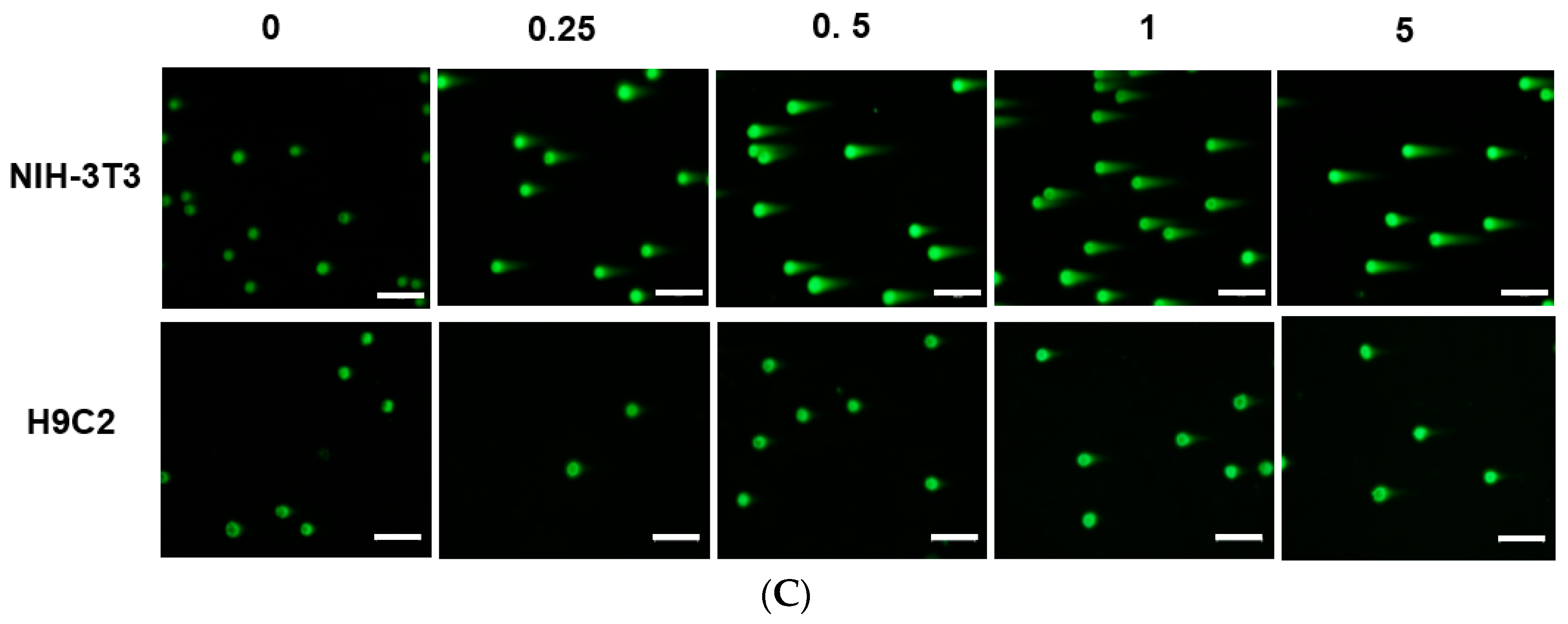
Although citron is well-known for its medicinal usage, we know little about its application as food and its bioactivities. Here, in an in vitro test, six different cells were used to measure the toxicity of citron extracts. Embryonic fibroblasts from mice (NIH-3T3), heart myoblasts from rats (H9C2), a human liver-cancer cell line (HepG2), and human epithelial colorectal adenocarcinoma cells (Caco-2) were selected to test the different effects. In addition, we also used two human-derived primary cells, human cardiac progenitor cells (hCPC) and human endothelial progenitor cells (hEPC), to examine the toxicity and make the results more credible. Our work showed that the cytotoxicity of citron extract had two aspects. One aspect was that the cytotoxicity was cell-type dependent (
Figure 2), selectively against primary hCPCs and hEPCs, especially in the citron
liposome group. The other was the genotoxicity (
Figure 5). No cytotoxicity was observed in NIH-3T3 and H9C2 cell lines, but citron
peptide can cause genotoxicity in those two cell lines. Interestingly, in the presence of liposome in citron
liposome, the gene damage disappeared. With the particles being in the nanometer scale, nanoparticles may penetrate biological membranes and have access to the cell. Nanoparticle can also have different biodistributions in the human body, perhaps because of different affinities with the diverse cell types or organs. In recent years, some studies have explored the nanotoxicity of nanoparticles [
21,
22]. In this view, we can conclude that citron extract behaved differently for different cell types. For instance, citron
liposome may show a higher affinity with hCPCs and hEPCs than other cell lines to cause cell damage (
Figure 2), and with high affinity for genes, citron
peptide can have a genotoxic effect in the H9C2 or NIH-3T3 cell lines (
Figure 5). We also discovered that citron extracts did not suppress human liver cancer cell lines or human epithelial colorectal adenocarcinoma cells, HepG2 and Caco-2.
The main purpose of this study is to identify the potential toxicity of citron nanoparticles (citronpeptide and citronliposome) in an in vitro system, and to provide accessible information for further industrial production and application. Many studies have shown that nanotoxicity can emerge with the decreased particle size to nanometer scale. Because the size of citronpeptide and citronliposome is between 100 and 200 nm, it is essential to examine their toxicity. Our results demonstrate that cytotoxicity increased in some cell lines, like hCPCs, hEPCs, and H9C2, after treatment with citronpeptide or citronliposome. Meanwhile, citronpeptide caused genotoxicity in the NIH-3T3 and H9C2 cell lines, but not cytotoxicity. These results clearly show the toxic effects of citron nanoparticles. In addition, although our studies provide information about the cytotoxicity of citron for applications in the food industry, more research is needed to reveal the mechanism before use in final food products.
4. Materials and Methods
4.1. Chemicals and Kits
Trizma base, N-laurylsarcosine, Triton X-100, boric acid, and 3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) were obtained from Sigma-Aldrich (Madison, WI, USA). The cell-culture medium and dish were acquired from Welgene (Daegu, Korea). Fetal bovine serum (FBS) was bought from Welgene (Daegu, Korea). The LIVE/DEAD® Viability/Cytotoxicity Assay Kit was purchased from Invitrogen (Carlsbad, CA, USA). The lactate dehydrogenase (LDH) Cytotoxicity Assay kit was obtained from Thermo Fisher Scientific (Waltham, MA, USA). All of the other reagents and chemicals were purchased from commercial companies.
4.2. Preparation of Samples
The method used for preparation of liposome has been described previously. Briefly, the 2% lecithin (Metarin P, Cargill Texturizing Solutions, Hamburg, Germany) was added to 0.5% peptide (Naticol 4000, Weishardt International, Graulhel, France) aqueous solution at a ratio of 1:1. Then, after homogenization for 3 min at 8000 rpm using a high-speed homogenizer (IKAT25 digital ULTRA-TURRAX®, Germany), the extracts were collected and homogenized again at 80 W for 3 min by using an ultrasonicator. The citron extract containing 50% citron and 50% sucrose was prepared by mixing with water (1:1 w/w) using a blender to make citron syrup for further study. The citronpeptide was obtained by mixing with fish skin peptide. Sixty grams of peptide were used per 0.5 L of citron extract. The citronliposome was obtained by mixing with fish-skin peptide and liposome. The ratio was 1 g liposome and 60 g fish-skin peptide per 0.5 L of citron extract. Finally, all of the samples were dried using a spray dryer (SD-1000, EYELA, Tokyo, Japan) to obtain a powder. The morphology and size of citron extracts were detected by using scanning electron microscope (SEM) and Nano Size analyzer.
4.3. Cell Culture
NIH-3T3 (mouse), H9C2 (rat), and HepG2 (human) were purchased from Korean Cell Line Bank (KCLB, Seoul, Korea) and cultured in high glucose Dulbecco’s Modified Eagle’s Medium (DMEM; Welgene, Daegu, Korea) containing 10% FBS and 1% penicillin/streptomycin (P/S, Welgene). Primary human EPCs and human CPCs were provided by S.M. Kwon (IRB No. 05-2-15-133, Pusan University, Pusan, Korea). The hEPCs were cultured in EC basal medium 2 (EBM-2MV, Lonza, Walkersville, MD, USA) containing 5% FBS, EGM-2-MV Bullet Kit, and 1% penicillin/streptomycin. The hCPCs were cultured in Ham’s F12 medium (Hyclone, Logan, UT, USA) containing 10% FBS, 1% penicillin/streptomycin, 0.005 U/mL human erythropoietin (hEPO, R & D system, Minneapolis, MN, USA), 5 ng/mL human basic fibroblast growth factor (hbFGF, PeproTech, Rocky Hill, NJ, USA), and 0.2 mM L-glutathione reduced (Sigma-Aldrich, St. Louis, MO, USA). All cell lines were cultured at 37 °C in a humidified incubator with 5% CO2 atmosphere. The logarithmic cells were harvested before performing each experiment.
4.4. MTT Assay
The cell viability was measured by employing an MTT assay as described previously [
23]. The cells were seeded into 96-well plates at a density of 1 × 10
5 cells per well and were cultured overnight. After incubation, the medium was replaced by a starvation medium containing 1% FBS for another 24 h. Then, the cells were treated with the desired concentrations of drugs for an additional 24 h. The supernatant was removed, and an MTT solution at a final concentration of 0.25 mg/mL was added in each well to label the cells. The plates were incubated in a humidified incubator for 4 h. Then, 100 μL of dimethyl sulfoxide (DMSO) was used to dissolve the formazan, and the cell viability was detected by measuring the optical density (OD) at the absorbance of 595 nm using BioTek Microplate Readers (Winooski, VT, USA).
4.5. Live/Dead Assay
To visualize the cell viability, the cells were labeled with calcein AM and ethidium homodimer (EthD-1) by using a Live/Dead assay kit. The cells were cultured in 48-well plates at a density of 1 × 105 cells per well for 24 h. After incubation with a starvation medium overnight, the various concentrations of drugs were added in each well and were incubated for the next 24 h. Then, the supernatant was discarded, and the cells were washed once using DPBS. A 500 μL cell-staining solution at a final concentration of 2 µM calcein AM and 4 µM EthD-1 in DPBS was added to each well, and the plates were incubated for 45 min at room temperature by blocking light. Finally, live and dead cells were visualized by using a fluorescence microscope (Nikon, ECLIPSE Ts2, Tokyo, Japan). The live cells are shown in green, the dead cells in red. The images merged by Image J software were used to show co-localization.
4.6. LDH Release Assay
Cell cytotoxicity was evaluated by measuring the enzymatic activity of released LDH from dead cells. Before the measurement, the cells were prepared, as described previously, and then incubated in various concentrations of drugs for 24 h. The plates were then centrifuged at 700× g for 5 min at room temperature using a Beckman Coulter Allegra® X-15R centrifuge. The supernatant was collected to measure the released LDH. The assay was performed according to the manufacturer’s instructions. The absorbance at 450 nm was detected to calculate the cell cytotoxicity.
4.7. Single-Cell Gel Electrophoresis Assay
A single-cell gel electrophoresis assay (comet assay) was carried out to measure the deoxyribonucleic acid (DNA) damage. The method used for this assay analysis has been described previously. Briefly, cells were first seeded into 48-well plates. After incubation with different concentrations of drugs for 24 h, the cells were harvested and mixed with 100 µL 0.7% low-melting-point agarose at a cell density of 2 × 105 per milliliter. Then, the cells were laid onto agarose-coated glass slides, which were stored at 4 °C for solidification until electrophoresis. Slides were placed in a lysis solution (2.5 M NaCl, 0.1 M EDTA, 10 mM Trizma base, 1% N-laurylsarcosine, 0.5% Triton X-100, and 10% DMSO, pH 10) for 1 h at 4 °C. After lysis, slides were submerged in an alkaline solution (300 mM NaOH, 1 mM EDTA, pH > 13) for 30 min at 4 °C. The slides were washed three times with electrophoresis buffer (89 Mm Trisbase, 89 mM boric acid, 2 mM EDTA, pH 8.3) and then subjected to electrophoresis at 25 V for 20 min using a RunOneTM Powder Supply. Then, slides were fixed with absolute ethanol and stained with DNA dye. The cell images were photographed using a fluorescence microscope and Casplab software was used to analysis the comet assay results.
4.8. Statistics
All data were analyzed by GraphPad Prism and shown as means ± standard deviation (SD). Comparisons between the groups were conducted by using two-way analysis of variance with Bonferroni post-tests and/or Student’s t test. Statistical significance was considered to be a p value less than 0.05.
5. Conclusions
In this study, toxicity studies were conducted to determine the possible toxic effects of nano-engineered citron samples in the cellular level and to provide credible information for the future application of citron. To make the result more convincing, two human primary cell lines were employed in our study. Our findings revealed that citronliposome could significantly cause cell damage in primary hEPC and hCPC cells, much greater than the slight damage caused by citron or citronpeptide. However, cytotoxicity was not observed from NIH-3T3, HepG2, and Caco-2 cell lines. In the genotoxicity test, significant toxic effects were not observed from hEPCs or hCPCs, followed by the treatment with citron, citronpeptide, or citronliposome. Minimal genotoxicity was detected from NIH-3T3 cells and in H9C2 cells after the treatment with citronpeptide, but disappeared in the presence of liposome in the citronliposome group. Overall, our findings provide important information for the further use of citron in the food industry. Moreover, in vitro study will be performed to further determine the toxicity in order to meet the final food products.