Overexpression of the Wild Soybean R2R3-MYB Transcription Factor GsMYB15 Enhances Resistance to Salt Stress and Helicoverpa Armigera in Transgenic Arabidopsis
Abstract
1. Introduction
2. Results
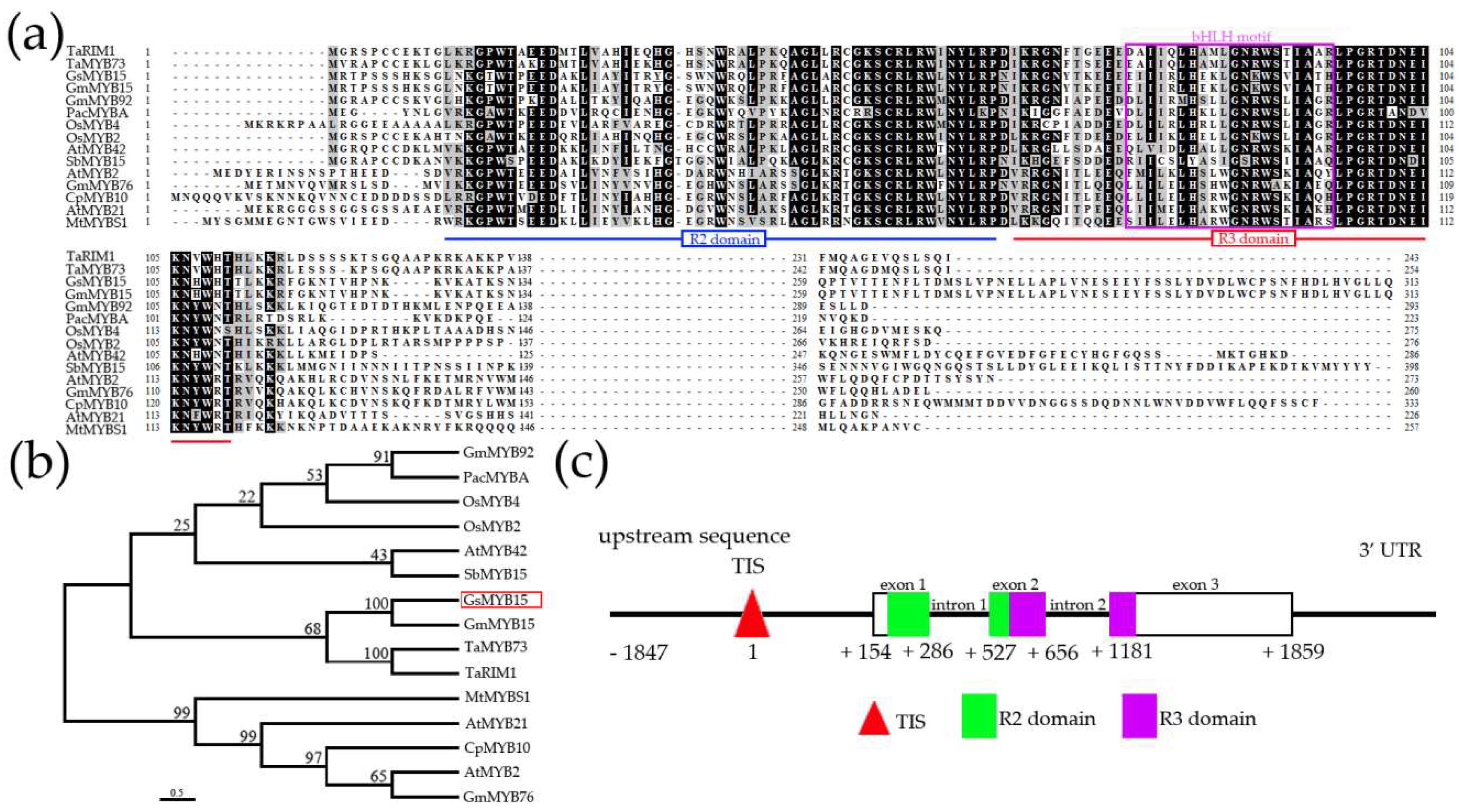
2.1. Clone and Sequence Analysis of GsMYB15
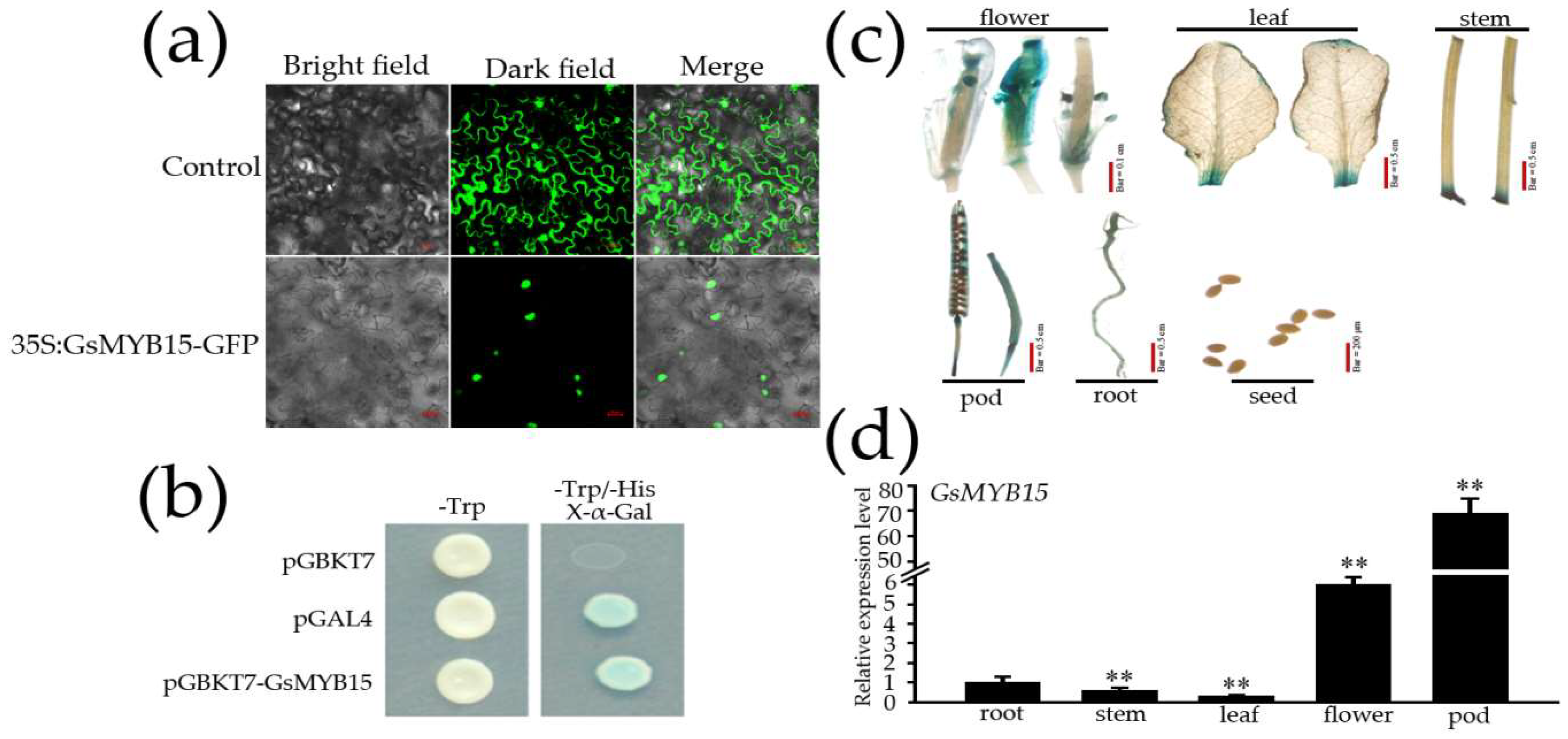
2.2. Sub-Cellular Localization, Transcriptional Activation Activity and Tissue-Specific Expression of GsMYB15
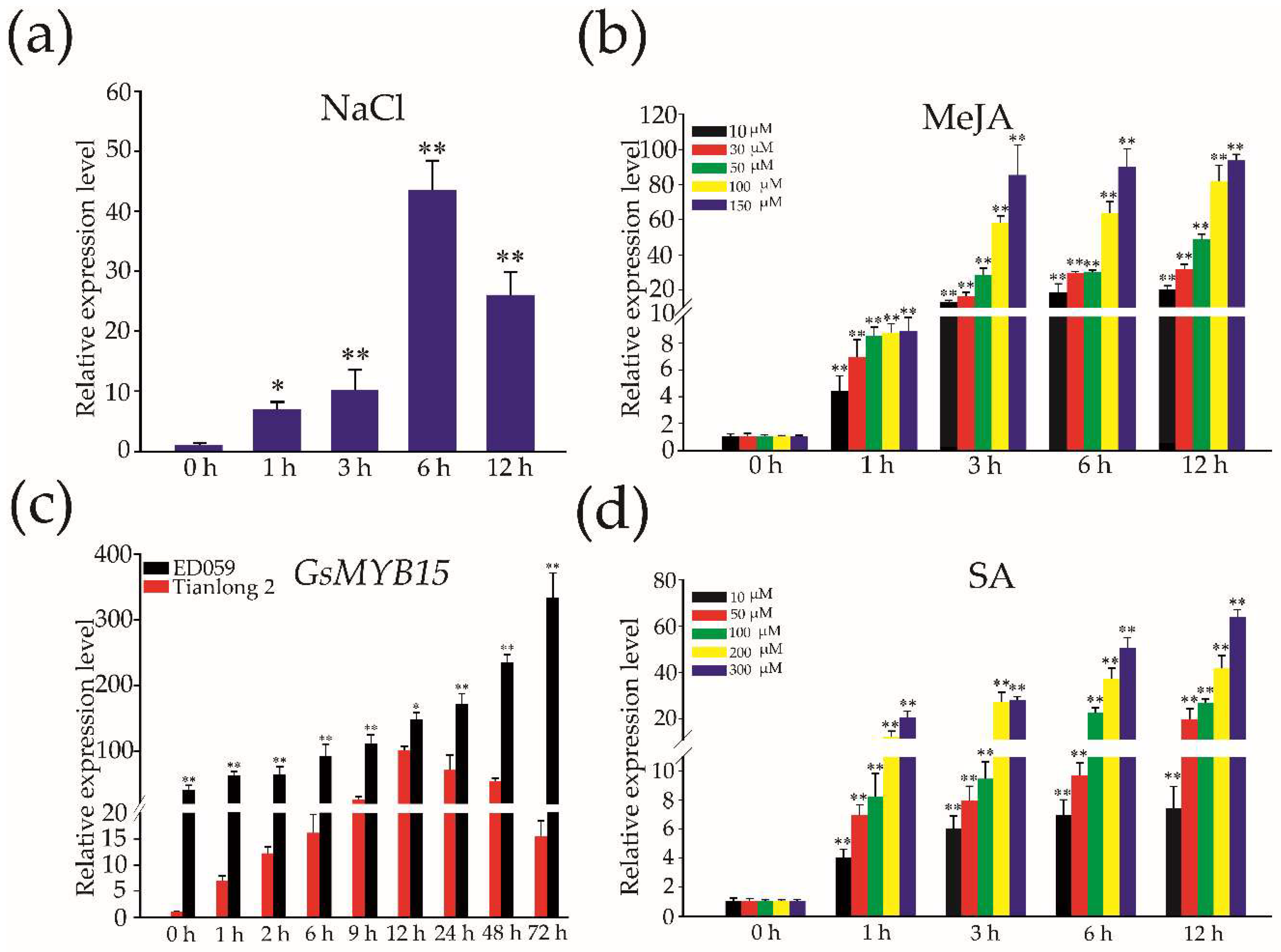
2.3. Expression of GsMYB15 in Soybean Leaves in Response to Salt, MeJA and SA Treatments and Insect Attacks
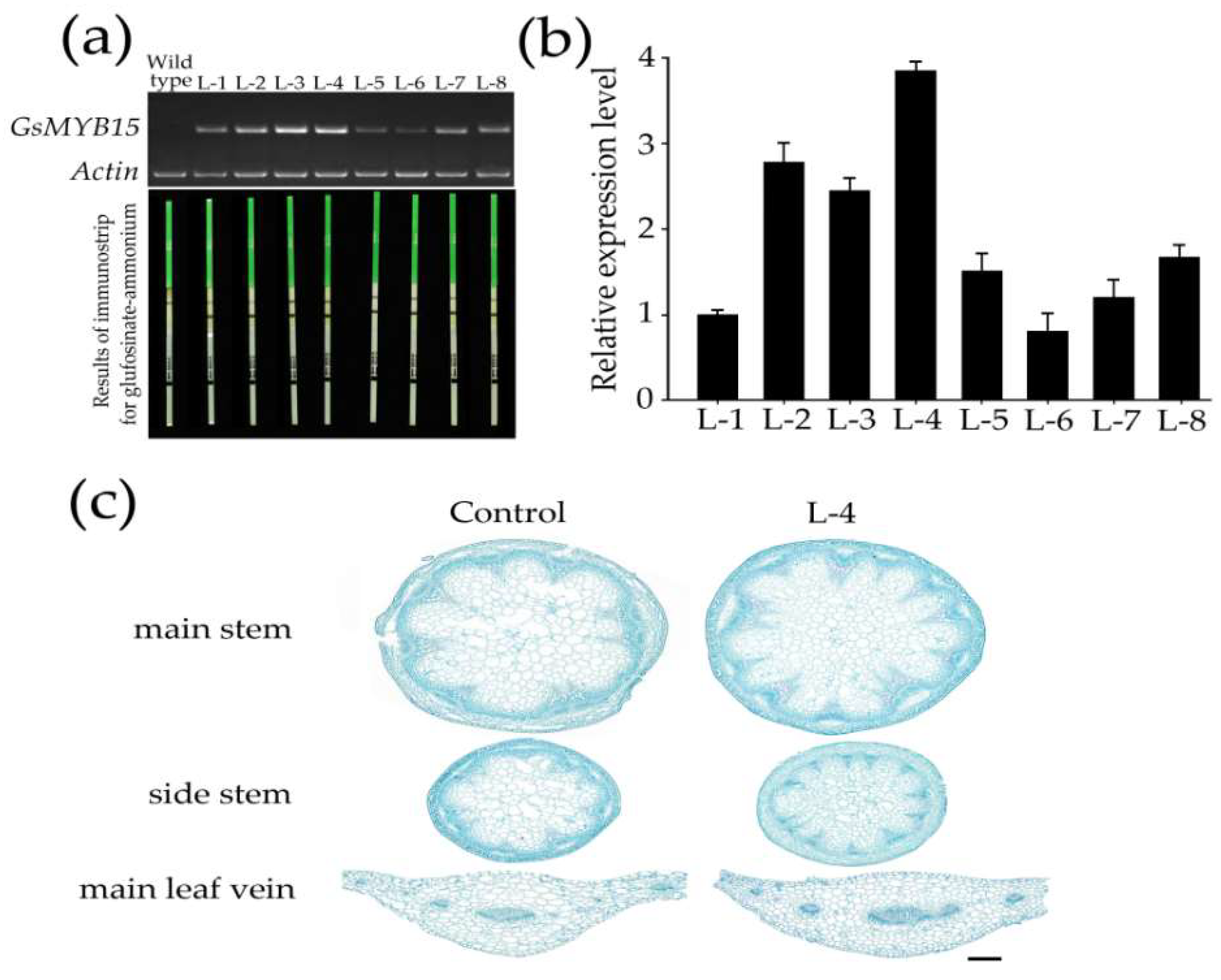
2.4. Physiological Changes in Arabidopsis Lines Overexpressing GsMYB15
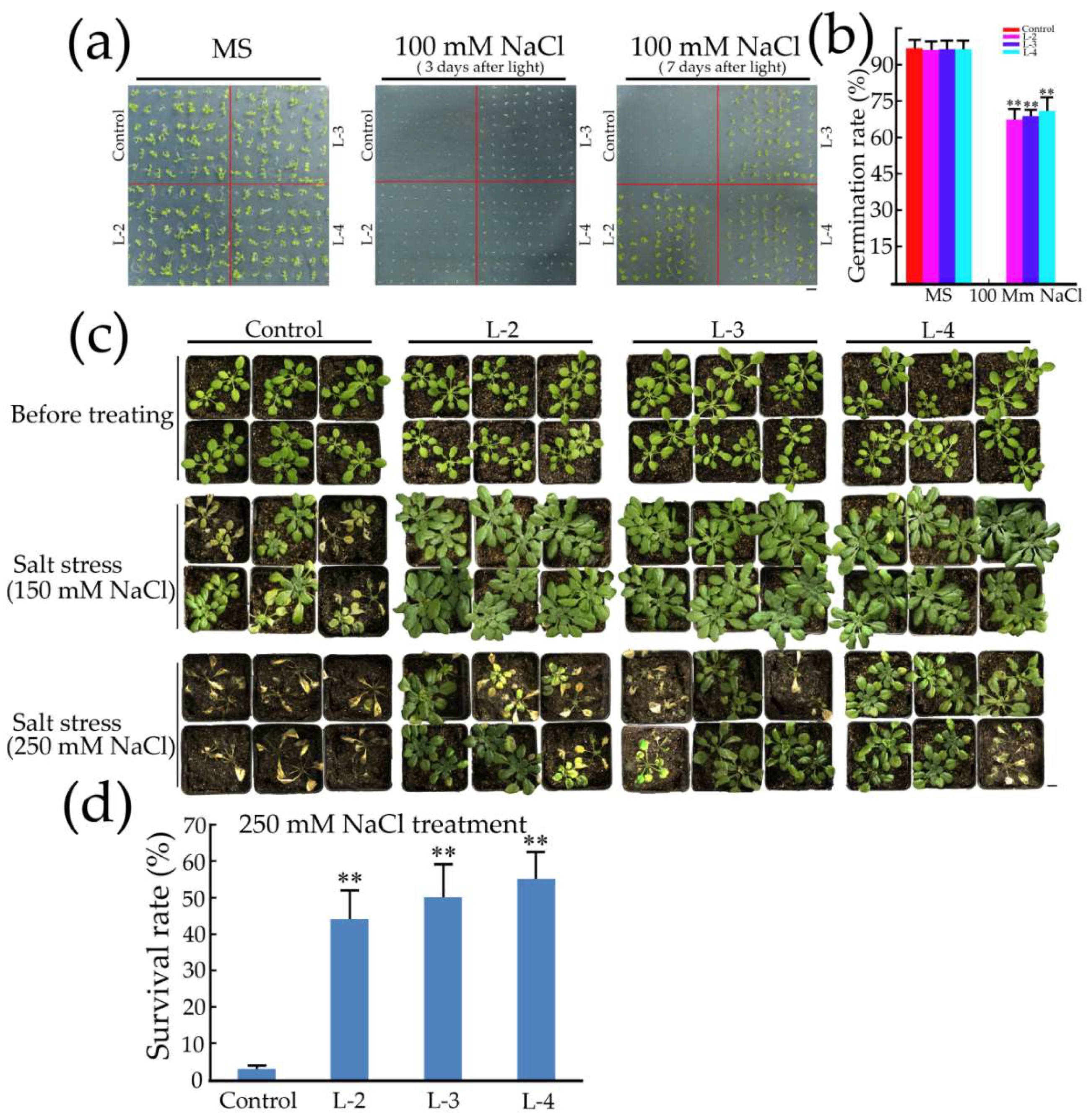
2.5. Overexpression of GsMYB15 Increased Salt Stress Tolerance in the Transgenic Arabidopsis Lines
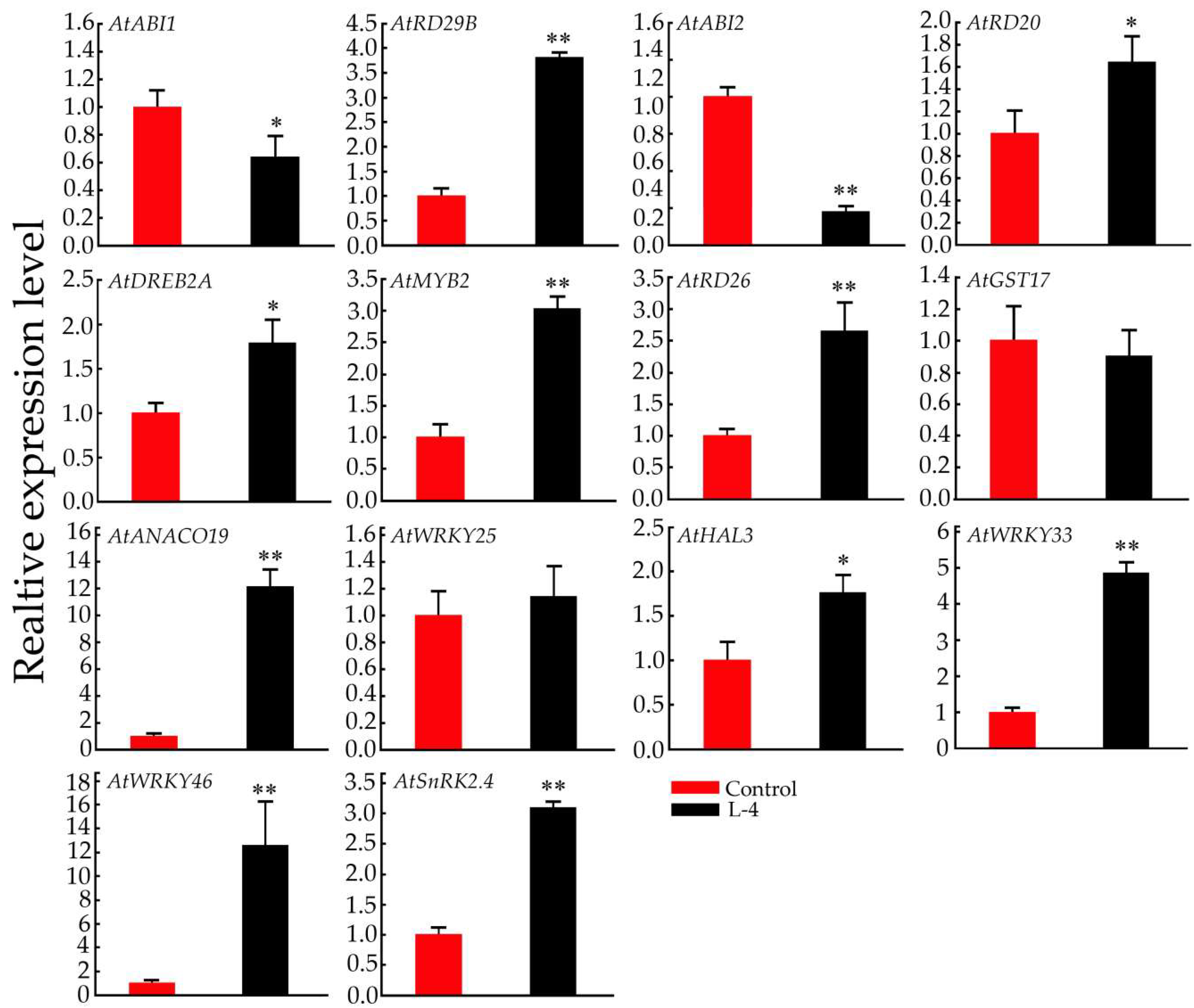
2.6. Expression Pattern Analysis of Salt-Related Genes in Transgenic Arabidopsis Plants
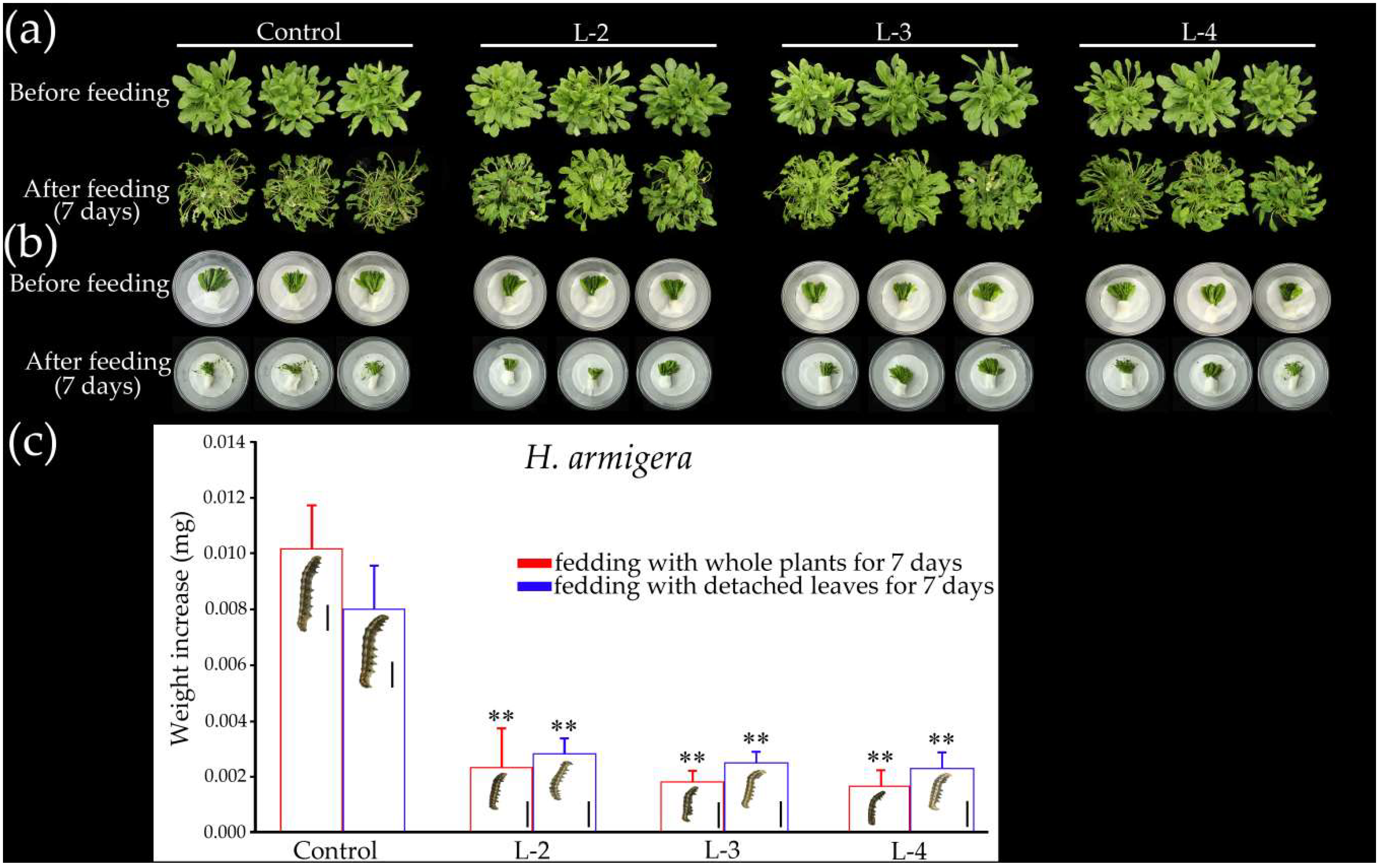
2.7. Overexpression of GsMYB15 Increased Insect Resistance in Transgenic Arabidopsis Plants
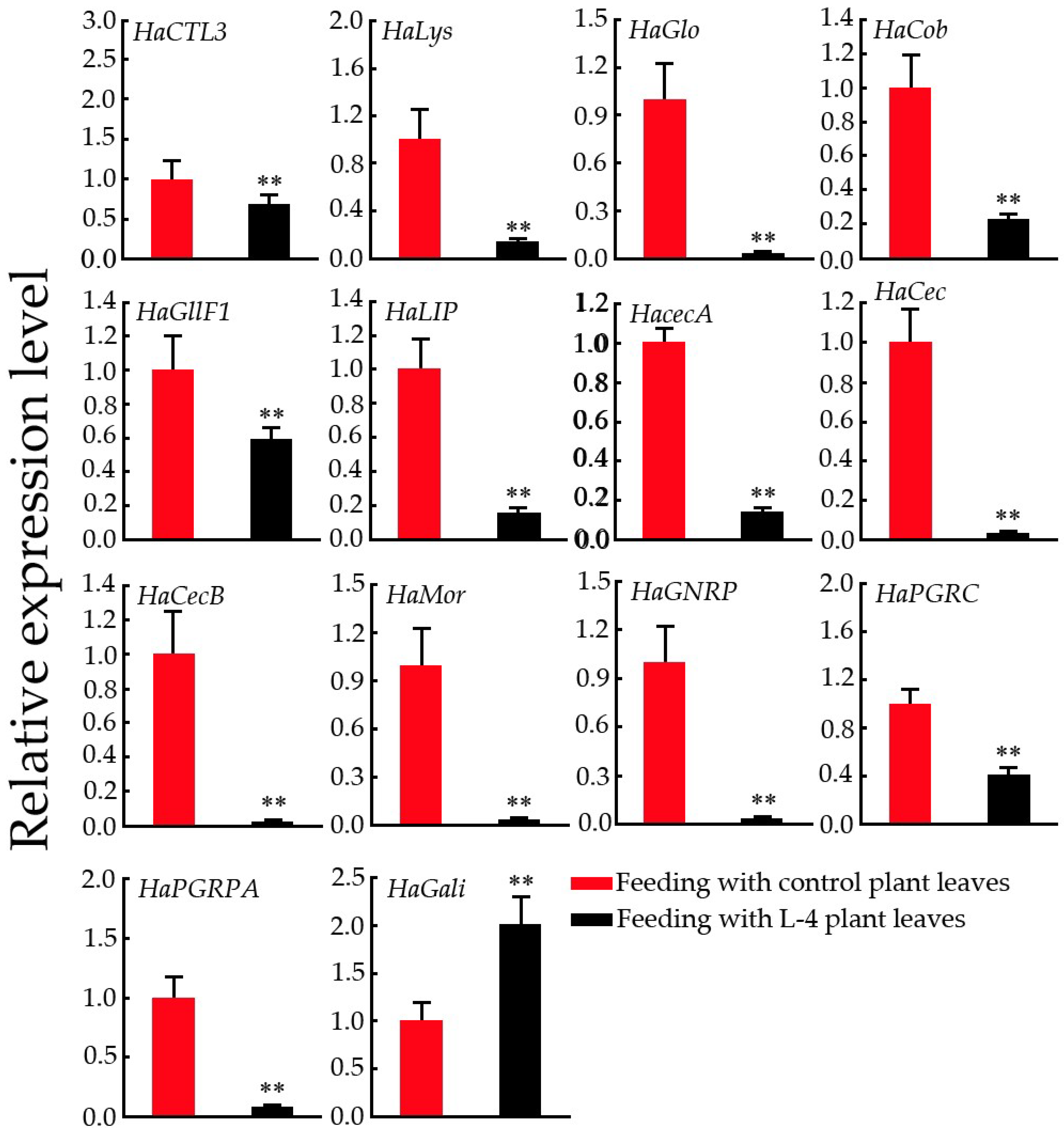
2.8. Expression Pattern Analysis of Immunity-Related Genes in H. armigera
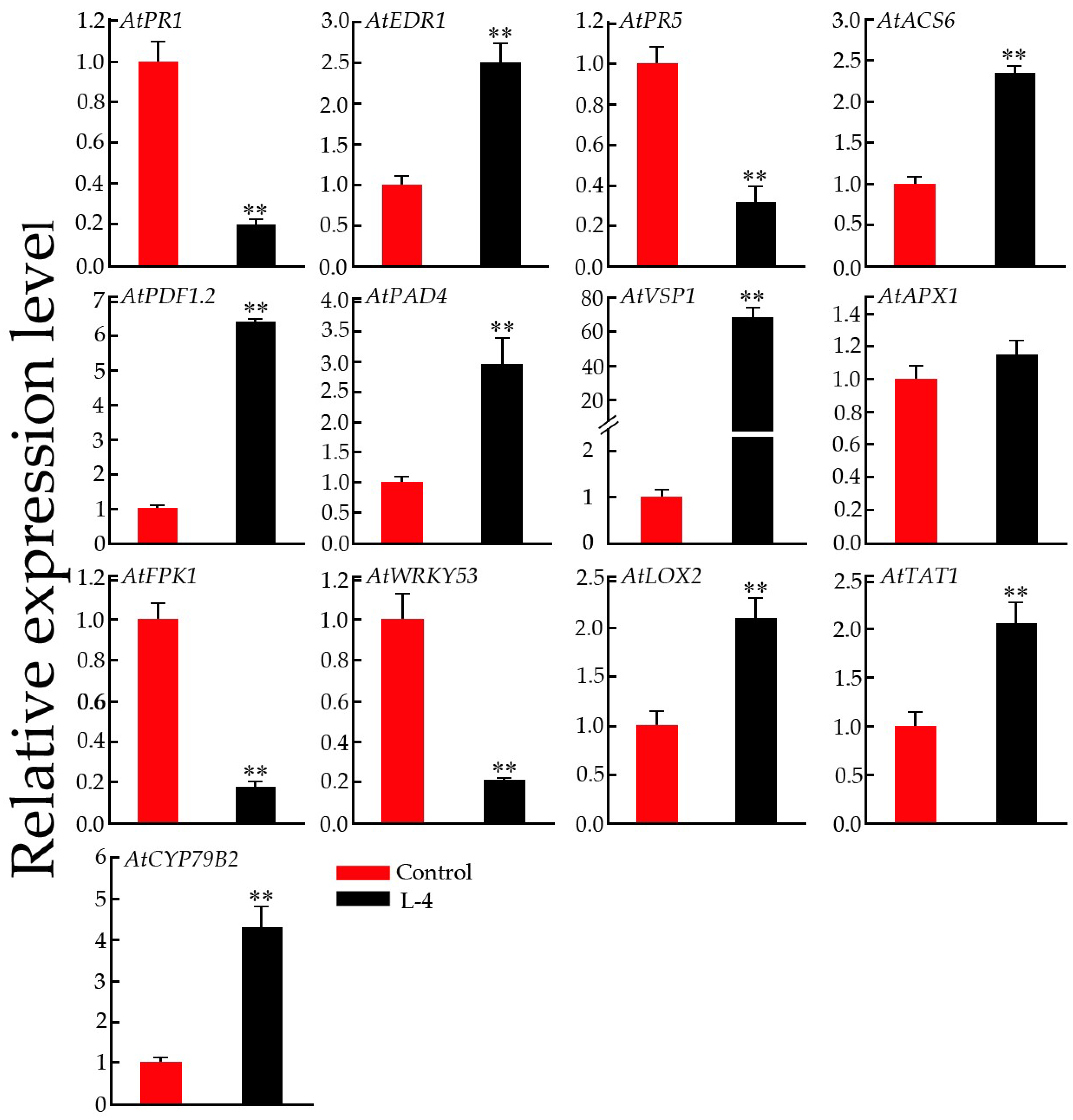
2.9. Expression Pattern Analysis of Defense-Related Genes in Transgenic Arabidopsis Plants
3. Discussion
3.1. GsMYB15 Can Respond to Salt Stress, MeJA and SA Treatment and Insect Attacks
3.2. GsMYB15 Plays a Positive Regulatory Role in Salt Stress Tolerance in Transgenic Arabidopsis Plants
3.3. GsMYB15 Enhances Herbivorous Insect Resistance in Transgenic Arabidopsis Plants
4. Materials and Methods
4.1. Soybean Plants and Treatments
4.2. Sequence Analysis
4.3. RNA Isolation and QPCR Analysis
4.4. Sub-Cellular Localization and Transcriptional Activation Activity Analysis of GsMYB15
4.5. GUS Staining and Expression Analysis
4.6. Generation of GsMYB15 Transgenic Arabidopsis Plants
4.7. Tissue Preparation
4.8. Analysis of Salinity Tolerance and Insect Resistance in Transgenic Arabidopsis Plants
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| MeJA | Methyl jasmonate |
| ABA | Abscisic acid |
| GUS | Beta-glucuronidase |
| TF | Transcription factor |
| SA | Salicylic Acid |
| bHLH | Basic helix-loop-helix |
| H. armigera | Helicoverpa armigera |
| OE | Overexpression |
References
- Zhu, J.K. Salt and drought stress signal transduction in plants. Annu. Rev. Plant Biol. 2002, 53, 247–273. [Google Scholar] [CrossRef] [PubMed]
- Howe, G.; Jander, G. Plant immunity to insect herbivores. Annu. Rev. Plant Biol. 2008, 59, 41–66. [Google Scholar] [CrossRef] [PubMed]
- Shen, X.J.; Guo, X.W.; Guo, X.; Zhao, D.; Zhao, W.; Chen, J.S.; Li, T.H. PacMYBA, a sweet cherry R2R3-MYB transcription factor, is a positive regulator of salt stress tolerance and pathogen resistance. Plant Physiol. Biochem. 2017, 112, 302–311. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.J.; Zhu, T. Networks of transcription factors with roles in environmental stress response. Trends Plant Sci. 2004, 9, 591–596. [Google Scholar] [CrossRef] [PubMed]
- Hirayama, T.; Shinozaki, K. Research on plant abiotic stress responses in the post-genome era: Past, present and future. Plant J. 2010, 61, 1041–1052. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.K. Abiotic stress signaling and responses in plants. Cell 2016, 167, 313–324. [Google Scholar] [CrossRef] [PubMed]
- Melcher, K.; Ng, L.M.; Zhou, X.E.; Soon, F.F.; Xu, Y.; Suino-Powell, K.M.; Park, S.Y.; Weiner, J.J.; Fujii, H.; Chinnusamy, V.; et al. A gate-latch-lock mechanism for hormone signalling by abscisic acid receptors. Nature 2009, 462, 602–608. [Google Scholar] [CrossRef]
- Walter, P.; Ron, D. The unfolded protein response: From stress pathway to homeostatic regulation. Science 2011, 334, 1081–1086. [Google Scholar] [CrossRef]
- Endler, A.; Kesten, C.; Schneider, R.; Zhang, Y.; Ivakov, A.; Froehlich, A.; Funke, N.; Persson, S. A mechanism for sustained cellulose synthesis during salt stress. Cell 2015, 162, 1353–1364. [Google Scholar] [CrossRef]
- Nakashima, K.; Ito, Y.; Yamaguchi-Shinozaki, K. Transcriptional regulatory networks in response to abiotic stresses in Arabidopsis and grasses. Plant Physiol. 2009, 149, 88–95. [Google Scholar] [CrossRef]
- Swarbreck, S.M.; Colaço, R.; Davies, J.M. Plant calcium-permeable channels. Plant Physiol. 2013, 163, 514–522. [Google Scholar] [CrossRef] [PubMed]
- Leng, P.; Yuan, B.; Guo, Y. The role of abscisic acid in fruit ripening and responses to abiotic stress. J. Exp. Bot. 2014, 65, 4577–4588. [Google Scholar] [CrossRef] [PubMed]
- Zheng, X.Z.; Kang, S.; Jing, Y.P.; Ren, Z.J.; Li, L.G.; Zhou, J.M.; Berkowitz, G.; Shi, J.S.; Fu, A.G.; Lan, W.Z.; et al. Danger-associated peptides close stomata by OST1-independent 3 activation of anion channels in guard cells. Plant Cell 2018, 30, 1132–1146. [Google Scholar] [CrossRef] [PubMed]
- Yang, D.H.; Hettenhausen, C.; Baldwin, I.T.; Wu, J.J. Silencing nicotiana attenuata calcium-dependent protein kinases, CDPK4 and CDPK5, strongly up-regulates wound- and herbivory-induced jasmonic acid accumulations. Plant Physiol. 2012, 159, 1591–1607. [Google Scholar] [CrossRef] [PubMed]
- Mao, Y.B.; Liu, Y.Q.; Chen, D.Y.; Chen, F.Y.; Fang, X.; Hong, G.J.; Wang, W.J.; Wang, W.J.; Chen, X.Y. Jasmonate response decay and defense metabolite accumulation contributes to age-regulated dynamics of plant insect resistance. Nat. Commun. 2015, 8, 13925. [Google Scholar] [CrossRef] [PubMed]
- Shen, X.J.; Guo, X.; Zhao, D.; Zhang, Q.; Jiang, Y.Z.; Wang, Y.T.; Peng, X.; Wei, Y.; Zhai, Z.F.; Zhao, W.; et al. Cloning and expression profiling of the PacSnRK2 and PacPP2C gene families during fruit development, ABA treatment, and dehydration stress in sweet cherry. Plant Physiol. Biochem. 2017, 119, 275–285. [Google Scholar] [CrossRef] [PubMed]
- Chico, J.M.; Fernández-Barbero, G.; Chini, A.; Fernández-Calvo, P.; Díez-Díaz, M.; Solano, R. Repression of Jasmonate-dependent defenses by shade involves differential regulation of protein stability of MYC transcription factors and their JAZ repressors in Arabidopsis. Plant Cell 2014, 26, 1967–1980. [Google Scholar] [CrossRef]
- Louis, J.; Basu, S.; Varsani, S.; Castano-Duque, L.; Jiang, V.; Williams, W.P.; Felton, G.W.; Luthe, D.S. Ethylene Contributes to maize insect resistance1-Mediated Maize Defense against the Phloem Sap-Sucking Corn Leaf Aphid. Plant Physiol. 2015, 169, 313–324. [Google Scholar] [CrossRef]
- Voght, S.P.; Fluegel, M.L.; Andrews, L.A.; Pallanck, L.J. Drosophila NPC1b Promotes an Early Step in Sterol Absorption from the Midgut Epithelium. Cell Metab. 2007, 5, 195–205. [Google Scholar] [CrossRef]
- Verma, V.; Ravindran, P.; Kumar, P.P. Plant hormone-mediated regulation of stress responses. BMC Plant Biol. 2016, 16, 86. [Google Scholar] [CrossRef]
- Hou, X.; Lee, L.Y.; Xia, K.; Yan, Y.; Yu, H. DELLAs modulate jasmonate signaling via competitive binding to JAZs. Dev. Cell 2010, 19, 884–894. [Google Scholar] [CrossRef] [PubMed]
- Choi, J.; Huh, S.U.; Kojima, M.; Sakakibara, H.; Paek, K.H.; Hwang, I. The cytokinin-activated transcription factor ARR2 promotes plant immunity via TGA3/NPR1-dependent salicylic acid signaling in Arabidopsis. Dev. Cell 2010, 19, 284–295. [Google Scholar] [CrossRef] [PubMed]
- Seo, P.J.; Park, C.M. MYB96-mediated abscisic acid signals induce pathogen resistance response by promoting salicylic acid biosynthesis in Arabidopsis. New Phytol. 2010, 186, 471–483. [Google Scholar] [CrossRef]
- Li, R.; Zhang, J.; Li, J.C.; Zhou, G.X.; Wang, Q.; Bian, W.B.; Matthias, E.; Lou, Y.G. Prioritizing plant defence over growth through WRKY regulation facilitates infestation by non-target herbivores. eLife 2015, 4, E04805. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Wang, J.; Zheng, Z.Y.; Fan, B.F.; Yu, J.Q.; Chen, Z.X. Characterization of the promoter and extended C-terminal domain of Arabidopsis WRKY33 and functional analysis of tomato WRKY33 homologues in plant stress responses. J. Exp. Bot. 2015, 66, 4567–4583. [Google Scholar] [CrossRef]
- Guo, J.P.; Xu, C.X.; Wu, D.; Zhao, Y.; Qiu, Y.F.; Wang, X.X.; OuYang, Y.D.; Cai, B.D.; Liu, X.; Jing, S.L.; et al. Bph6 encodes an exocyst-localized protein and confers broad resistance to planthoppers in rice. Nat. Genet. 2018, 50, 297–306. [Google Scholar] [CrossRef] [PubMed]
- Zhong, Y.J.; Wang, Y.G.; Guo, J.F.; Zhu, X.L.; Shi, J.; He, Q.J.; Liu, Y.; Wu, Y.R.; Zhang, L.; Lv, Q.D.; et al. Rice SPX6 negatively regulates the phosphate starvation response through suppression of the transcription factor PHR2. New Phytol. 2018, 219, 135–148. [Google Scholar] [CrossRef] [PubMed]
- Dubos, C.; Stracke, R.; Grotewold, E.; Weisshaar, B.; Martin, C.; Lepiniec, L. MYB transcription factors in Arabidopsis. Trends Plant Sci. 2010, 15, 573–581. [Google Scholar] [CrossRef]
- Wang, T.; Tohge, T.; Ivakov, A.; Mueller-Roeber, B.; Fernie, A.R.; Mutwil, M.; Schippers, J.H.; Persson, S. Salt-related MYB1 coordinates abscisic acid biosynthesis and signaling during salt stress in Arabidopsis. Plant Physiol. 2015, 169, 1027–1041. [Google Scholar] [CrossRef]
- Wei, Q.H.; Zhang, F.; Sun, F.S.; Luo, Q.C.; Wang, R.B.; Chen, M.J.; Chen, M.J.; Chang, J.L.; Yang, G.X.; He, G.Y. A wheat MYB transcriptional repressor TaMyb1D regulates phenylpropanoid metabolism and enhances tolerance to drought and oxidative stresses in transgenic tobacco plants. Plant Sci. 2017, 265, 112–123. [Google Scholar] [CrossRef]
- Seo, P.J.; Lee, S.B.; Suh, M.C.; Park, M.J.; Go, Y.S.; Park, C.M. The MYB96 Transcription factor regulates cuticular wax biosynthesis under drought conditions in Arabidopsis. Plant Cell 2011, 23, 1138–1152. [Google Scholar] [CrossRef] [PubMed]
- Zhai, Y.; Li, P.; Mei, Y.; Chen, M.Y.; Chen, X.C.; Xu, H.; Zhou, X.A.; Dong, H.S.; Zhang, C.L.; Jiang, W.H. Three MYB genes co-regulate the phloem-based defence against English grain aphid in wheat. J. Exp. Bot. 2017, 15, 4153–4169. [Google Scholar] [CrossRef] [PubMed]
- Onkokesung, N.; Reichelt, M.; van Doorn, A.; Schuurink, R.C.; van Loon, J.J.; Dicke, M. Modulation of flavonoid metabolites in Arabidopsis thaliana through overexpression of the MYB75 transcription factor: Role of kaempferol-3,7dirhamnoside in resistance to the specialist insect herbivore Pieris brassicae. J. Exp. Bot. 2014, 65, 2203–2217. [Google Scholar] [CrossRef] [PubMed]
- Mourtzinis, S.; Borg, B.S.; Naeve, S.L.; Osthus, J.; Conley, S.P. Characterizing Soybean Meal Value Variation across the United States: A Swine Case Study. Agron. J. 2018, 110, 2343–2349. [Google Scholar] [CrossRef]
- Olmstead, J.; Brummer, E.C. Benefits and barriers to perennial forage crops in Iowa corn and soybean rotations. Renew. Agric. Food Syst. 2008, 23, 97–107. [Google Scholar] [CrossRef]
- Ullah, A.; Manghwar, H.; Shaban, M.; Khan, A.H.; Akbar, A.; Ali, U.; Ali, E.; Fahad, S. Phytohormones enhanced drought tolerance in plants: A coping strategy. Environ. Sci. Pollut. Res. 2018, 25, 33103–33118. [Google Scholar] [CrossRef] [PubMed]
- Parmar, N.; Singh, K.H.; Sharma, D.; Singh, L.; Kumar, P.; Nanjundan, J.; Khan, Y.J.; Chauhan, D.K.; Thakur, A.K. Genetic engineering strategies for biotic and abiotic stress tolerance and quality enhancement in horticultural crops: A comprehensive review. 3 Biotech 2017, 7, 239. [Google Scholar] [CrossRef]
- Espley, R.V.; Hellens, R.P.; Putterill, J.; Stevenson, D.E.; Kutty-Amma, S.; Allan, A.C. Red colouration in apple fruit is due to the activity of the MYB transcription factor, MdMYB10. Plant J. 2007, 49, 414–427. [Google Scholar] [CrossRef]
- Jakab, G.; Ton, J.; Flors, V.; Zimmerli, L.; Metraux, J.P.; Mauch-Mani, B. Enhancing Arabidopsis salt and drought stress tolerance by chemical priming for its abscisic acid responses. Plant Physiol. 2005, 139, 267–274. [Google Scholar] [CrossRef]
- Chen, J.H.; Jiang, H.W.; Hsieh, E.J.; Chen, H.Y.; Chien, C.T.; Hsieh, H.L.; Lin, T.P. Drought and salt stress tolerance of Arabidopsis glutathione S-transferase U17 knockout mutant are attributed to the combined effect of glutathione and abscisic acid. Plant Physiol. 2011, 158, 340–351. [Google Scholar] [CrossRef]
- Browse, J. Jasmonate Passes Muster: A receptor and targets for the defense hormone. Annu. Rev. Plant Biol. 2009, 60, 183–205. [Google Scholar] [CrossRef] [PubMed]
- Yan, C.; Fan, M.; Yang, M.; Zhao, J.J.; Zhang, W.B.; Su, Y.; Xiao, L.L.; Deng, H.T.; Xie, D.X. Injury Activates Ca2+/Calmodulin-Dependent Phosphorylation of JAV1-JAZ8-WRKY51 Complex for Jasmonate Biosynthesis. Mol. Cell 2018, 70, 136–149. [Google Scholar] [CrossRef] [PubMed]
- Spoel, S.H.; Johnson, J.S.; Dong, X. Regulation of tradeoffs between plant defenses against pathogens with different life styles. Proc. Natl. Acad. Sci. USA 2007, 104, 18842–18847. [Google Scholar] [CrossRef]
- Abe, H.A.; Urao, T.U.; Ito, T.; Seki, M.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Arabidopsis AtMYC2 (bHLH) and AtMYB2 (MYB) Function as transcriptional activators in abscisic acid signaling. Plant Cell 2003, 15, 63–78. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.Q.; Deyholos, M.K. Functional characterization of Arabidopsis NaCl-inducible WRKY25 and WRKY33 transcription factors in abiotic stresses. Plant Mol. Biol. 2009, 69, 91–105. [Google Scholar] [CrossRef]
- Chung, Y.; Kwon, S.I.; Choe, S.H. Antagonistic regulation of Arabidopsis growth by brassinosteroids and abiotic stresses. Mol. Cells 2014, 37, 795–803. [Google Scholar] [CrossRef]
- Pruthvi, V.; Narasimhan, R.; Nataraja, K.N. Simultaneous expression of abiotic stress responsive transcription factors, AtDREB2A, AtHB7 and AtABF3 improves salinity and drought tolerance in peanut (Arachis hypogaea L.). PLoS ONE 2014, 9, E111152. [Google Scholar] [CrossRef]
- Ding, Z.J.; Yan, J.Y.; Li, C.X.; Li, G.X.; Wu, Y.R.; Zheng, S.J. Transcription factor WRKY46 modulates the development of Arabidopsis lateral roots in osmotic/salt stress conditions via regulation of ABA signaling and auxin homeostasis. Plant J. 2015, 84, 56–69. [Google Scholar] [CrossRef]
- Wu, J.Q.; Baldwin, I.T. New insights into plant responses to the attack from insect herbivores. Annu. Rev. Genet. 2010, 44, 1–24. [Google Scholar] [CrossRef]
- An, J.P.; Li, R.; Qu, F.J.; You, C.X.; Wang, X.F.; Hao, Y.J. R2R3-MYB transcription factor MdMYB23 is involved in the cold tolerance and proanthocyanidin accumulation in apple. Plant J. 2018, 96, 562–577. [Google Scholar] [CrossRef]
- Xu, F.C.; Liu, H.L.; Xu, Y.Y.; Zhao, J.R.; Guo, Y.W.; Long, L.; Gao, W.; Song, C. Heterogeneous expression of the cotton R2R3-MYB transcription factor GbMYB60 increases salt sensitivity in transgenic Arabidopsis. Plant Cell Tissue Organ Cult. 2017, 133, 15–25. [Google Scholar] [CrossRef]
- Xie, Y.P.; Chen, P.X.; Yan, Y.; Bao, C.N.; Li, X.W.; Wang, L.P.; Shen, X.X.; Li, H.Y.; Liu, X.F.; Niu, C.D.; et al. An atypical R2R3 MYB transcription factor increases cold hardiness by CBF-dependent and CBF-independent pathways in apple. New Phytol. 2017, 218, 201–218. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.L.; Ling, H.; Ma, J.J.; Chen, Y.; Su, Y.C.; Lin, Q.L.; Gao, S.W.; Wang, H.B.; Que, Y.X.; Xu, L.P. A sugarcane R2R3-MYB transcription factor gene is alternatively spliced during drought stress. Sci. Rep. 2017, 7, 2045–2322. [Google Scholar] [CrossRef] [PubMed]
- Li, T.; Zhang, X.Y.; Huang, Y.; Xu, Z.S.; Wang, F.; Xiong, A.S. An R2R3-MYB transcription factor, S1MYB28, involved in the regulation of TYLCV infection in tomato. Sci. Hortic. 2018, 237, 192–200. [Google Scholar] [CrossRef]
- Spoel, S.H.; Koornneef, A.; Claessens, S.M.C.; Korzelius, J.P.; Van Pelt, J.A.; Mueller, M.J.; Bechala, A.J.; Metraux, J.P.; Brown, R.; Kazan, K.; et al. NPR1 Modulates Cross-Talk between Salicylate- and Jasmonate-Dependent defense pathways through a novel function in the cytosol. Plant Cell 2003, 15, 760–770. [Google Scholar] [CrossRef]
- Wasternack, C. Jasmonates: An update on biosynthesis, signal transduction and action in plant stress response, growth and development. Ann. Bot. 2007, 100, 681–697. [Google Scholar] [CrossRef]
- Koo, A.J.; Gao, X.; Jones, A.D.; Howe, G.A. A rapid wound signal activates the systemic synthesis of bioactive jasmonates in Arabidopsis. Plant J. 2009, 59, 974–986. [Google Scholar] [CrossRef]
- Tamura, K.; Peterson, D.; Peterson, N.; Stecher, G.; Nei, M.; Kumar, S. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 2011, 28, 2731–2739. [Google Scholar] [CrossRef]
- Lescot, M.; Déhais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Peer, Y.V. PlantCARE, a database of plant cis-acting regulator elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef]
- Chenna, R. Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res. 2003, 31, 3497–3500. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−∆∆Ct Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Shen, X.; Zhao, K.; Liu, L.; Zhang, K.; Yuan, H.; Liao, X.; Wang, Q.; Guo, X.W.; Li, F.; Li, T.H. A role for PacMYBA in ABA-regulated anthocyanin biosynthesis in red-colored sweet cherry cv. Hong Deng (Prunus avium L.). Plant Cell Physiol. 2014, 55, 862–880. [Google Scholar] [CrossRef]
- Bai, G.; Yang, D.H.; Zhao, Y.; Ha, S.; Yang, F.; Ma, J.; Gao, X.S.; Wang, Z.M.; Zhu, J.K. Interactions between soybean ABA receptors and type 2C protein phosphatases. Plant Mol. Biol. 2013, 83, 651–664. [Google Scholar] [CrossRef]
- Steven, J.C.; Andrew, F.B. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998, 16, 735–743. [Google Scholar] [CrossRef]
- Kerk, N.M.; Ceserani, T.; Tausta, S.L.; Sussex, I.M.; Nelson, T.M. Laser capture microdissection of cells from plant tissues. Plant Physiol. 2003, 132, 27–35. [Google Scholar] [CrossRef]
| Motif | Stand | Distance from ATG | Sequence | Function |
|---|---|---|---|---|
| ARE | - + | 377 1389 | TGGTTT TGGTTT | Cis-acting regulatory element essential for the anaerobic induction |
| O2-site | - - - | 1049 278 407 | GATGATGTTG TTGAGGATGT GTTGACGTGA | Cis-acting regulatory element involved in zein metabolism regulation |
| CGTCA-motif | + - + | 278 407 1119 | CGTCA CGTCA CGTCA | Cis-acting regulatory element involved in the MeJA-responsiveness |
| GARE-motif | - | 56 | TCTGTTT | Gibberellin response element |
| LTR | + | 1135 | CCGAAA | Cis-acting element involved in low-temperature responsiveness |
| HSE | + | 1460 | AAAAAATTTC | Cis-acting element involved in heat stress responsiveness |
| TGACG-motif | - - + | 1049 278 407 | TGACG TGACG TGACG | Cis-acting regulatory element involved in the MeJA-responsiveness |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shen, X.-J.; Wang, Y.-Y.; Zhang, Y.-X.; Guo, W.; Jiao, Y.-Q.; Zhou, X.-A. Overexpression of the Wild Soybean R2R3-MYB Transcription Factor GsMYB15 Enhances Resistance to Salt Stress and Helicoverpa Armigera in Transgenic Arabidopsis. Int. J. Mol. Sci. 2018, 19, 3958. https://doi.org/10.3390/ijms19123958
Shen X-J, Wang Y-Y, Zhang Y-X, Guo W, Jiao Y-Q, Zhou X-A. Overexpression of the Wild Soybean R2R3-MYB Transcription Factor GsMYB15 Enhances Resistance to Salt Stress and Helicoverpa Armigera in Transgenic Arabidopsis. International Journal of Molecular Sciences. 2018; 19(12):3958. https://doi.org/10.3390/ijms19123958
Chicago/Turabian StyleShen, Xin-Jie, Yan-Yan Wang, Yong-Xing Zhang, Wei Guo, Yong-Qing Jiao, and Xin-An Zhou. 2018. "Overexpression of the Wild Soybean R2R3-MYB Transcription Factor GsMYB15 Enhances Resistance to Salt Stress and Helicoverpa Armigera in Transgenic Arabidopsis" International Journal of Molecular Sciences 19, no. 12: 3958. https://doi.org/10.3390/ijms19123958
APA StyleShen, X.-J., Wang, Y.-Y., Zhang, Y.-X., Guo, W., Jiao, Y.-Q., & Zhou, X.-A. (2018). Overexpression of the Wild Soybean R2R3-MYB Transcription Factor GsMYB15 Enhances Resistance to Salt Stress and Helicoverpa Armigera in Transgenic Arabidopsis. International Journal of Molecular Sciences, 19(12), 3958. https://doi.org/10.3390/ijms19123958




