Loss of Mitochondrial AAA Proteases AFG3L2 and YME1L Impairs Mitochondrial Structure and Respiratory Chain Biogenesis
Abstract
1. Introduction
2. Results and Discussion
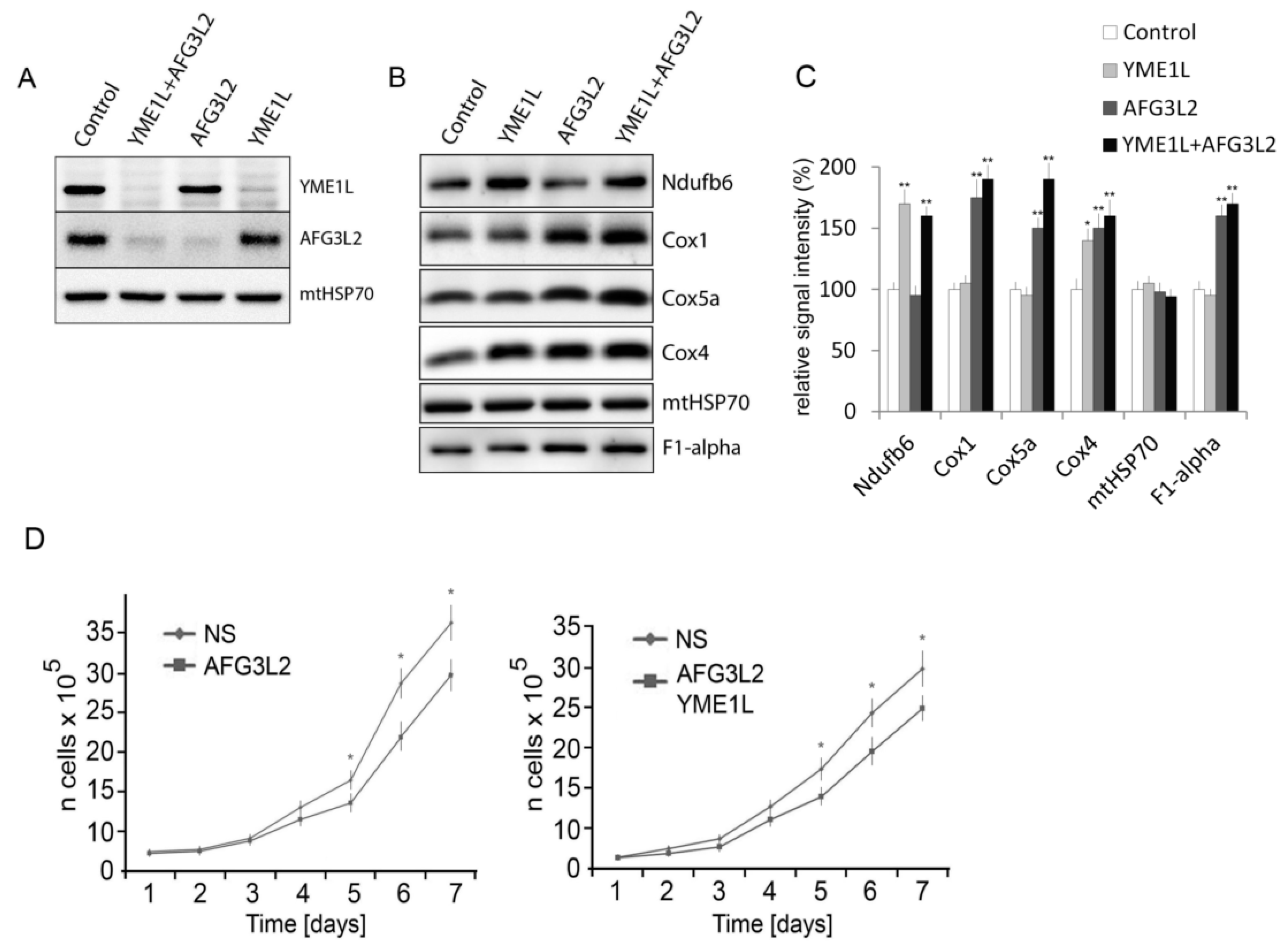
2.1. Knockdown of AFG3L2 and YME1L Leads to Increased Accumulation of Complex I, IV and V Subunits
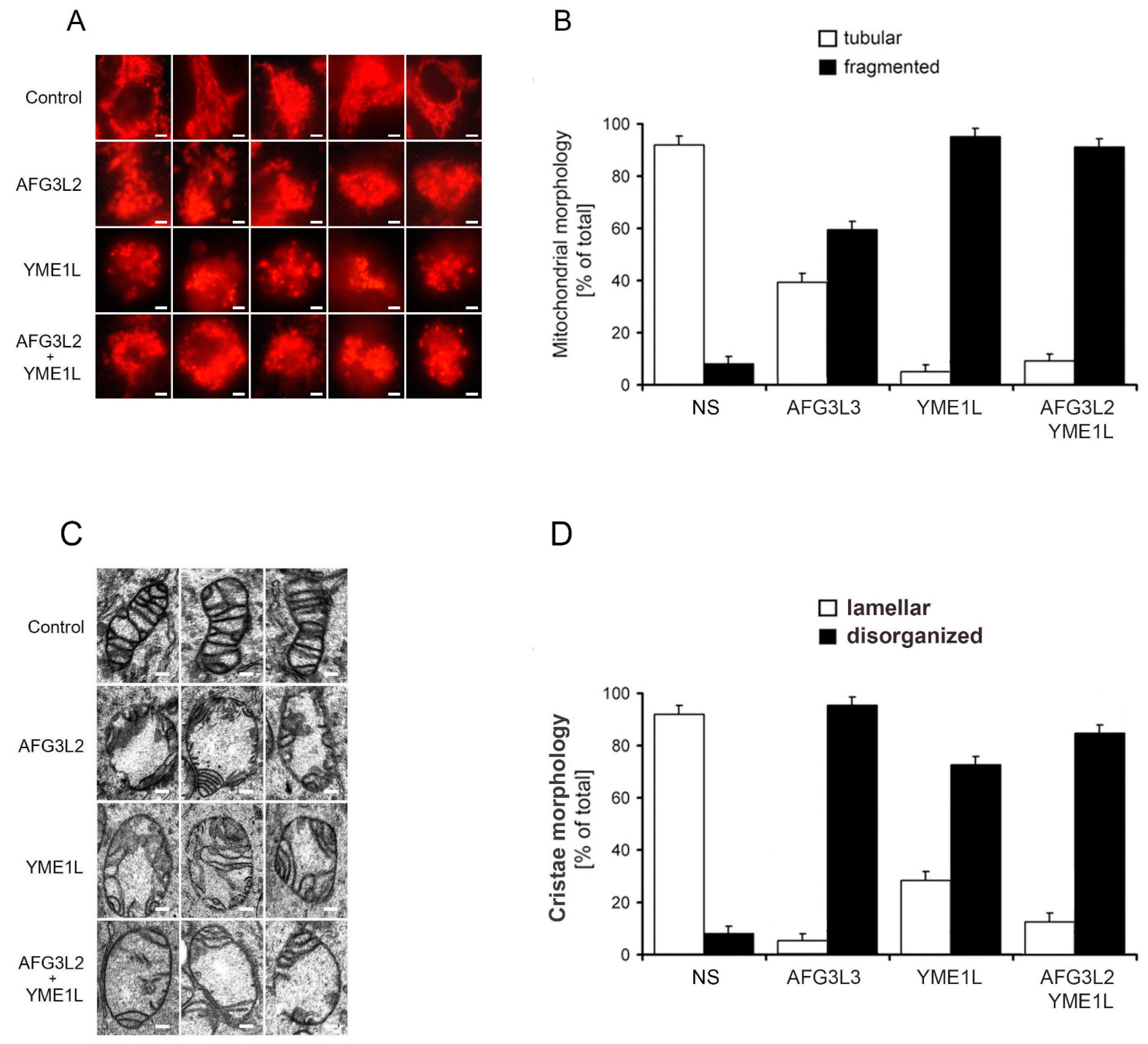
2.2. Loss of AFG3L2 and/or YME1L Leads to Mitochondrial Fragmentation and Cristae Depletion and Disorganization
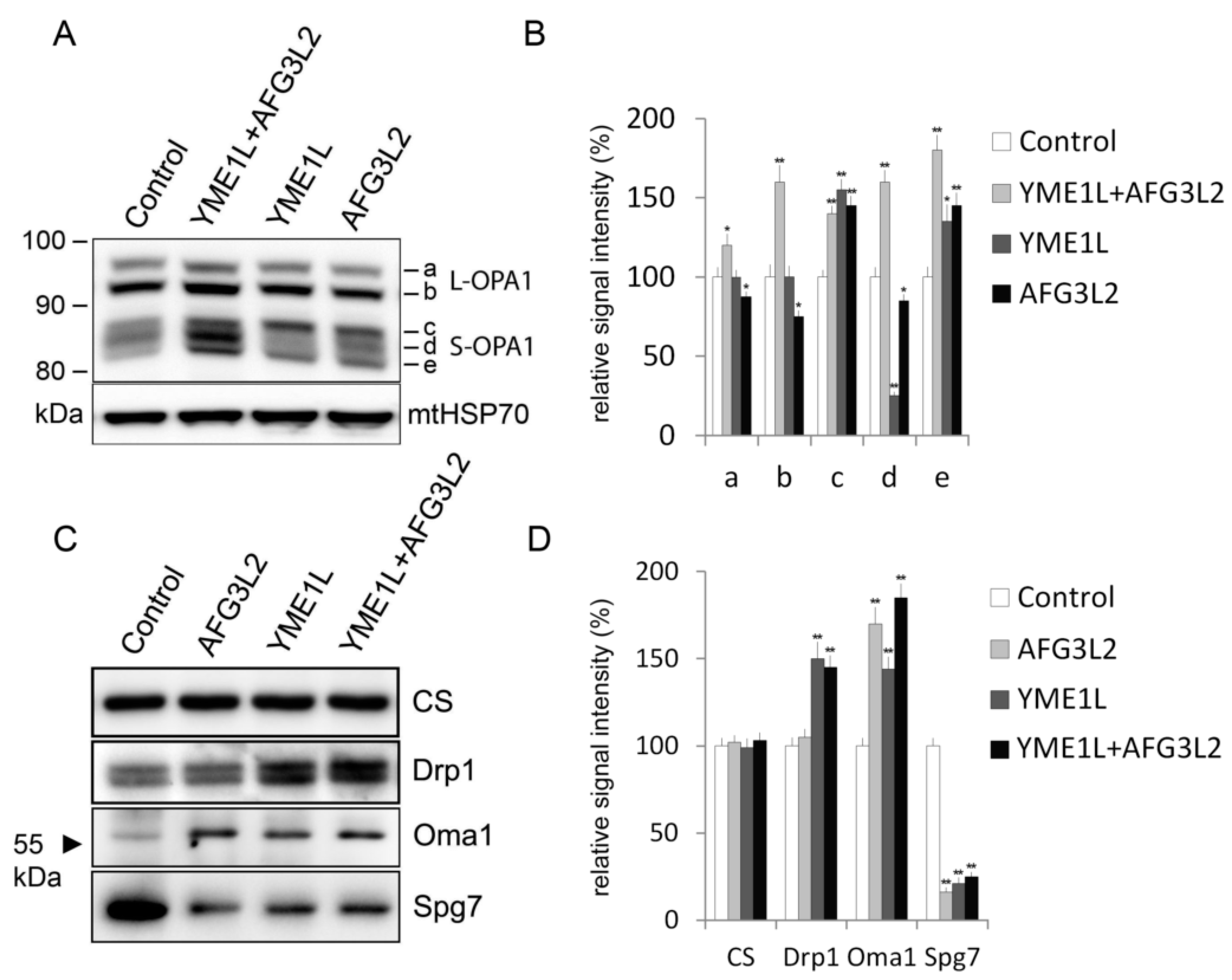
2.3. Loss of AFG3L2 and/or YME1L Leads to OPA1 Upregulation, Accumulation of Short OPA1 Forms, Elevated 60 kDa Oma1, and Reduced SPG7
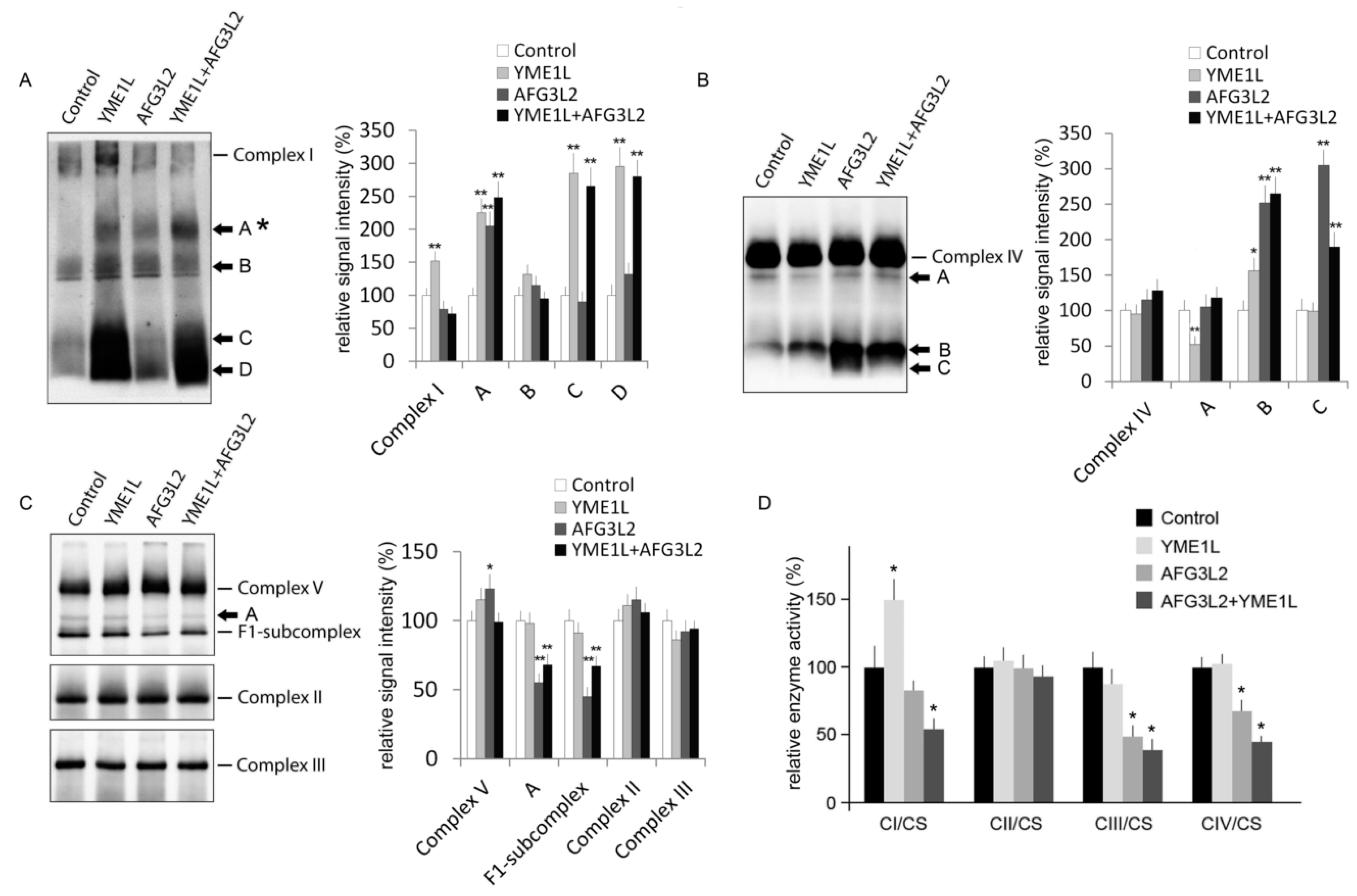
2.4. AFG3L2/YME1L KD Cells Show Reduced Complex I Holoenzyme and Impaired Activity of Complexes I, III, and IV
3. Materials and Methods
3.1. Cell Culture and Transfection
3.2. shRNA, ORF Constructs, and Mutagenesis
3.3. Epifluorescence and Electron Microscopy
3.4. Electrophoresis and Western Blotting
3.5. Antibodies
3.6. Mitochondrial Isolation and Subfractionation
3.7. Assessment of Cell Proliferation
3.8. Assesment of Mitochondrial Morphology and Ultrastructure
3.9. Enzyme Activity Assays
3.10. Statistical Analysis
Author Contributions
Funding
Conflicts of Interest
References
- Glynn, S.E. Multifunctional Mitochondrial AAA Proteases. Front. Mol. Biosci. 2017, 4, 34. [Google Scholar] [CrossRef] [PubMed]
- Pickles, S.; Vigie, P.; Youle, R.J. Mitophagy and Quality Control Mechanisms in Mitochondrial Maintenance. Curr. Biol. 2018, 28, R170–R185. [Google Scholar] [CrossRef] [PubMed]
- Morciano, G.; Pedriali, G.; Sbano, L.; Iannitti, T.; Giorgi, C.; Pinton, P. Intersection of mitochondrial fission and fusion machinery with apoptotic pathways: Role of Mcl-1. Biol. Cell 2016, 108, 279–293. [Google Scholar] [CrossRef] [PubMed]
- Levytskyy, R.M.; Bohovych, I.; Khalimonchuk, O. Metalloproteases of the Inner Mitochondrial Membrane. Biochemistry 2017, 56, 4737–4746. [Google Scholar] [CrossRef] [PubMed]
- Koppen, M.; Langer, T. Protein degradation within mitochondria: Versatile activities of AAA proteases and other peptidases. Crit. Rev. Biochem. Mol. Biol. 2007, 42, 221–242. [Google Scholar] [CrossRef] [PubMed]
- Voos, W.; Jaworek, W.; Wilkening, A.; Bruderek, M. Protein quality control at the mitochondrion. Essays Biochem. 2016, 60, 213–225. [Google Scholar] [CrossRef] [PubMed]
- Stiburek, L.; Cesnekova, J.; Kostkova, O.; Fornuskova, D.; Vinsova, K.; Wenchich, L.; Houstek, J.; Zeman, J. YME1L controls the accumulation of respiratory chain subunits and is required for apoptotic resistance, cristae morphogenesis, and cell proliferation. Mol. Biol. Cell 2012, 23, 1010–1023. [Google Scholar] [CrossRef] [PubMed]
- Ruan, Y.; Li, H.; Zhang, K.; Jian, F.; Tang, J.; Song, Z. Loss of Yme1L perturbates mitochondrial dynamics. Cell Death Dis. 2013, 4, e896. [Google Scholar] [CrossRef]
- Rainbolt, T.K.; Atanassova, N.; Genereux, J.C.; Wiseman, R.L. Stress-regulated translational attenuation adapts mitochondrial protein import through Tim17A degradation. Cell Metab. 2013, 18, 908–919. [Google Scholar] [CrossRef]
- Rainbolt, T.K.; Lebeau, J.; Puchades, C.; Wiseman, R.L. Reciprocal Degradation of YME1L and OMA1 Adapts Mitochondrial Proteolytic Activity during Stress. Cell Rep. 2016, 14, 2041–2049. [Google Scholar] [CrossRef] [PubMed]
- Cesnekova, J.; Rodinova, M.; Hansikova, H.; Houstek, J.; Zeman, J.; Stiburek, L. The mammalian homologue of yeast Afg1 ATPase (lactation elevated 1) mediates degradation of nuclear-encoded complex IV subunits. Biochem. J. 2016, 473, 797–804. [Google Scholar] [CrossRef] [PubMed]
- Hornig-Do, H.T.; Tatsuta, T.; Buckermann, A.; Bust, M.; Kollberg, G.; Rötig, A.; Hellmich, M.; Nijtmans, L.; Wiesner, R.J. Nonsense mutations in the COX1 subunit impair the stability of respiratory chain complexes rather than their assembly. EMBO J. 2012, 31, 1293–1307. [Google Scholar] [CrossRef] [PubMed]
- Richter, U.; Lahtinen, T.; Marttinen, P.; Suomi, F.; Battersby, B.J. Quality control of mitochondrial protein synthesis is required for membrane integrity and cell fitness. J. Cell Biol. 2015, 211, 373–389. [Google Scholar] [CrossRef]
- Tsai, C.W.; Wu, Y.; Pao, P.-C.; Phillips, C.B.; Williams, C.; Miller, C.; Ranaghan, M.; Tsai, M.-F. Proteolytic control of the mitochondrial calcium uniporter complex. Proc. Natl. Acad. Sci. USA 2017, 114, 4388–4393. [Google Scholar] [CrossRef] [PubMed]
- Consolato, F.; Maltecca, F.; Tulli, S.; Sambri, I.; Casari, G. m-AAA and i-AAA complexes coordinate to regulate OMA1, the stress-activated supervisor of mitochondrial dynamics. J. Cell Sci. 2018, 131. [Google Scholar] [CrossRef]
- Shanmughapriya, S.; Rajan, S.; Hoffman, N.E.; Tomar, D.; Nemani, N.; Madesh, M. SPG7 Is an Essential and Conserved Component of the Mitochondrial Permeability Transition Pore. Mol. Cell 2015, 60, 47–62. [Google Scholar] [CrossRef]
- Magri, S.; Fracasso, V.; Plumari, M.; Alfei, E.; Ghezzi, D.; Gellera, C.; Rusmini, P.; Poletti, A.; Bella, D.D.; Elia, A.E.; et al. Concurrent AFG3L2 and SPG7 mutations associated with syndromic parkinsonism and optic atrophy with aberrant OPA1 processing and mitochondrial network fragmentation. Hum. Mutat. 2018, 39. [Google Scholar] [CrossRef]
- Hartmann, B.; Wai, T.; Hu, H.; MacVicar, T.; Musante, L.; Fischer-Zirnsak, B.; Stenzel, W.; Gräf, R.; van den Heuvel, L.; Ropers, H.H.; et al. Homozygous YME1L1 mutation causes mitochondriopathy with optic atrophy and mitochondrial network fragmentation. Elife 2016, 5. [Google Scholar] [CrossRef]
- Sprenger, H.G.; Wani, G.; Hesseling, A.; König, T.; Patron, M.; MacVicar, T.; Ahola, S.; Wai, T.; Barth, E.; Rugarli, E.I.; et al. Loss of the mitochondrial i-AAA protease YME1L leads to ocular dysfunction and spinal axonopathy. EMBO Mol. Med. 2018. [Google Scholar] [CrossRef] [PubMed]
- Cesnekova, J.; Spacilova, J.; Hansikova, H.; Houstek, J.; Zeman, J.; Stiburek, L. LACE1 interacts with p53 and mediates its mitochondrial translocation and apoptosis. Oncotarget 2016, 7, 47687–47698. [Google Scholar] [CrossRef]
- Stiburek, L.; Hansikova, H.; Tesarova, M.; Cerna, L.; Zeman, J. Biogenesis of eukaryotic cytochrome c oxidase. Physiol. Res. 2006, 55 (Suppl. 2), S27–S41. [Google Scholar]
- Baker, M.J.; Lampe, P.A.; Stojanovski, D.; Korwitz, A.; Anand, R.; Tatsuta, T.; Langer, T. Stress-induced OMA1 activation and autocatalytic turnover regulate OPA1-dependent mitochondrial dynamics. EMBO J. 2014, 33, 578–593. [Google Scholar] [CrossRef] [PubMed]
- Ehses, S.; Raschke, I.; Mancuso, G.; Bernacchia, A.; Geimer, S.; Tondera, D.; Martinou, J.-C.; Westermann, B.; Rugarli, E.I.; Langer, T. Regulation of OPA1 processing and mitochondrial fusion by m-AAA protease isoenzymes and OMA1. J. Cell Biol. 2009, 187, 1023–1036. [Google Scholar] [CrossRef] [PubMed]
- Twig, G.; Elorza, A.; Molina, A.J.A.; Mohamed, H.; Wikstrom, J.D.; Walzer, G.; Stiles, L.; Haigh, S.E.; Katz, S.; Las, G.; et al. Fission and selective fusion govern mitochondrial segregation and elimination by autophagy. EMBO J. 2008, 27, 433–446. [Google Scholar] [CrossRef]
- Patten, D.A.; Wong, J.; Khacho, M.; Soubannier, V.; Mailloux, R.J.; Pilon-Larose, K.; MacLaurin, J.G.; Park, D.S.; McBride, H.M.; Trinkle-Mulcahy, L.; et al. OPA1-dependent cristae modulation is essential for cellular adaptation to metabolic demand. EMBO J. 2014, 33, 2676–2691. [Google Scholar] [CrossRef] [PubMed]
- Del Dotto, V.; Mishra, P.; Vidoni, S.; Fogazza, M.; Maresca, A.; Caporali, L.; McCaffery, J.M.; Cappelletti, M.; Baruffini, E.; Lenaers, G.; et al. OPA1 Isoforms in the Hierarchical Organization of Mitochondrial Functions. Cell Rep. 2017, 19, 2557–2571. [Google Scholar] [CrossRef]
- Frezza, C.; Cipolat, S.; de Brito, O.M.; Micaroni, M.; Beznoussenko, G.V.; Rudka, T.; Bartoli, D.; Polishuck, R.S.; Danial, N.N.; De Strooper, B.; et al. OPA1 controls apoptotic cristae remodeling independently from mitochondrial fusion. Cell 2006, 126, 177–189. [Google Scholar] [CrossRef]
- Meeusen, S.; DeVay, R.; Block, J.; Cassidy-Stone, A.; Wayson, S.; McCaffery, J.M.; Nunnari, J. Mitochondrial inner-membrane fusion and crista maintenance requires the dynamin-related GTPase Mgm1. Cell 2006, 127, 383–395. [Google Scholar] [CrossRef]
- Anand, R.; Wai, T.; Baker, M.J.; Kladt, N.; Schauss, A.C.; Rugarli, E.; Langer, T. The i-AAA protease YME1L and OMA1 cleave OPA1 to balance mitochondrial fusion and fission. J. Cell Biol. 2014, 204, 919–929. [Google Scholar] [CrossRef]
- MacVicar, T.; Langer, T. OPA1 processing in cell death and disease—The long and short of it. J. Cell Sci. 2016, 129, 2297–2306. [Google Scholar] [CrossRef]
- Head, B.; Griparic, L.; Amiri, M.; Gandre-Babbe, S.; van der Bliek, A.M. Inducible proteolytic inactivation of OPA1 mediated by the OMA1 protease in mammalian cells. J. Cell Biol. 2009, 187, 959–966. [Google Scholar] [CrossRef]
- Maltecca, F.; De Stefani, D.; Cassina, L.; Consolato, F.; Wasilewski, M.; Scorrano, L.; Rizzuto, R.; Casari, G. Respiratory dysfunction by AFG3L2 deficiency causes decreased mitochondrial calcium uptake via organellar network fragmentation. Hum. Mol. Genet. 2012, 21, 3858–3870. [Google Scholar] [CrossRef] [PubMed]
- Nan, J.; Hu, H.; Sun, Y.; Zhu, L.; Wang, Y.; Zhong, Z.; Zhao, J.; Zhang, N.; Wang, Y.; Wang, Y.; et al. TNFR2 Stimulation Promotes Mitochondrial Fusion via Stat3- and NF-kB-Dependent Activation of OPA1 Expression. Circ. Res. 2017, 121, 392–410. [Google Scholar] [CrossRef]
- Zhang, K.; Li, H.; Song, Z. Membrane depolarization activates the mitochondrial protease OMA1 by stimulating self-cleavage. EMBO Rep. 2014, 15, 576–585. [Google Scholar] [CrossRef]
- Chang, C.R.; Blackstone, C. Dynamic regulation of mitochondrial fission through modification of the dynamin-related protein Drp1. Ann. N. Y. Acad. Sci. 2010, 1201, 34–39. [Google Scholar] [CrossRef]
- Koppen, M.; Bonn, F.; Ehses, S.; Langer, T. Autocatalytic processing of m-AAA protease subunits in mitochondria. Mol. Biol. Cell 2009, 20, 4216–4224. [Google Scholar] [CrossRef] [PubMed]
- Atorino, L.; Silvestri, L.; Koppen, M.; Cassina, L.; Ballabio, A.; Marconi, R.; Langer, T.; Casari, G. Loss of m-AAA protease in mitochondria causes complex I deficiency and increased sensitivity to oxidative stress in hereditary spastic paraplegia. J. Cell Biol. 2003, 163, 777–787. [Google Scholar] [CrossRef] [PubMed]
- Maltecca, F.; Aghaie, A.; Schroeder, D.G.; Cassina, L.; Taylor, B.A.; Phillips, S.J.; Malaguti, M.; Previtali, S.; Guénet, J.-L.; Quattrini, A.; et al. The mitochondrial protease AFG3L2 is essential for axonal development. J. Neurosci. 2008, 28, 2827–2836. [Google Scholar] [CrossRef]
- Luft, J.H. Permanganate: A new fixative for electron microscopy. J. Biophys. Biochem. Cytol. 1956, 2, 799–802. [Google Scholar] [CrossRef] [PubMed]
- Stiburek, L.; Fornuskova, D.; Wenchich, L.; Pejznochova, M.; Hansikova, H.; Zeman, J. Knockdown of human Oxa1l impairs the biogenesis of F1Fo-ATP synthase and NADH:ubiquinone oxidoreductase. J. Mol. Biol. 2007, 374, 506–516. [Google Scholar] [CrossRef]
- Vaseva, A.V.; Moll, U.M. Identification of p53 in mitochondria. Methods Mol. Biol. 2013, 962, 75–84. [Google Scholar] [PubMed]
- Stiburek, L.; Vesela, K.; Hansikova, H.; Pecina, P.; Tesarova, M.; Cerna, L.; Houstek, J.; Zeman, J. Tissue-specific cytochrome c oxidase assembly defects due to mutations in SCO2 and SURF1. Biochem. J. 2005, 392 Pt 3, 625–632. [Google Scholar] [CrossRef]
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cesnekova, J.; Rodinova, M.; Hansikova, H.; Zeman, J.; Stiburek, L. Loss of Mitochondrial AAA Proteases AFG3L2 and YME1L Impairs Mitochondrial Structure and Respiratory Chain Biogenesis. Int. J. Mol. Sci. 2018, 19, 3930. https://doi.org/10.3390/ijms19123930
Cesnekova J, Rodinova M, Hansikova H, Zeman J, Stiburek L. Loss of Mitochondrial AAA Proteases AFG3L2 and YME1L Impairs Mitochondrial Structure and Respiratory Chain Biogenesis. International Journal of Molecular Sciences. 2018; 19(12):3930. https://doi.org/10.3390/ijms19123930
Chicago/Turabian StyleCesnekova, Jana, Marie Rodinova, Hana Hansikova, Jiri Zeman, and Lukas Stiburek. 2018. "Loss of Mitochondrial AAA Proteases AFG3L2 and YME1L Impairs Mitochondrial Structure and Respiratory Chain Biogenesis" International Journal of Molecular Sciences 19, no. 12: 3930. https://doi.org/10.3390/ijms19123930
APA StyleCesnekova, J., Rodinova, M., Hansikova, H., Zeman, J., & Stiburek, L. (2018). Loss of Mitochondrial AAA Proteases AFG3L2 and YME1L Impairs Mitochondrial Structure and Respiratory Chain Biogenesis. International Journal of Molecular Sciences, 19(12), 3930. https://doi.org/10.3390/ijms19123930




