Oviposition by Spodoptera exigua on Solanum dulcamara Alters the Plant’s Response to Herbivory and Impairs Larval Performance
Abstract
1. Introduction
2. Results
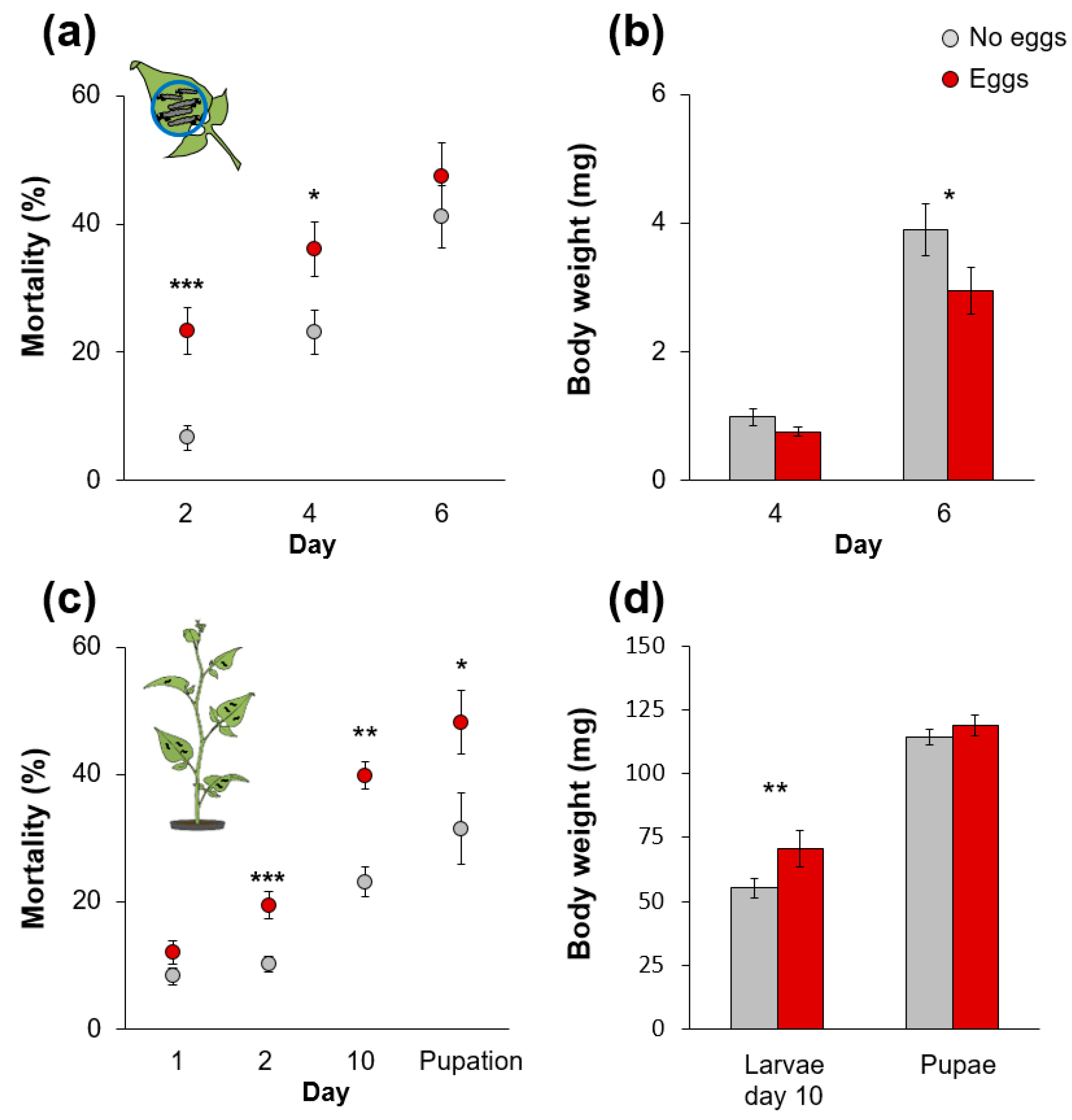
2.1. Moth Oviposition on S. dulcamara Impairs Performance of Subsequently Feeding Larvae
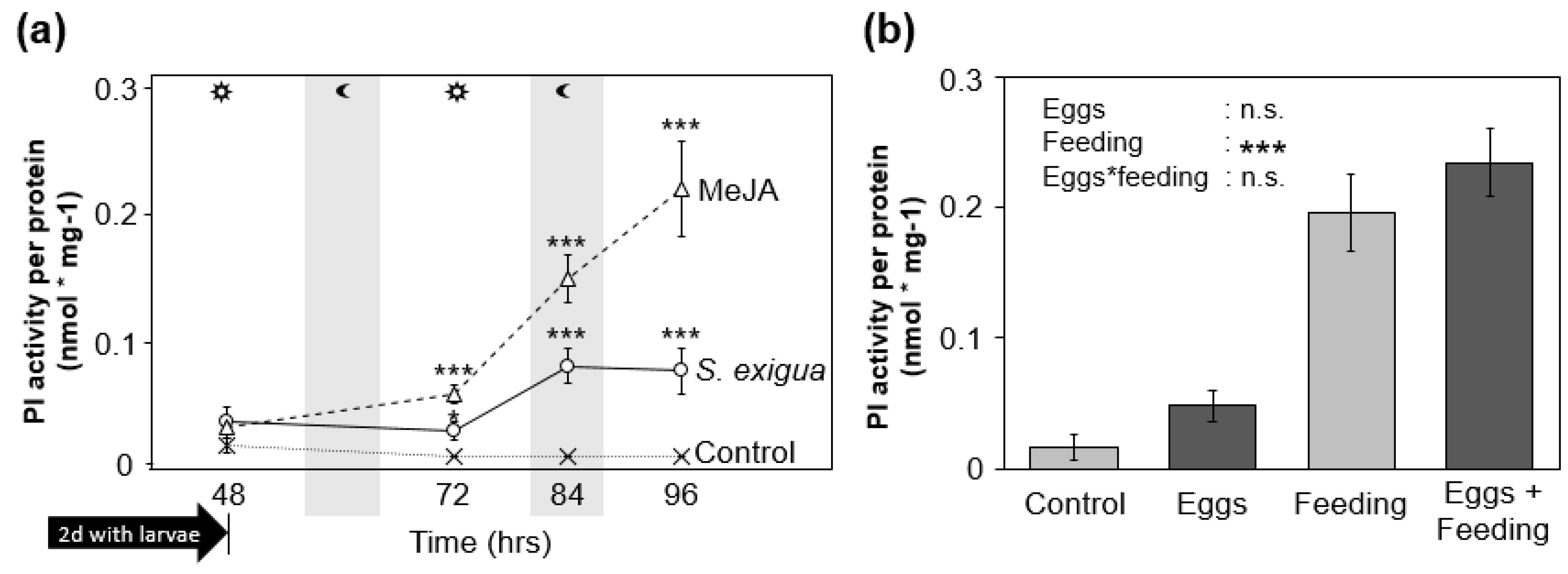
2.2. Feeding-Induced PI Activity in S. dulcamara Is Not Altered by Prior Oviposition
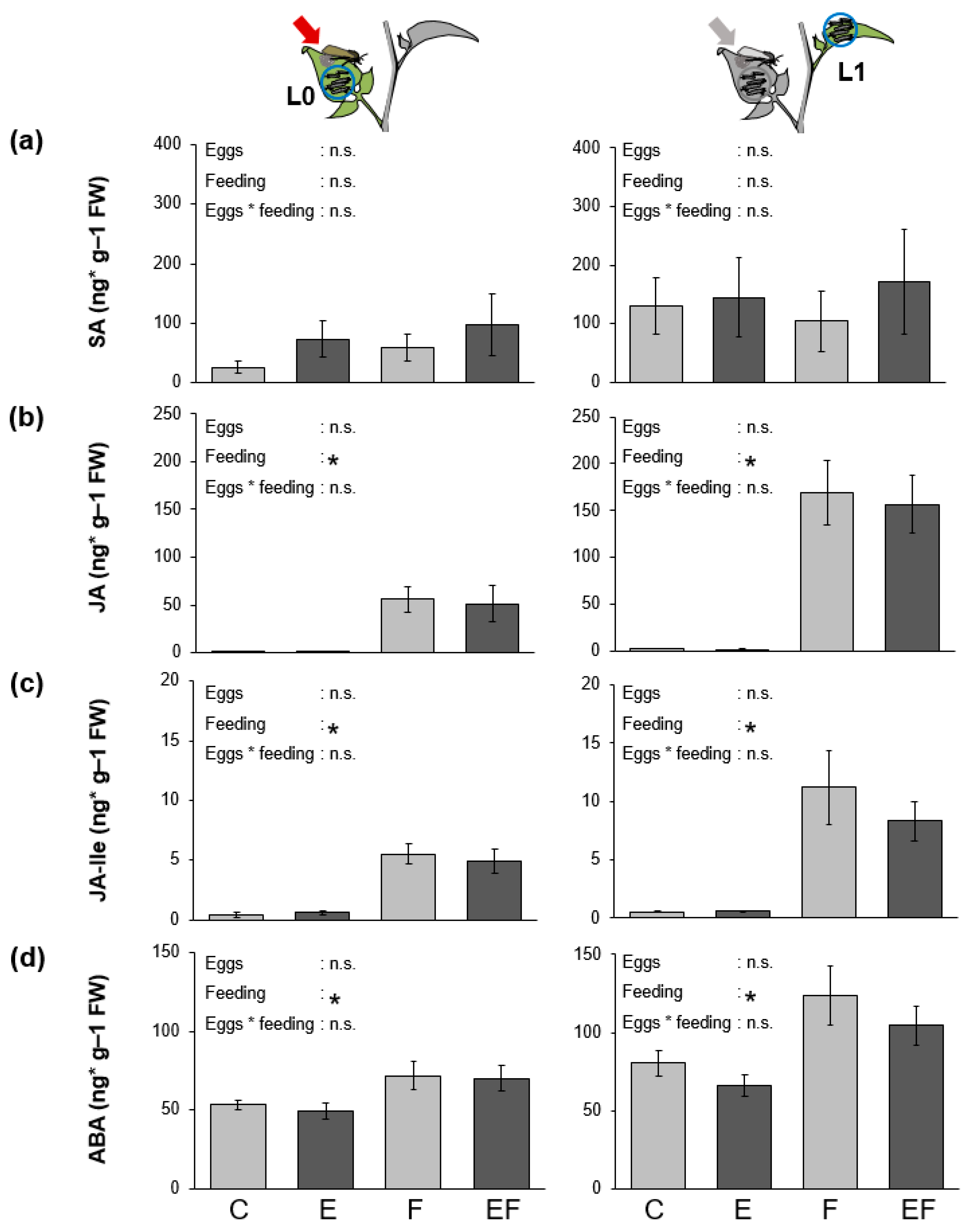
2.3. S. dulcamara’s Phytohormonal Response to Larval Feeding and Oviposition
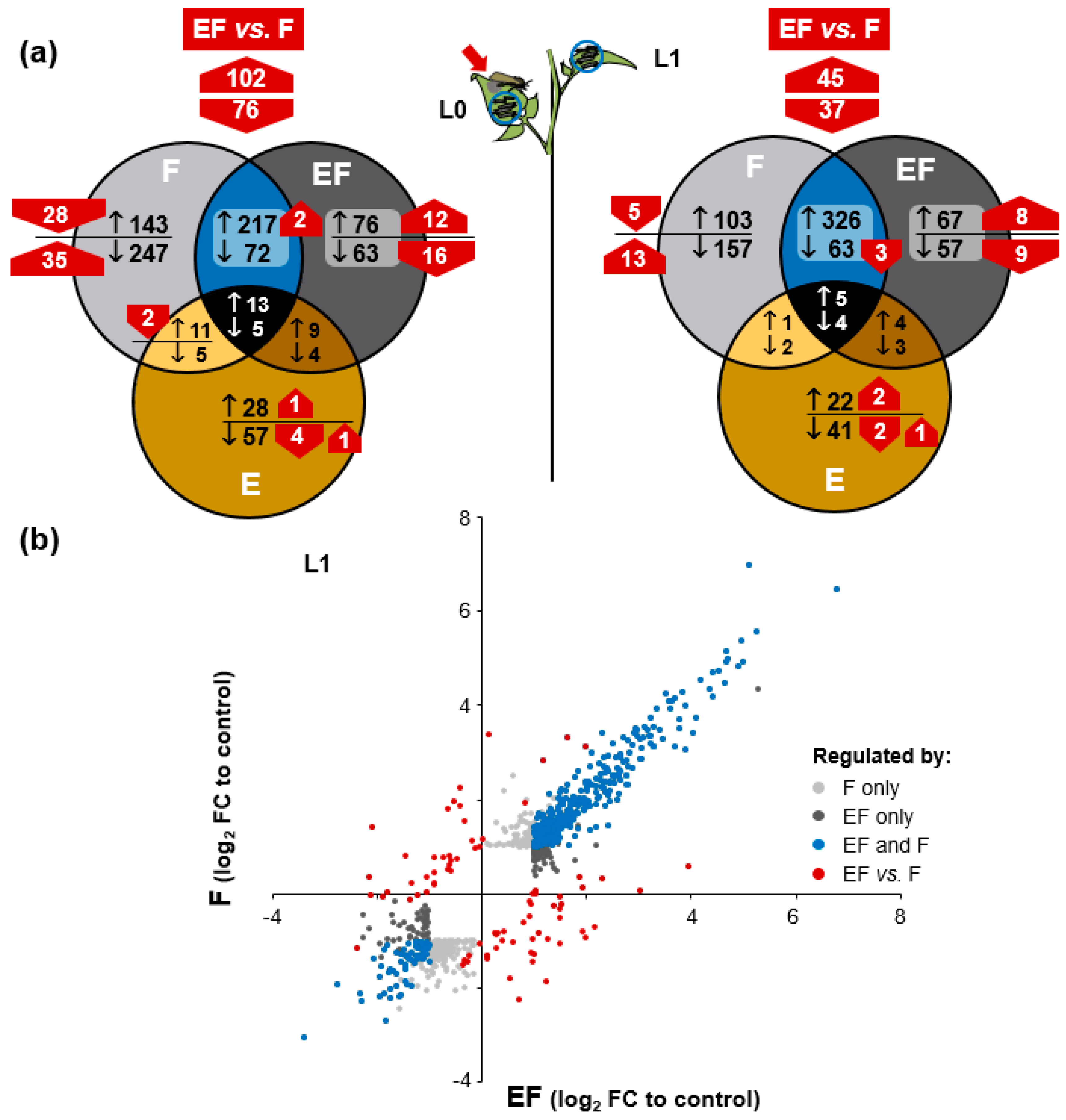
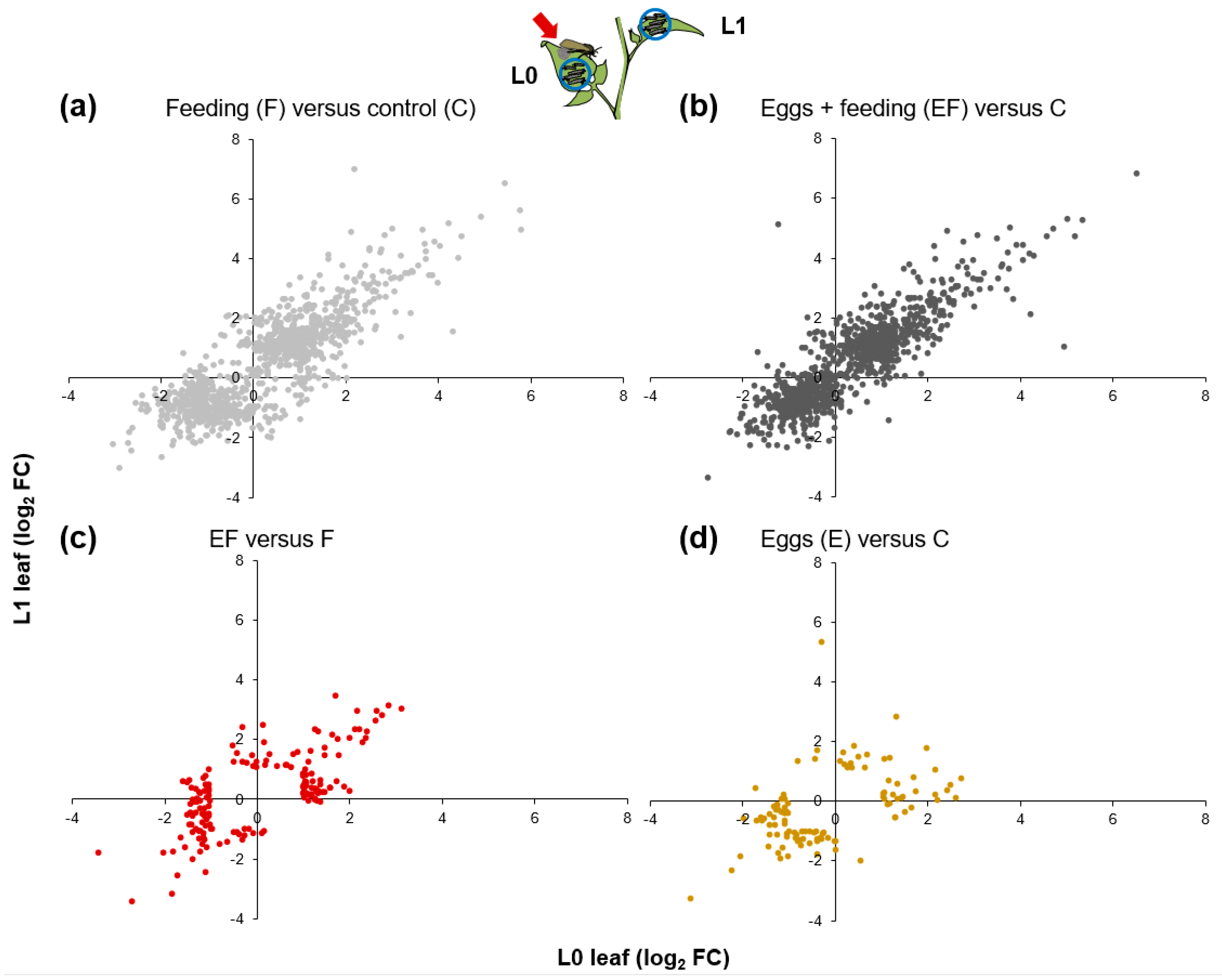
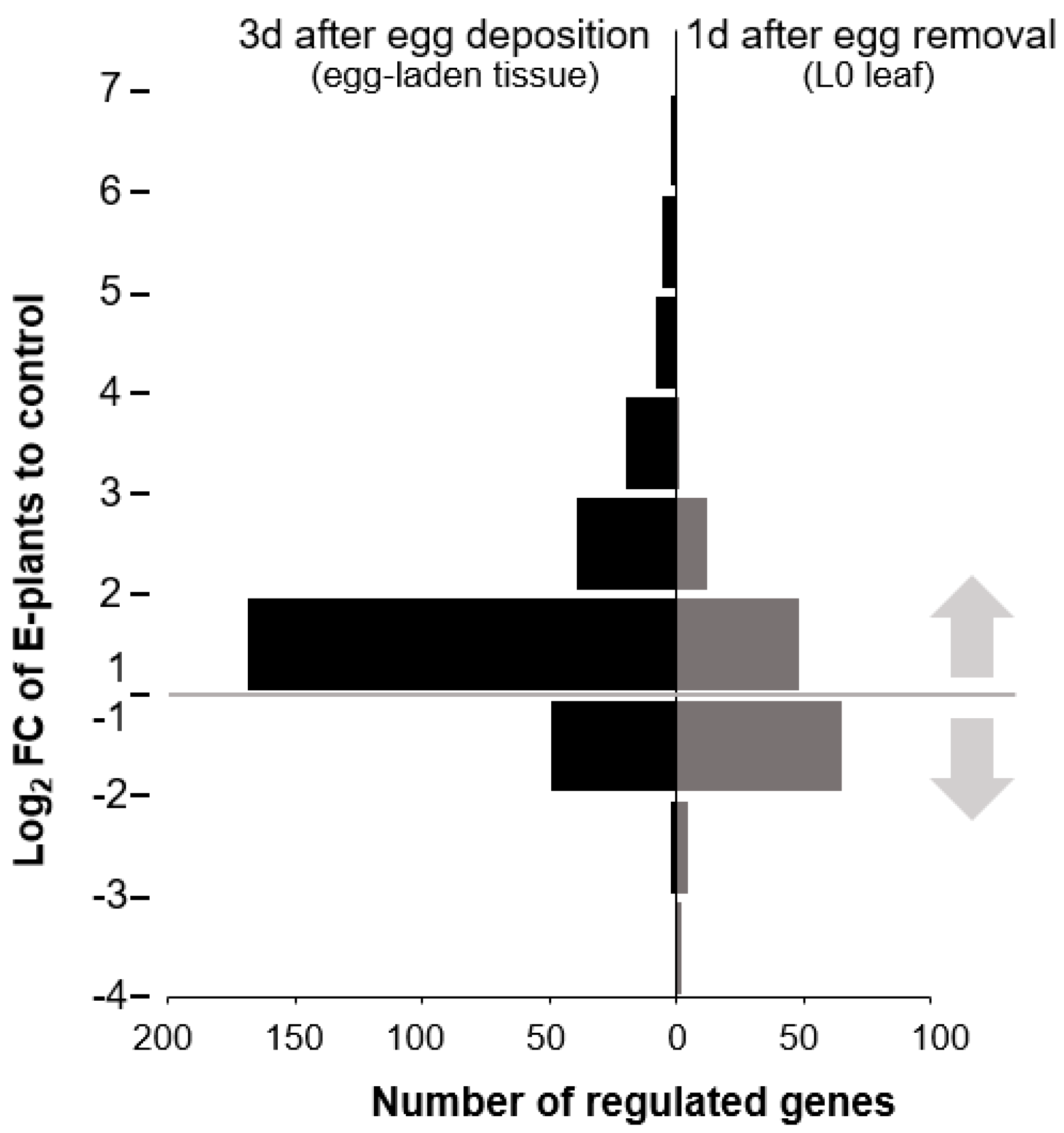
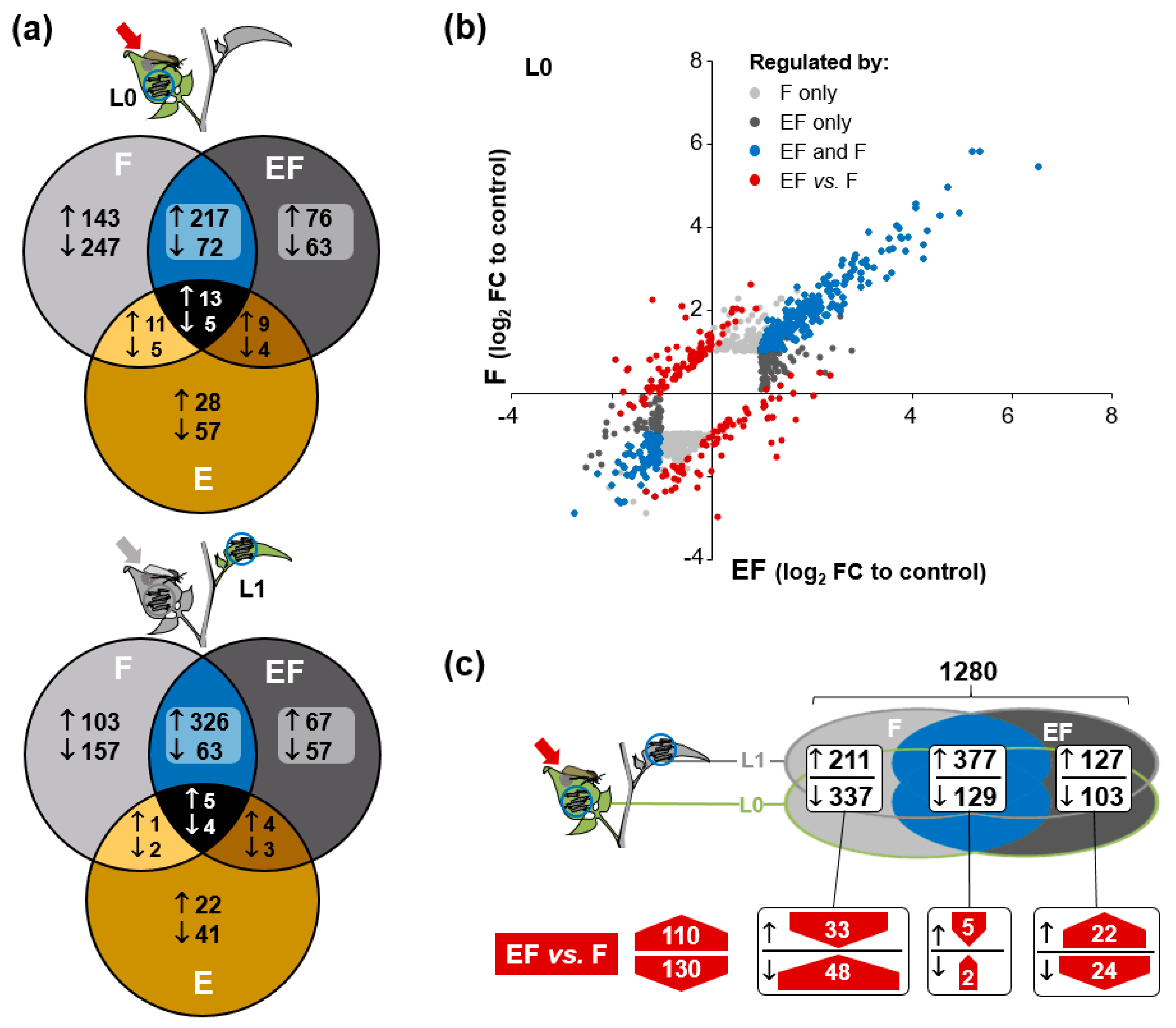
2.4. Oviposition Modifies S. dulcamara’s Transcriptional Response to Larval Feeding
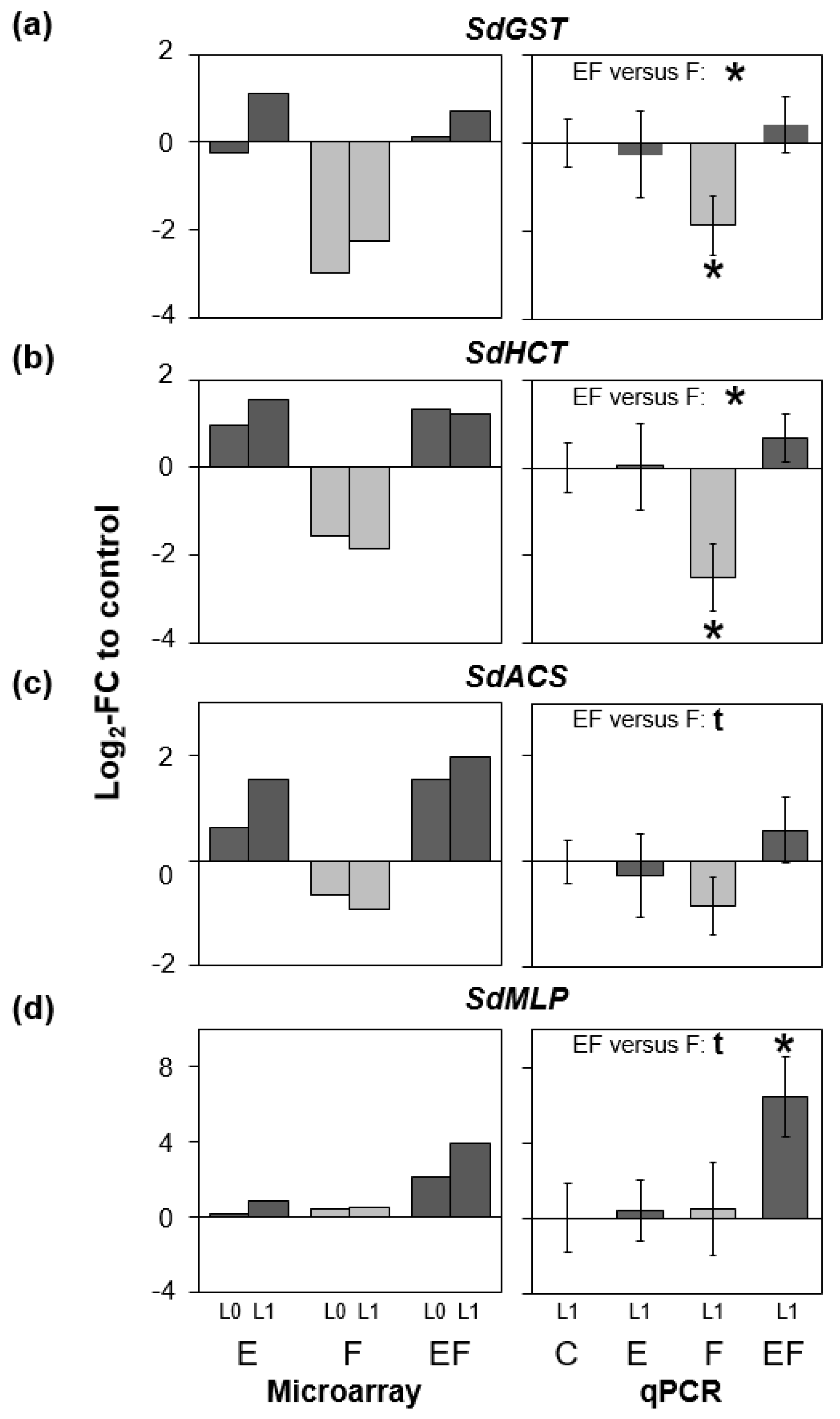
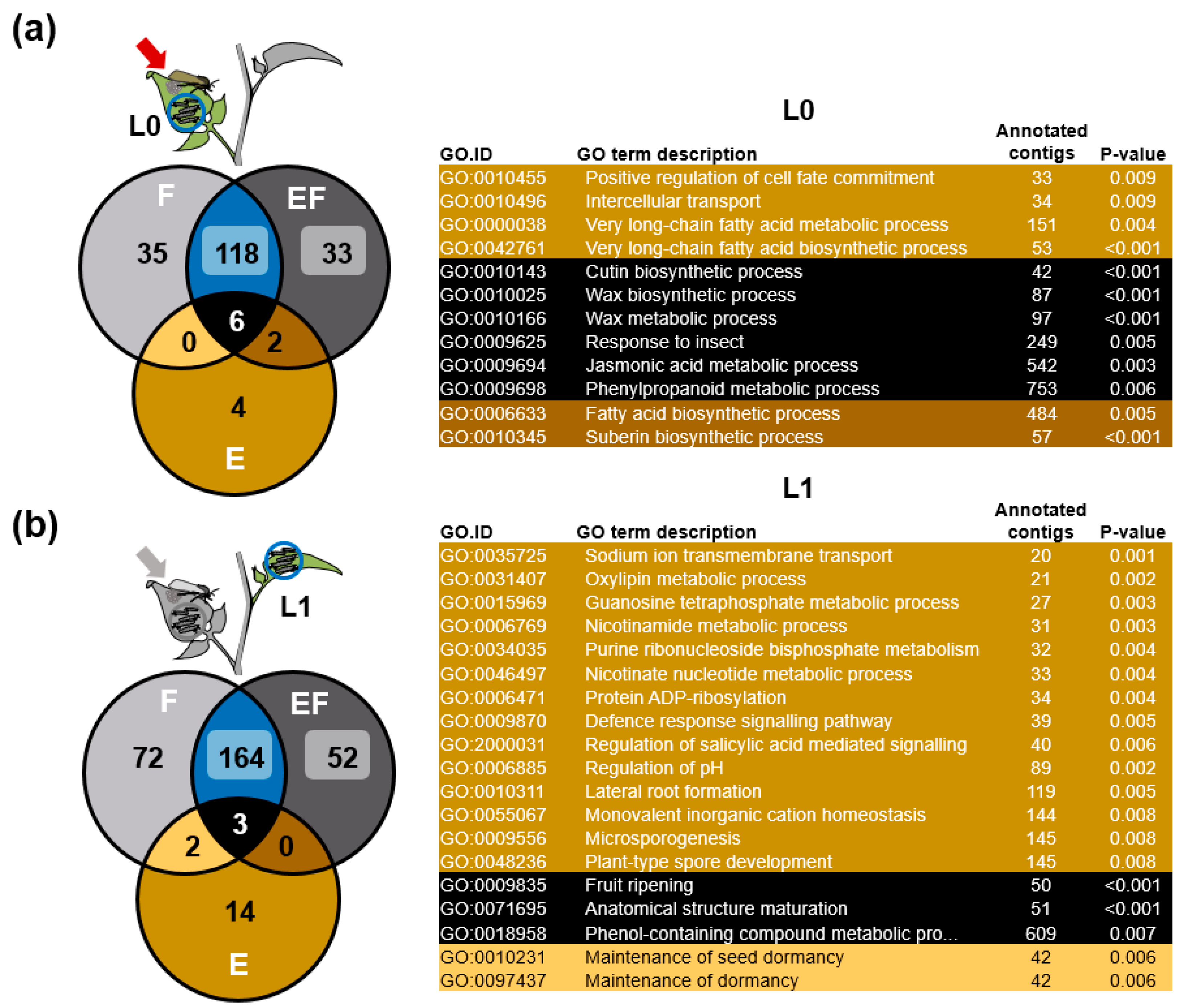
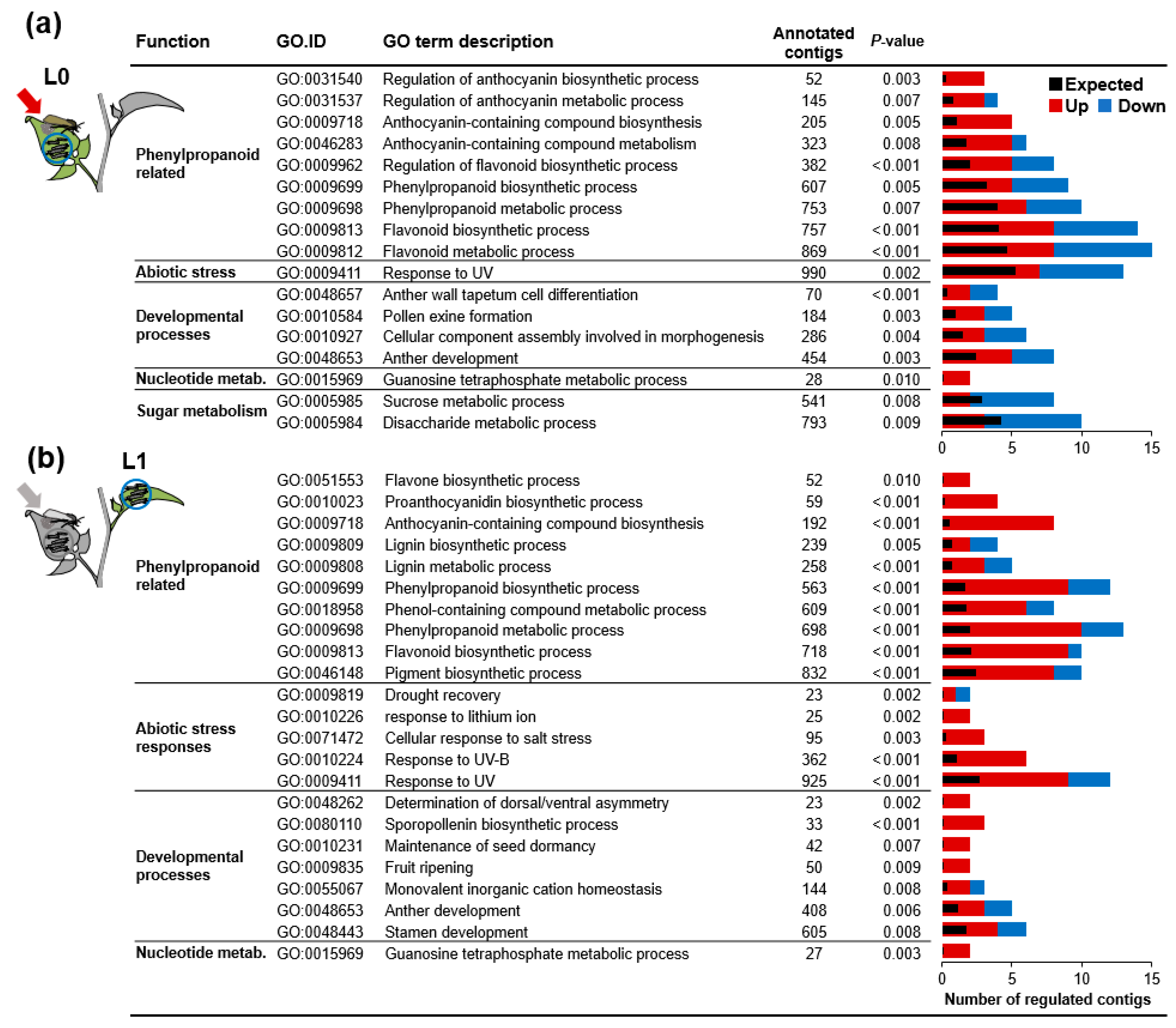
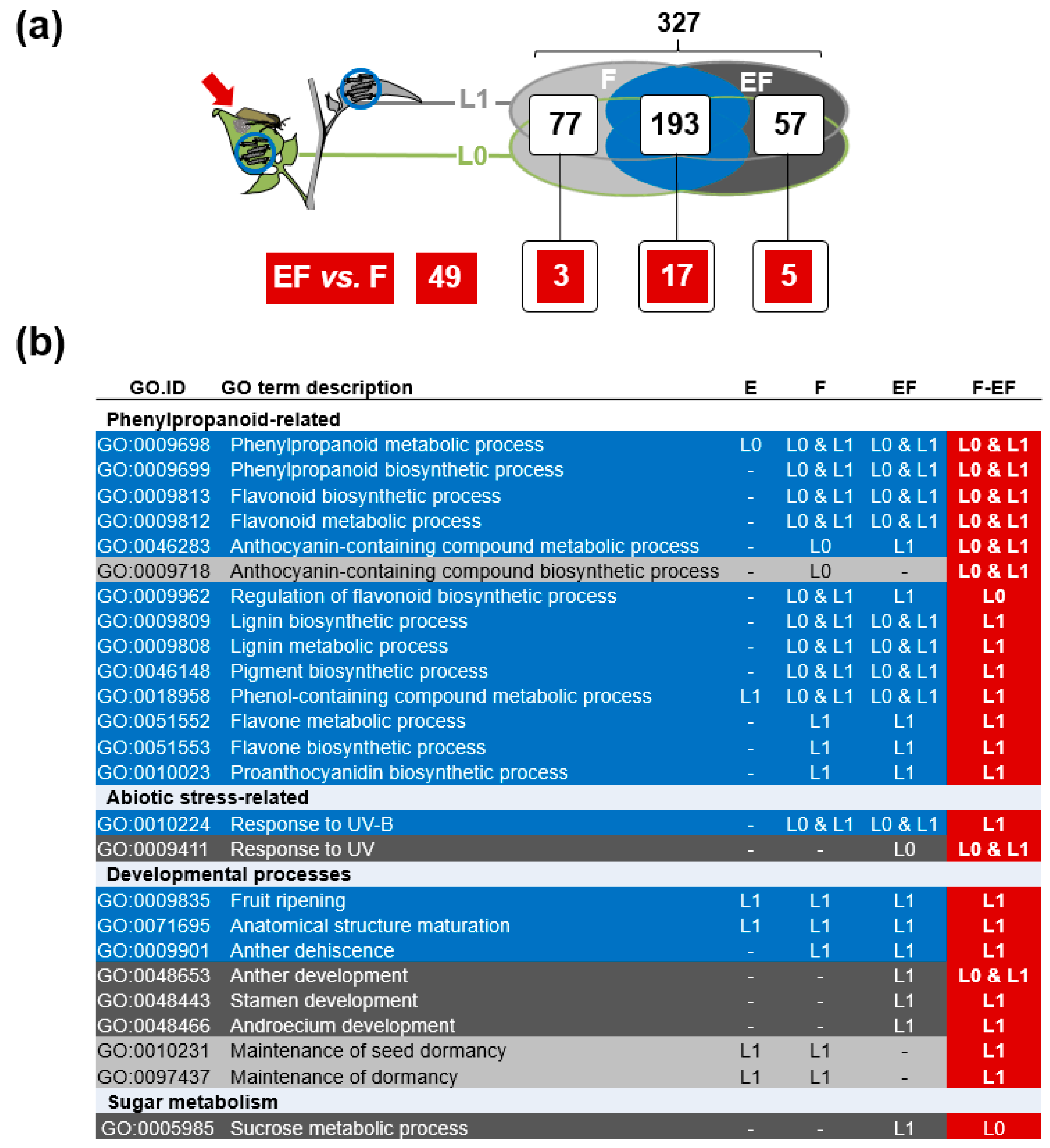
2.5. Genes of Phenylpropanoid-Related Pathways Were More Strongly Expressed in Previously Oviposited Plants
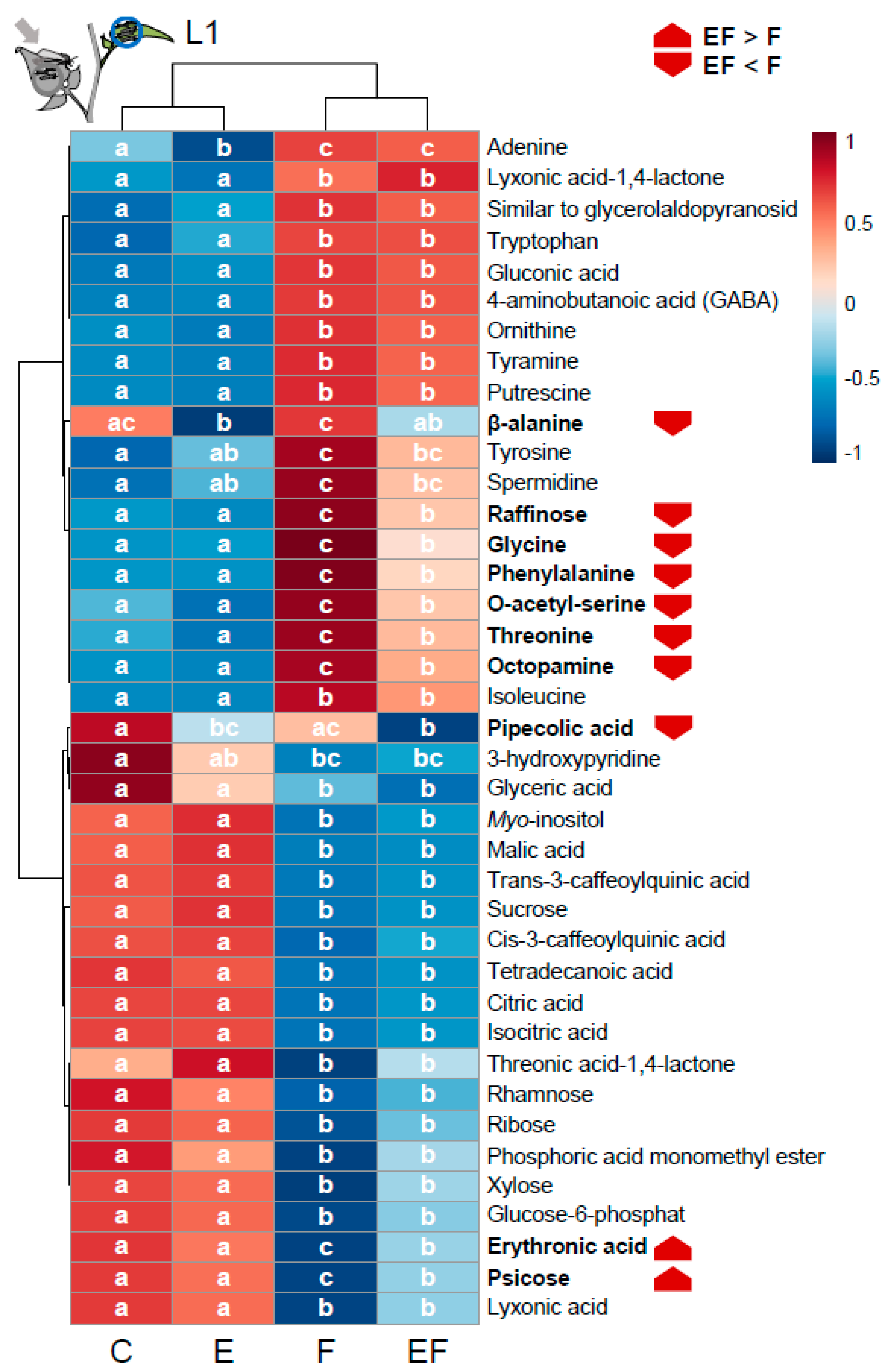
2.6. Oviposition Alters S. dulcamara’s Metabolic Response to Larval Herbivory
3. Discussion
3.1. Inferior Performance of S. exigua Larvae on Oviposited S. dulcamara
3.2. S. dulcamara’s PI Activity Does Not Explain the Effects of Oviposition on S. exigua Larvae
3.3. No Effects of Oviposition on S. dulcamara’s Phytohormonal Response to One Day of Larval Feeding
3.4. Oviposition Altered S. dulcamara’s Transcriptional Response to Larval Feeding
3.5. Oviposition Altered the Transcriptional Regulation of Phenylpropanoids in Response to Larval Feeding
3.6. Oviposition-Mediated Modifications of Feeding-Induced Changes of Amino Acid and Sugar Metabolism
3.7. Conclusion
4. Materials and Methods
4.1. Plants and Insects
4.2. Oviposition and Herbivory Treatments
4.3. Larval Performance and Feeding Damage
4.4. Induction of PI Activity by Larval Feeding
4.5. Extraction and Quantification of PI Activity.
4.6. RNA Extraction and Microarray Analysis
4.7. Reverse Transcription and qPCR Analysis
4.8. Phytohormone Extraction and Quantification
4.9. Non-Targeted Metabolome Analysis
4.10. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| ABA | Abscisic acid |
| GABA | 4-amino butanoic acid |
| FC | Fold-change |
| GC-MS | Gas chromatography coupled to mass spectrometry |
| GLMM | generalized linear mixed model |
| GO | Gene ontology |
| GST | Glutathione S-transferase |
| HQT | Hydroxycinnamoyl-CoA quinate transferase |
| JA | Jasmonic acid |
| JA-Ile | Jasmonic acid isoleucine |
| MDPI | Multidisciplinary Digital Publishing Institute |
| MeJA | Methyl jasmonate |
| LMM | Linear mixed models |
| LSD | Least significant difference |
| PI | Protease inhibitor |
| SA | Salicylic acid |
| SE | Standard error |
| TOF-MS | Time-of-flight mass spectrometry |
Appendix A

References
- Steppuhn, A.; Baldwin, I.T. Induced defenses and the cost-benefit paradigm. In Induced Plant Resistance to Herbivory; Schaller, A., Ed.; Springer: Dordrecht, The Netherlands, 2008; pp. 61–83. [Google Scholar] [CrossRef]
- Frost, C.J.; Mescher, M.C.; Carlson, J.E.; De Moraes, C.M. Plant defense priming against herbivores: Getting ready for a different battle. Plant Physiol. 2008, 146, 818–824. [Google Scholar] [CrossRef] [PubMed]
- Gális, I.; Gaquerel, E.; Pandey, S.P.; Baldwin, I.T. Molecular mechanisms underlying plant memory in JA-mediated defence responses. Plant Cell Environ. 2009, 32, 617–627. [Google Scholar] [CrossRef] [PubMed]
- Heil, M.; Kost, C. Priming of indirect defences. Ecol. Lett. 2006, 9, 813–817. [Google Scholar] [CrossRef] [PubMed]
- Hilker, M.; Fatouros, N.E. Resisting the onset of herbivore attack: Plants perceive and respond to insect eggs. Curr. Opin. Plant Biol. 2016, 32, 9–16. [Google Scholar] [CrossRef] [PubMed]
- Geuss, D.; Stelzer, S.; Lortzing, T.; Steppuhn, A. Solanum dulcamara’s response to eggs of an insect herbivore comprises ovicidal hydrogen peroxide production. Plant Cell Environ. 2017, 40, 2663–2677. [Google Scholar] [CrossRef] [PubMed]
- Seino, Y.; Suzuki, Y.; Sogawa, K. An ovicidal substance produced by rice plants in response to oviposition by the whitebacked planthopper, Sogatella furcifera (HORVATH) (Homoptera: Delphacidae). Appl. Entomol. Zool. 1996, 31, 467–473. [Google Scholar] [CrossRef]
- Blenn, B.; Bandoly, M.; Kuffner, A.; Otte, T.; Geiselhardt, S.; Fatouros, N.E.; Hilker, M. Insect egg deposition induces indirect defense and epicuticular wax changes in Arabidopsis thaliana. J. Chem. Ecol. 2012, 38, 882–892. [Google Scholar] [CrossRef] [PubMed]
- Fatouros, N.E.; Broekgaarden, C.; Bukovinszkine’Kiss, G.; van Loon, J.J.A.; Mumm, R.; Huigens, M.E.; Dicke, M.; Hilker, M. Male-derived butterfly anti-aphrodisiac mediates induced indirect plant defense. Proc. Natl. Acad. Sci. USA 2008, 105, 10033–10038. [Google Scholar] [CrossRef] [PubMed]
- Austel, N.; Eilers, E.J.; Meiners, T.; Hilker, M. Elm leaves “warned” by insect egg deposition reduce survival of hatching larvae by a shift in their quantitative leaf metabolite pattern. Plant Cell Environ. 2016, 39, 366–376. [Google Scholar] [CrossRef]
- Bandoly, M.; Hilker, M.; Steppuhn, A. Oviposition by Spodoptera exigua on Nicotiana attenuata primes induced plant defense against larval herbivory. Plant J. 2015, 83, 661–672. [Google Scholar] [CrossRef]
- Beyaert, I.; Köpke, D.; Stiller, J.; Hammerbacher, A.; Yoneya, K.; Schmidt, A.; Gershenzon, J.; Hilker, M. Can insect egg deposition ‘warn’ a plant of future feeding damage by herbivorous larvae? Proc. R. Soc. B 2012, 279, 101–108. [Google Scholar] [CrossRef] [PubMed]
- Geiselhardt, S.; Yoneya, K.; Blenn, B.; Drechsler, N.; Gershenzon, J.; Kunze, R.; Hilker, M. Egg laying of cabbage white butterfly (Pieris brassicae) on Arabidopsis thaliana affects subsequent performance of the larvae. PLoS ONE 2013, 8, e59661. [Google Scholar] [CrossRef] [PubMed]
- Pashalidou, F.G.; Fatouros, N.E.; van Loon, J.J.A.; Dicke, M.; Gols, R. Plant-mediated effects of butterfly egg deposition on subsequent caterpillar and pupal development, across different species of wild Brassicaceae. Ecol. Entomol. 2015, 40, 444–450. [Google Scholar] [CrossRef]
- Onkokesung, N.; Gaquerel, E.; Kotkar, H.; Kaur, H.; Baldwin, I.T.; Galis, I. MYB8 controls inducible phenolamide levels by activating three novel hydroxycinnamoyl-coenzyme A:polyamine transferases in Nicotiana attenuata. Plant Physiol. 2012, 158, 389–407. [Google Scholar] [CrossRef] [PubMed]
- Schäfer, M.; Brütting, C.; Xu, S.; Ling, Z.; Steppuhn, A.; Baldwin, I.T.; Schuman, M.C. NaMYB8 regulates distinct, optimally gei herbivore defense traits. J. Integr. Plant Biol. 2017, 59, 844–850. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Tooker, J.F.; Luthe, D.S.; De Moraes, C.M.; Felton, G.W. Insect eggs can enhance wound response in plants: A study system of tomato Solanum lycopersicum L. and Helicoverpa zea Boddie. PLoS ONE 2012, 7, e37420. [Google Scholar] [CrossRef] [PubMed]
- Lortzing, T.; Steppuhn, A. Jasmonate signalling in plants shapes plant-insect interaction ecology. Curr. Opin. Insect Sci. 2016, 14, 32–39. [Google Scholar] [CrossRef] [PubMed]
- Bruessow, F.; Gouhier-Darimont, C.; Buchala, A.; Metraux, J.P.; Reymond, P. Insect eggs suppress plant defence against chewing herbivores. Plant J. 2010, 62, 876–885. [Google Scholar] [CrossRef]
- Pieterse, C.M.J.; Van der Does, D.; Zamioudis, C.; Leon-Reyes, A.; Van Wees, S.C.M. Hormonal modulation of plant immunity. Annu. Rev. Cell. Dev. B. 2012, 28, 489–521. [Google Scholar] [CrossRef]
- Thaler, J.S.; Humphrey, P.T.; Whiteman, N.K. Evolution of jasmonate and salicylate signal crosstalk. Trends Plant Sci. 2012, 17, 260–270. [Google Scholar] [CrossRef]
- Lortzing, V.; Oberlander, J.; Lortzing, T.; Tohge, T.; Steppuhn, A.; Kunze, R.; Hilker, M. Insect egg deposition renders plant defense against hatching larvae more effective in a salicylic acid-dependent manner. Plant Cell Environ. 2018. [Google Scholar] [CrossRef] [PubMed]
- Bonnet, C.; Lassueur, S.; Ponzio, C.; Gols, R.; Dicke, M.; Reymond, P. Combined biotic stresses trigger similar transcriptomic responses but contrasting resistance against a chewing herbivore in Brassica nigra. BMC Plant Biol. 2017, 17, 127. [Google Scholar] [CrossRef] [PubMed]
- Firtzlaff, V.; Oberländer, J.; Geiselhardt, S.; Hilker, M.; Kunze, R. Pre-exposure of Arabidopsis to the abiotic or biotic environmental stimuli “chilling” or “insect eggs” exhibits different transcriptomic responses to herbivory. Sci. Rep. 2016, 6, 28544. [Google Scholar] [CrossRef] [PubMed]
- Little, D.; Gouhier-Darimont, C.; Bruessow, F.; Reymond, P. Oviposition by pierid butterflies triggers defense responses in Arabidopsis. Plant Physiol. 2007, 143, 784–800. [Google Scholar] [CrossRef] [PubMed]
- Altmann, S.; Muino, J.M.; Lortzing, V.; Brandt, R.; Himmelbach, A.; Altschmied, L.; Hilker, M. Transcriptomic basis for reinforcement of elm anti-herbivore defense mediated by insect egg deposition. Mol. Ecol. 2018. [Google Scholar] [CrossRef]
- Drok, S.; Bandoly, M.; Stelzer, S.; Lortzing, T.; Steppuhn, A. Moth oviposition shapes the species-specific transcriptional and phytohormonal response of Nicotiana attenuata to larval feeding. Sci. Rep. 2018, 8, 10249. [Google Scholar] [CrossRef]
- Nguyen, D.; D’Agostino, N.; Tytgat, T.; Sun, P.; Lortzing, T.; Visser, E.; Cristescu, S.; Steppuhn, A.; Mariani, C.; van Dam, N.; et al. Drought and flooding have distinct effects on herbivore-induced responses and resistance in Solanum dulcamara. Plant Cell Environ. 2016, 39, 1485–1499. [Google Scholar] [CrossRef]
- Viswanathan, D.V.; Lifchits, O.A.; Thaler, J.S. Consequences of sequential attack for resistance to herbivores when plants have specific induced responses. Oikos 2007, 116, 1389–1399. [Google Scholar] [CrossRef]
- Smits, P.H.; van Velden, M.C.; van de Vrie, M.; Vlak, J.M. Feeding and dispersion of Spodoptera exigua larvae and its relevance for control with a nuclear polyhedrosis virus. Entomol. Exp. Appl. 1987, 43, 67–72. [Google Scholar] [CrossRef]
- Underwood, N. Density dependence in insect performance within individual plants: Induced resistance to Spodoptera exigua in tomato. Oikos 2010, 119, 1993–1999. [Google Scholar] [CrossRef]
- McNutt, D.W.; Samuelson, K.; Underwood, N. Pathways for plant-mediated negative feedback to insect herbivores: Accounting for non-linear effects of larval density on plant quality and quantity. Entomol. Exp. Appl. 2017, 162, 93–104. [Google Scholar] [CrossRef]
- Hartl, M.; Giri, A.P.; Kaur, H.; Baldwin, I.T. Serine protease inhibitors specifically defend Solanum nigrum against generalist herbivores but do not influence plant growth and development. Plant Cell 2010, 22, 4158–4175. [Google Scholar] [CrossRef] [PubMed]
- Jongsma, M.A.; Bakker, P.L.; Visser, B.; Stiekema, W.J. Trypsin inhibitor activity in mature tobacco and tomato plants is mainly induced locally in response to insect attack, wounding and virus infection. Planta 1994, 195, 29–35. [Google Scholar] [CrossRef]
- Van Dam, N.M.; Horn, M.; Mares, M.; Baldwin, I.T. Ontogeny constrains systemic protease inhibitor response in Nicotiana attenuata. J. Chem. Ecol. 2001, 27, 547–568. [Google Scholar] [CrossRef] [PubMed]
- Calf, O.W.; Huber, H.; Peters, J.L.; Weinhold, A.; van Dam, N.M. Glycoalkaloid composition explains variation in slug resistance in Solanum dulcamara. Oecologia 2018, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Simmonds, M.S.J. Flavonoid–insect interactions: Recent advances in our knowledge. Phytochemstry 2003, 64, 21–30. [Google Scholar] [CrossRef]
- Dixon, R.A.; Xie, D.-Y.; Sharma, S.B. Proanthocyanidins—A final frontier in flavonoid research? New Phytol. 2005, 165, 9–28. [Google Scholar] [CrossRef] [PubMed]
- Bandoly, M.; Grichnik, R.; Hilker, M.; Steppuhn, A. Priming of anti-herbivore defence in Nicotiana attenuata by insect oviposition: Herbivore specific effects. Plant Cell Environ. 2016, 39, 848–859. [Google Scholar] [CrossRef] [PubMed]
- Schwachtje, J.; Baldwin, I.T. Why does herbivore attack reconfigure primary metabolism? Plant Physiol. 2008, 146, 845–851. [Google Scholar] [CrossRef]
- Maeda, H.; Dudareva, N. The shikimate pathway and aromatic amino acid biosynthesis in plants. Annu. Rev. Plant Biol. 2012, 63, 73–105. [Google Scholar] [CrossRef]
- Vogt, T. Phenylpropanoid biosynthesis. Mol. Plant 2010, 3, 2–20. [Google Scholar] [CrossRef] [PubMed]
- Ramaswamy, N.K.; Behere, A.G.; Nair, P.M. A novel pathway for the synthesis of solanidine in the isolated chloroplast from greening potatoes. Eur. J. Biochem. 1976, 67, 275–282. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.-C.; Sulli, M.; Fu, D.-Q. The role of phytochromes in regulating biosynthesis of sterol glycoalkaloid in eggplant leaves. PLoS ONE 2017, 12, e0189481. [Google Scholar] [CrossRef] [PubMed]
- Schwachtje, J.; Fischer, A.; Erban, A.; Kopka, J. Primed primary metabolism in systemic leaves: A functional systems analysis. Sci. Rep. 2018, 8, 216. [Google Scholar] [CrossRef] [PubMed]
- Lortzing, T.; Firtzlaff, V.; Nguyen, D.; Rieu, I.; Stelzer, S.; Schad, M.; Kallarackal, J.; Steppuhn, A. Transcriptomic responses of Solanum dulcamara to natural and simulated herbivory. Mol. Ecol. Resour. 2017, 17, e196–e211. [Google Scholar] [CrossRef] [PubMed]
- Babst, B.A.; Ferrieri, R.A.; Thorpe, M.R.; Orians, C.M. Lymantria dispar herbivory induces rapid changes in carbon transport and partitioning in Populus nigra. Entomol. Exp. Appl. 2008, 128, 117–125. [Google Scholar] [CrossRef]
- Schwachtje, J.; Minchin, P.E.H.; Jahnke, S.; van Dongen, J.T.; Schittko, U.; Baldwin, I.T. SNF1-related kinases allow plants to tolerate herbivory by allocating carbon to roots. Proc. Natl. Acad. Sci. USA 2006, 103, 12935–12940. [Google Scholar] [CrossRef]
- Fait, A.; Fromm, H.; Walter, D.; Galili, G.; Fernie, A.R. Highway or byway: The metabolic role of the GABA shunt in plants. Trends Plant Sci. 2008, 13, 14–19. [Google Scholar] [CrossRef]
- Scholz, S.S.; Reichelt, M.; Mekonnen, D.W.; Ludewig, F.; Mithöfer, A. Insect herbivory-eicited GABA accumulation in plants is a wound-induced, direct, systemic, and jasmonate-independent defense response. Front. Plant Sci. 2015, 6. [Google Scholar] [CrossRef]
- Huang, X.P.; Renwick, J.A. Chemical and experiential basis for rejection of Tropaeolum majus by Pieris rapae larvae. J. Chem. Ecol. 1995, 21, 1601–1617. [Google Scholar] [CrossRef]
- Leiss, K.A.; Maltese, F.; Choi, Y.H.; Verpoorte, R.; Klinkhamer, P.G.L. Identification of chlorogenic acid as a resistance factor for thrips in Chrysanthemum. Plant Physiol. 2009, 150, 1567–1575. [Google Scholar] [CrossRef] [PubMed]
- Mallikarjuna, N.; Kranthi, K.R.; Jadhav, D.R.; Kranthi, S.; Chandra, S. Influence of foliar chemical compounds on the development of Spodoptera litura (Fab.) in interspecific derivatives of groundnut. J. Appl. Entomol. 2004, 128, 321–328. [Google Scholar] [CrossRef]
- Bernsdorff, F.; Döring, A.-C.; Gruner, K.; Schuck, S.; Bräutigam, A.; Zeier, J. Pipecolic acid orchestrates plant systemic acquired resistance and defense priming via salicylic acid-dependent and -independent pathways. Plant Cell 2016, 28, 102–129. [Google Scholar] [CrossRef] [PubMed]
- Hilfiker, O.; Groux, R.; Bruessow, F.; Kiefer, K.; Zeier, J.; Reymond, P. Insect eggs induce a systemic acquired resistance in Arabidopsis. Plant J. 2014, 80, 1085–1094. [Google Scholar] [CrossRef] [PubMed]
- Ritchie, M.E.; Phipson, B.; Wu, D.; Hu, Y.; Law, C.W.; Shi, W.; Smyth, G.K. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015, 43, e47. [Google Scholar] [CrossRef] [PubMed]
- Alexa, A.; Rahnenführer, J. Bioconductor—TopGo. Available online: https://www.bioconductor.org/ packages/release/bioc/html/topGO.html; http://cobra20.fhcrc.org/packages/release/bioc/html/ topGO.html (accessed on 30 November 2016).
- Erban, A.; Schauer, N.; Fernie, A.R.; Kopka, J. Nonsupervised construction and application of mass spectral and retention time index libraries from time-of-flight gas chromatography-mass spectrometry metabolite profiles. In Metabolomics; Humana Press; Humana Press Inc.: Totowa, NJ, USA, 2007; pp. 19–38. [Google Scholar]
- Dethloff, F.; Erban, A.; Orf, I.; Alpers, J.; Fehrle, I.; Beine-Golovchuk, O.; Schmidt, S.; Schwachtje, J.; Kopka, J. Profiling methods to identify cold-regulated primary metabolites using gas chromatography coupled to mass spectrometry. In Plant Cold Acclimation; Humana Press: New York, NY, USA, 2014; pp. 171–197. [Google Scholar]
- Luedemann, A.; Strassburg, K.; Erban, A.; Kopka, J. TagFinder for the quantitative analysis of gas chromatography—Mass spectrometry (GC-MS)-based metabolite profiling experiments. Bioinformatics 2008, 24, 732–737. [Google Scholar] [CrossRef] [PubMed]
- Hummel, J.; Strehmel, N.; Selbig, J.; Walther, D.; Kopka, J. Decision tree supported substructure prediction of metabolites from GC-MS profiles. Metabolomics 2010, 6, 322–333. [Google Scholar] [CrossRef] [PubMed]
- Kopka, J.; Schauer, N.; Krueger, S.; Birkemeyer, C.; Usadel, B.; Bergmüller, E.; Dörmann, P.; Weckwerth, W.; Gibon, Y.; Stitt, M.; et al. GMD@CSB.DB: The Golm Metabolome Database. Bioinformatics 2005, 21, 1635–1638. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing, version 3.1.2; R Foundation for Statistical Computing: Vienna, Austria, 2014. [Google Scholar]
- Xia, J.; Wishart, D.S. Using MetaboAnalyst 3.0 for comprehensive metabolomics data analysis. Curr. Protoc. Bioinform. 2016, 55, 14.10.1–14.10.91. [Google Scholar] [CrossRef]
- Bates, D.; Maechler, M.; Bolker, B.; Walker, S. Lme4: Linear Mixed-Effects Models Using Eigen and S4. R package version 1.1-7. 2014. [Google Scholar]
- Oliveros, J. Venny. An Interactive Tool for Comparing Lists with Venn’s Diagrams. Available online: http://bioinfogp.cnb.csic.es/tools/venny/index.html (accessed on 27 October 2018).
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Geuss, D.; Lortzing, T.; Schwachtje, J.; Kopka, J.; Steppuhn, A. Oviposition by Spodoptera exigua on Solanum dulcamara Alters the Plant’s Response to Herbivory and Impairs Larval Performance. Int. J. Mol. Sci. 2018, 19, 4008. https://doi.org/10.3390/ijms19124008
Geuss D, Lortzing T, Schwachtje J, Kopka J, Steppuhn A. Oviposition by Spodoptera exigua on Solanum dulcamara Alters the Plant’s Response to Herbivory and Impairs Larval Performance. International Journal of Molecular Sciences. 2018; 19(12):4008. https://doi.org/10.3390/ijms19124008
Chicago/Turabian StyleGeuss, Daniel, Tobias Lortzing, Jens Schwachtje, Joachim Kopka, and Anke Steppuhn. 2018. "Oviposition by Spodoptera exigua on Solanum dulcamara Alters the Plant’s Response to Herbivory and Impairs Larval Performance" International Journal of Molecular Sciences 19, no. 12: 4008. https://doi.org/10.3390/ijms19124008
APA StyleGeuss, D., Lortzing, T., Schwachtje, J., Kopka, J., & Steppuhn, A. (2018). Oviposition by Spodoptera exigua on Solanum dulcamara Alters the Plant’s Response to Herbivory and Impairs Larval Performance. International Journal of Molecular Sciences, 19(12), 4008. https://doi.org/10.3390/ijms19124008





