Functional Annotation of Bacterial Signal Transduction Systems: Progress and Challenges
Abstract
1. Introduction
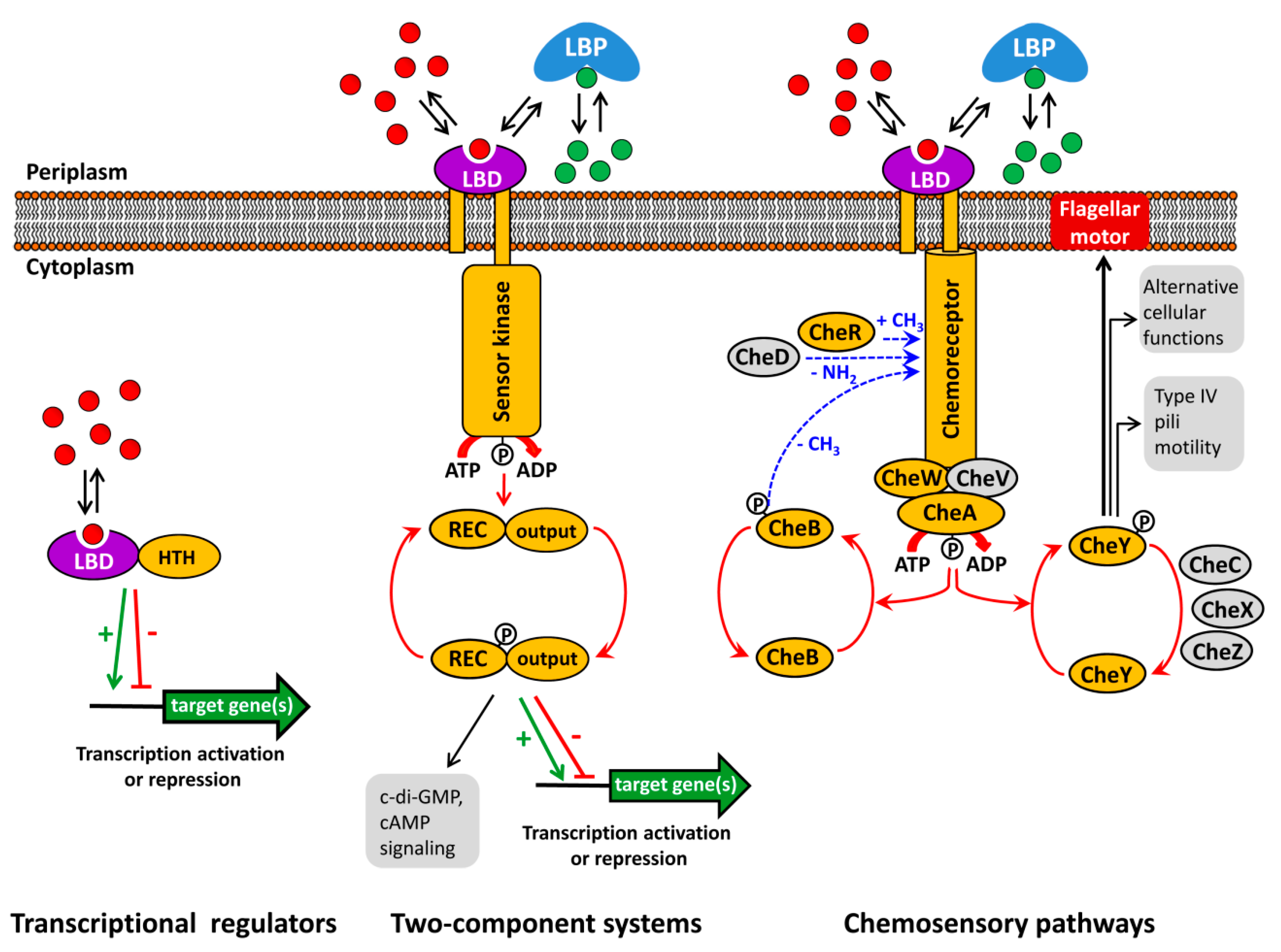
2. Functional Annotation Using Genetic Approaches
2.1. Multiple Receptors
2.2. Chemotaxis Is Induced or Repressed by the Cognate Ligands
2.3. Energy Taxis May Mask Chemotaxis
3. Functional Annotation Using Thermal Shift Assays of Purified Recombinant Sensor Proteins
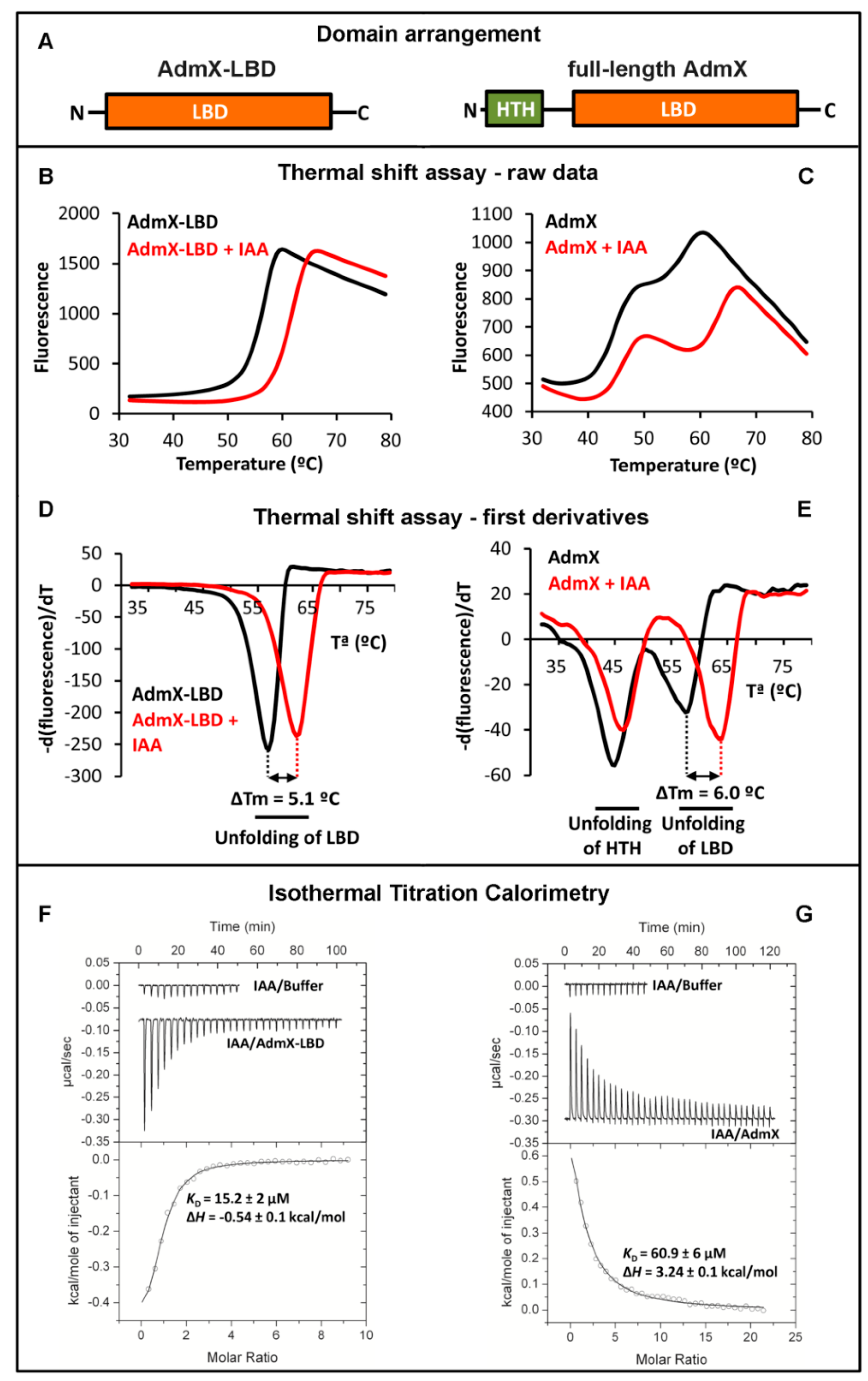
3.1. The Case of The Transcriptional Regulator AdmX
3.1.1. Thermal Shift Assays with AdmX and AdmX-LBD
3.1.2. Study of Ligand Binding to AdmX and AdmX-LBD by Isothermal Titration Calorimetry
3.1.3. The Identification of the AdmX Signal Molecule Permits to Elucidate the Molecular Mechanism and Physiological Relevance
3.2. Functional Annotation of Chemoreceptors Using Thermal Shift Assays
4. Three-Dimensional Structural Information Provides Clues on Ligands Recognized by Homologous Sensor Proteins
5. Protein Structure Based Virtual Ligand Screening
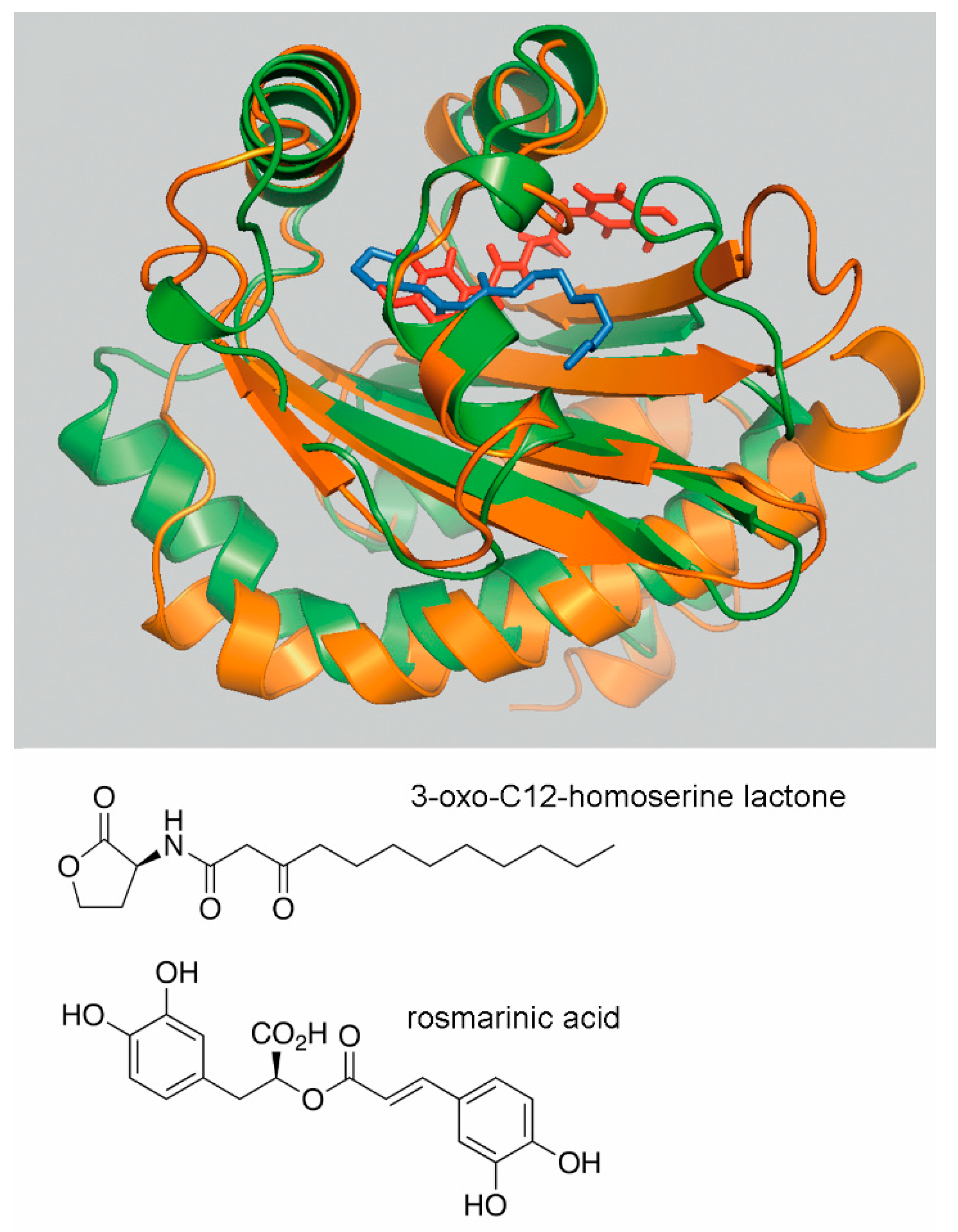
5.1. The Identification of Rosmarinic Acid as Plant Derived Quorum Sensing Agonist
5.1.1. Identification of Rosmarinic Acid as RhlR Ligand
5.1.2. Assessment of the Global Role of Rosmarinic Acid
5.2. Further Examples that Illustrate the Usefulness of Virtual Ligand Screening to Identify Protein Ligands
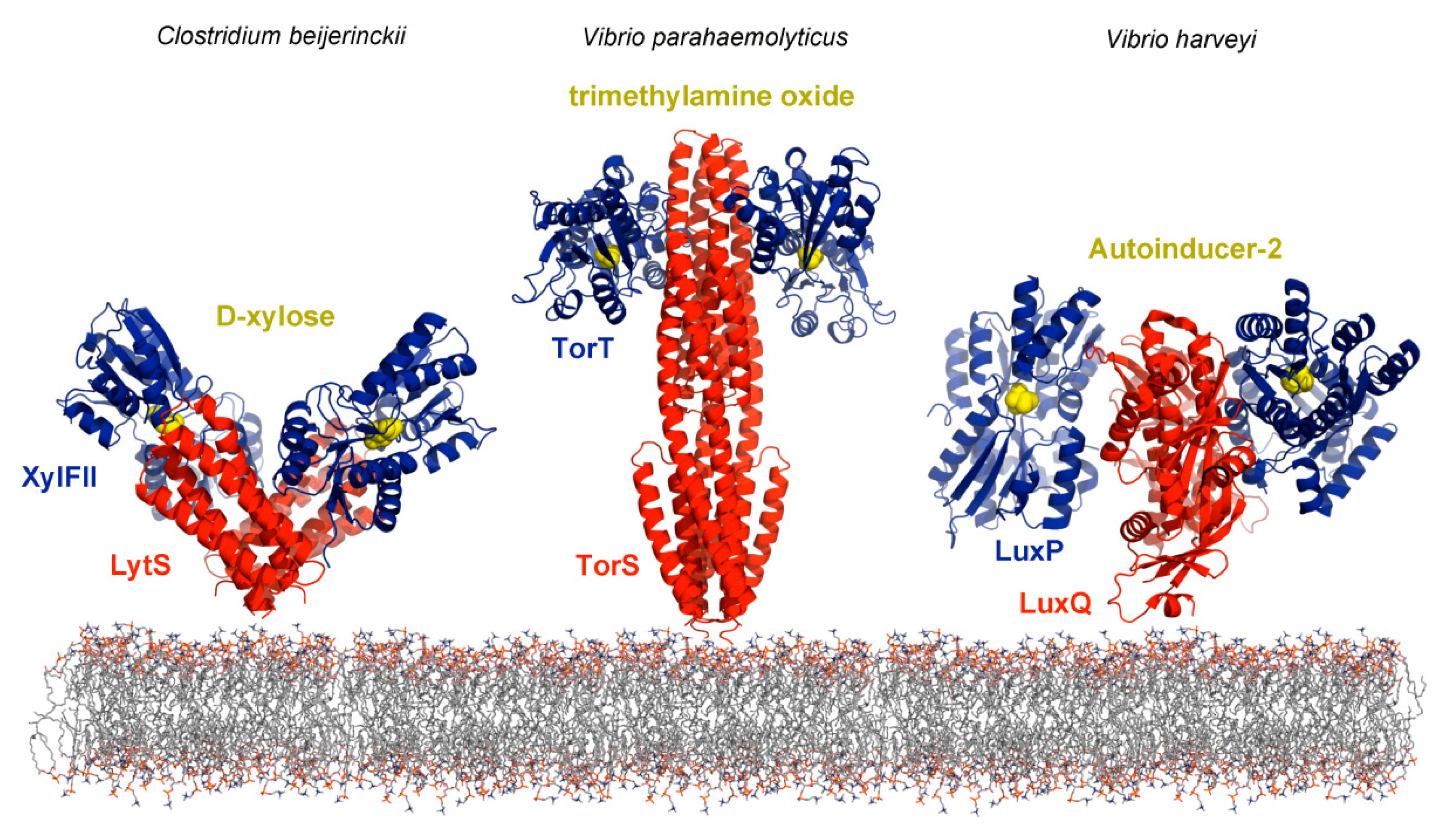
6. The Challenge: Signal Input Through Ligand Binding Proteins
6.1. Sensor Protein Activation by the Binding of Ligand Binding Proteins
6.2. Identification of Ligand Binding Proteins That Interact With Sensor Proteins
7. Concluding Remarks
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| 4HB | 4-helix bundle |
| HBM | Helical Bimodular |
| HTH | Helix-turn-helix motif containing DNA binding domain |
| IAA | Indole-3-acetic acid |
| ITC | Isothermal Titration Calorimetry |
| LBD | Ligand binding domain |
| Pi | Inorganic phosphate |
| QS | Quorum sensing |
| RA | Rosmarinic acid |
| REC | Receiver domain |
| TCS | Two-component system |
| Tm | Melting temperature |
References
- Ulrich, L.E.; Koonin, E.V.; Zhulin, I.B. One-component systems dominate signal transduction in prokaryotes. Trends Microbiol. 2005, 13, 52–56. [Google Scholar] [CrossRef] [PubMed]
- Wuichet, K.; Zhulin, I.B. Origins and diversification of a complex signal transduction system in prokaryotes. Sci. Signal. 2010, 3, ra50. [Google Scholar] [CrossRef] [PubMed]
- Jacob-Dubuisson, F.; Mechaly, A.; Betton, J.M.; Antoine, R. Structural insights into the signalling mechanisms of two-component systems. Nat. Rev. Microbiol. 2018, 16, 585–593. [Google Scholar] [CrossRef] [PubMed]
- Groisman, E.A. Feedback Control of Two-Component Regulatory Systems. Annu. Rev. Microbiol. 2016, 70, 103–124. [Google Scholar] [CrossRef] [PubMed]
- Zschiedrich, C.P.; Keidel, V.; Szurmant, H. Molecular Mechanisms of Two-Component Signal Transduction. J. Mol. Biol. 2016, 428, 3752–3775. [Google Scholar] [CrossRef] [PubMed]
- Galperin, M.Y. Diversity of structure and function of response regulator output domains. Curr. Opin. Microbiol. 2010, 13, 150–159. [Google Scholar] [CrossRef] [PubMed]
- Ortega, A.; Zhulin, I.B.; Krell, T. Sensory Repertoire of Bacterial Chemoreceptors. Microbiol. Mol. Biol. Rev. 2017, 81. [Google Scholar] [CrossRef] [PubMed]
- Galperin, M.Y. Structural classification of bacterial response regulators: Diversity of output domains and domain combinations. J. Bacteriol. 2006, 188, 4169–4182. [Google Scholar] [CrossRef] [PubMed]
- Pham, H.T.; Parkinson, J.S. Phenol sensing by Escherichia coli chemoreceptors: A nonclassical mechanism. J. Bacteriol. 2011, 193, 6597–6604. [Google Scholar] [CrossRef] [PubMed]
- Yuan, J.; Jin, F.; Glatter, T.; Sourjik, V. Osmosensing by the bacterial PhoQ/PhoP two-component system. Proc. Natl. Acad. Sci. USA 2017, 114, 10792–10798. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.C.; Morgan, L.K.; Godakumbura, P.; Kenney, L.J.; Anand, G.S. The inner membrane histidine kinase EnvZ senses osmolality via helix-coil transitions in the cytoplasm. EMBO J. 2012, 31, 2648–2659. [Google Scholar] [CrossRef] [PubMed]
- Petrova, O.E.; Sauer, K. Dispersion by Pseudomonas aeruginosa requires an unusual posttranslational modification of BdlA. Proc. Natl. Acad. Sci. USA 2012, 109, 16690–16695. [Google Scholar] [CrossRef] [PubMed]
- Somavanshi, R.; Ghosh, B.; Sourjik, V. Sugar Influx Sensing by the Phosphotransferase System of Escherichia coli. PLoS Biol. 2016, 14, e2000074. [Google Scholar] [CrossRef] [PubMed]
- Balleza, E.; Lopez-Bojorquez, L.N.; Martinez-Antonio, A.; Resendis-Antonio, O.; Lozada-Chavez, I.; Balderas-Martinez, Y.I.; Encarnacion, S.; Collado-Vides, J. Regulation by transcription factors in bacteria: Beyond description. FEMS Microbiol. Rev. 2009, 33, 133–151. [Google Scholar] [CrossRef] [PubMed]
- Gushchin, I.; Melnikov, I.; Polovinkin, V.; Ishchenko, A.; Yuzhakova, A.; Buslaev, P.; Bourenkov, G.; Grudinin, S.; Round, E.; Balandin, T.; et al. Mechanism of transmembrane signaling by sensor histidine kinases. Science 2017, 356, eaah6345. [Google Scholar] [CrossRef] [PubMed]
- Ottemann, K.M.; Xiao, W.; Shin, Y.K.; Koshland, D.E., Jr. A piston model for transmembrane signaling of the aspartate receptor. Science 1999, 285, 1751–1754. [Google Scholar] [CrossRef] [PubMed]
- Bi, S.; Pollard, A.M.; Yang, Y.; Jin, F.; Sourjik, V. Engineering Hybrid Chemotaxis Receptors in Bacteria. ACS Synth. Biol. 2016, 5, 989–1001. [Google Scholar] [CrossRef] [PubMed]
- Reyes-Darias, J.A.; Yang, Y.; Sourjik, V.; Krell, T. Correlation between signal input and output in PctA and PctB amino acid chemoreceptor of Pseudomonas aeruginosa. Mol. Microbiol. 2015, 96, 513–525. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, T.; Phadtare, S.; Inouye, M. The design and development of Tar-EnvZ chimeric receptors. Methods Enzymol. 2007, 423, 166–183. [Google Scholar] [PubMed]
- Ward, S.M.; Delgado, A.; Gunsalus, R.P.; Manson, M.D. A NarX-Tar chimera mediates repellent chemotaxis to nitrate and nitrite. Mol. Microbiol. 2002, 44, 709–719. [Google Scholar] [CrossRef] [PubMed]
- Krell, T. Tackling the bottleneck in bacterial signal transduction research: High-throughput identification of signal molecules. Mol. Microbiol. 2015, 96, 685–688. [Google Scholar] [CrossRef] [PubMed]
- Francis, V.I.; Stevenson, E.C.; Porter, S.L. Two-component systems required for virulence in Pseudomonas aeruginosa. FEMS Microbiol. Lett. 2017, 364. [Google Scholar] [CrossRef] [PubMed]
- Reyes-Darias, J.A.; Garcia, V.; Rico-Jimenez, M.; Corral-Lugo, A.; Lesouhaitier, O.; Juarez-Hernandez, D.; Yang, Y.; Bi, S.; Feuilloley, M.; Munoz-Rojas, J.; et al. Specific gamma-aminobutyrate chemotaxis in pseudomonads with different lifestyle. Mol. Microbiol. 2015, 97, 488–501. [Google Scholar] [CrossRef] [PubMed]
- Grimm, A.C.; Harwood, C.S. NahY, a catabolic plasmid-encoded receptor required for chemotaxis of Pseudomonas putida to the aromatic hydrocarbon naphthalene. J. Bacteriol. 1999, 181, 3310–3316. [Google Scholar] [PubMed]
- Luu, R.A.; Kootstra, J.D.; Nesteryuk, V.; Brunton, C.N.; Parales, J.V.; Ditty, J.L.; Parales, R.E. Integration of chemotaxis, transport and catabolism in Pseudomonas putida and identification of the aromatic acid chemoreceptor PcaY. Mol. Microbiol. 2015, 96, 134–147. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Wood, P.L.; Parales, J.V.; Parales, R.E. Chemotaxis to pyrimidines and identification of a cytosine chemoreceptor in Pseudomonas putida. J. Bacteriol. 2009, 191, 2909–2916. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.; Kato, J.; Kuroda, A.; Ikeda, T.; Takiguchi, N.; Ohtake, H. Identification and characterization of two chemotactic transducers for inorganic phosphate in Pseudomonas aeruginosa. J. Bacteriol. 2000, 182, 3400–3404. [Google Scholar] [CrossRef] [PubMed]
- Hida, A.; Oku, S.; Nakashimada, Y.; Tajima, T.; Kato, J. Identification of boric acid as a novel chemoattractant and elucidation of its chemoreceptor in Ralstonia pseudosolanacearum Ps29. Sci. Rep. 2017, 7, 8609. [Google Scholar] [CrossRef] [PubMed]
- Lacal, J.; Garcia-Fontana, C.; Munoz-Martinez, F.; Ramos, J.L.; Krell, T. Sensing of environmental signals: Classification of chemoreceptors according to the size of their ligand binding regions. Environ. Microbiol. 2010, 12, 2873–2884. [Google Scholar] [CrossRef] [PubMed]
- Alexandre, G.; Greer-Phillips, S.; Zhulin, I.B. Ecological role of energy taxis in microorganisms. FEMS Microbiol. Rev. 2004, 28, 113–126. [Google Scholar] [CrossRef] [PubMed]
- Ni, B.; Huang, Z.; Wu, Y.F.; Fan, Z.; Jiang, C.Y.; Liu, S.J. A novel chemoreceptor MCP2983 from Comamonas testosteroni specifically binds to cis-aconitate and triggers chemotaxis towards diverse organic compounds. Appl. Microbiol. Biotechnol. 2015, 99, 2773–2781. [Google Scholar] [CrossRef] [PubMed]
- Hida, A.; Tajima, T.; Kato, J. Two citrate chemoreceptors involved in chemotaxis to citrate and/or citrate-metal complexes in Ralstonia pseudosolanacearum. J. Biosci. Bioeng. 2018. [Google Scholar] [CrossRef] [PubMed]
- Corral-Lugo, A.; Matilla, M.A.; Martín-Mora, D.; Silva Jiménez, H.; Mesa Torres, N.; Kato, J.; Hida, A.; Oku, S.; Conejero-Muriel, M.; Gavira, J.A.; et al. High-affinity chemotaxis to histamine mediated by the TlpQ chemoreceptor of the human pathogen Pseudomonas aeruginosa. Mbio 2018, 9, e01894-18. [Google Scholar] [CrossRef] [PubMed]
- Feng, H.; Zhang, N.; Du, W.; Zhang, H.; Liu, Y.; Fu, R.; Shao, J.; Zhang, G.; Shen, Q.R.; Zhang, R. Identification of chemotaxis compounds in root exudates and their sensing chemoreceptors in plant growth-promoting rhizobacteria Bacillus amyloliquefaciens SQR9. Mol. Plant Microbe Interact. 2018. [Google Scholar] [CrossRef] [PubMed]
- Taguchi, K.; Fukutomi, H.; Kuroda, A.; Kato, J.; Ohtake, H. Genetic identification of chemotactic transducers for amino acids in Pseudomonas aeruginosa. Microbiology 1997, 143, 3223–3229. [Google Scholar] [CrossRef] [PubMed]
- Rico-Jimenez, M.; Munoz-Martinez, F.; Garcia-Fontana, C.; Fernandez, M.; Morel, B.; Ortega, A.; Ramos, J.L.; Krell, T. Paralogous chemoreceptors mediate chemotaxis towards protein amino acids and the non-protein amino acid gamma-aminobutyrate (GABA). Mol. Microbiol. 2013, 88, 1230–1243. [Google Scholar] [CrossRef] [PubMed]
- Oku, S.; Komatsu, A.; Tajima, T.; Nakashimada, Y.; Kato, J. Identification of chemotaxis sensory proteins for amino acids in Pseudomonas fluorescens Pf0-1 and their involvement in chemotaxis to tomato root exudate and root colonization. Microbes Environ. 2012, 27, 462–469. [Google Scholar] [CrossRef] [PubMed]
- Webb, B.A.; Hildreth, S.; Helm, R.F.; Scharf, B.E. Sinorhizobium meliloti chemoreceptor McpU mediates chemotaxis toward host plant exudates through direct proline sensing. Appl. Environ. Microbiol. 2014, 80, 3404–3415. [Google Scholar] [CrossRef] [PubMed]
- Webb, B.A.; Karl Compton, K.; Castaneda Saldana, R.; Arapov, T.D.; Keith Ray, W.; Helm, R.F.; Scharf, B.E. Sinorhizobium meliloti chemotaxis to quaternary ammonium compounds is mediated by the chemoreceptor McpX. Mol. Microbiol. 2017, 103, 333–346. [Google Scholar] [CrossRef] [PubMed]
- Oku, S.; Komatsu, A.; Nakashimada, Y.; Tajima, T.; Kato, J. Identification of Pseudomonas fluorescens Chemotaxis Sensory Proteins for Malate, Succinate, and Fumarate, and Their Involvement in Root Colonization. Microbes Environ. 2014, 29, 413–419. [Google Scholar] [CrossRef] [PubMed]
- Pineda-Molina, E.; Reyes-Darias, J.A.; Lacal, J.; Ramos, J.L.; Garcia-Ruiz, J.M.; Gavira, J.A.; Krell, T. Evidence for chemoreceptors with bimodular ligand-binding regions harboring two signal-binding sites. Proc. Natl. Acad. Sci. USA 2012, 109, 18926–18931. [Google Scholar] [CrossRef] [PubMed]
- Martin-Mora, D.; Reyes-Darias, J.A.; Ortega, A.; Corral-Lugo, A.; Matilla, M.A.; Krell, T. McpQ is a specific citrate chemoreceptor that responds preferentially to citrate/metal ion complexes. Environ. Microbiol. 2016, 18, 3284–3295. [Google Scholar] [CrossRef] [PubMed]
- Lacal, J.; Garcia-Fontana, C.; Callejo-Garcia, C.; Ramos, J.L.; Krell, T. Physiologically relevant divalent cations modulate citrate recognition by the McpS chemoreceptor. J. Mol. Recognit. 2011, 24, 378–385. [Google Scholar] [CrossRef] [PubMed]
- Rico-Jimenez, M.; Reyes-Darias, J.A.; Ortega, A.; Diez Pena, A.I.; Morel, B.; Krell, T. Two different mechanisms mediate chemotaxis to inorganic phosphate in Pseudomonas aeruginosa. Sci. Rep. 2016, 6, 28967. [Google Scholar] [CrossRef] [PubMed]
- Bains, M.; Fernandez, L.; Hancock, R.E. Phosphate starvation promotes swarming motility and cytotoxicity of Pseudomonas aeruginosa. Appl. Environ. Microbiol. 2012, 78, 6762–6768. [Google Scholar] [CrossRef] [PubMed]
- Alexandre, G. Coupling metabolism and chemotaxis-dependent behaviours by energy taxis receptors. Microbiology 2010, 156, 2283–2293. [Google Scholar] [CrossRef] [PubMed]
- Rabinovitch-Deere, C.A.; Parales, R.E. Three types of taxis used in the response of Acidovorax sp. strain JS42 to 2-nitrotoluene. Appl. Environ. Microbiol. 2012, 78, 2306–2315. [Google Scholar] [CrossRef] [PubMed]
- Alvarez-Ortega, C.; Harwood, C.S. Identification of a malate chemoreceptor in Pseudomonas aeruginosa by screening for chemotaxis defects in an energy taxis-deficient mutant. Appl. Environ. Microbiol. 2007, 73, 7793–7795. [Google Scholar] [CrossRef] [PubMed]
- Simeonov, A. Recent developments in the use of differential scanning fluorometry in protein and small molecule discovery and characterization. Expert. Opin. Drug Discov. 2013, 8, 1071–1082. [Google Scholar] [CrossRef] [PubMed]
- Cimmperman, P.; Baranauskiene, L.; Jachimoviciute, S.; Jachno, J.; Torresan, J.; Michailoviene, V.; Matuliene, J.; Sereikaite, J.; Bumelis, V.; Matulis, D. A quantitative model of thermal stabilization and destabilization of proteins by ligands. Biophys J. 2008, 95, 3222–3231. [Google Scholar] [CrossRef] [PubMed]
- Fernandez, M.; Morel, B.; Corral-Lugo, A.; Krell, T. Identification of a chemoreceptor that specifically mediates chemotaxis toward metabolizable purine derivatives. Mol. Microbiol. 2016, 99, 34–42. [Google Scholar] [CrossRef] [PubMed]
- Pantoliano, M.W.; Petrella, E.C.; Kwasnoski, J.D.; Lobanov, V.S.; Myslik, J.; Graf, E.; Carver, T.; Asel, E.; Springer, B.A.; Lane, P.; et al. High-density miniaturized thermal shift assays as a general strategy for drug discovery. J. Biomol. Screen 2001, 6, 429–440. [Google Scholar] [CrossRef] [PubMed]
- McKellar, J.L.; Minnell, J.J.; Gerth, M.L. A high-throughput screen for ligand binding reveals the specificities of three amino acid chemoreceptors from Pseudomonas syringae pv. actinidiae. Mol. Microbiol. 2015, 96, 694–707. [Google Scholar] [CrossRef] [PubMed]
- Fernandez, M.; Ortega, A.; Rico-Jimenez, M.; Martin-Mora, D.; Daddaoua, A.; Matilla, M.A.; Krell, T. High-Throughput Screening to Identify Chemoreceptor Ligands. Methods Mol. Biol. 2018, 1729, 291–301. [Google Scholar] [PubMed]
- Ehrhardt, M.K.G.; Warring, S.L.; Gerth, M.L. Screening Chemoreceptor-Ligand Interactions by High-Throughput Thermal-Shift Assays. Methods Mol. Biol. 2018, 1729, 281–290. [Google Scholar] [PubMed]
- Matilla, M.A.; Daddaoua, A.; Chini, A.; Morel, B.; Krell, T. An auxin controls bacterial antibiotics production. Nucleic Acids Res. 2018. [Google Scholar] [CrossRef]
- Matilla, M.A.; Drew, A.; Udaondo, Z.; Krell, T.; Salmond, G.P. Genome Sequence of Serratia plymuthica A153, a Model Rhizobacterium for the Investigation of the Synthesis and Regulation of Haterumalides, Zeamine, and Andrimid. Genome Announc 2016, 4. [Google Scholar] [CrossRef] [PubMed]
- Matilla, M.A.; Nogellova, V.; Morel, B.; Krell, T.; Salmond, G.P. Biosynthesis of the acetyl-CoA carboxylase-inhibiting antibiotic, andrimid, in Serratia is regulated by Hfq and the LysR-type transcriptional regulator, AdmX. Environ. Microbiol. 2016. [Google Scholar] [CrossRef] [PubMed]
- Leyser, O. Auxin Signaling. Plant Physiol. 2018, 176, 465–479. [Google Scholar] [CrossRef] [PubMed]
- Corral-Lugo, A.; de la Torre, J.; Matilla, M.A.; Fernandez, M.; Morel, B.; Espinosa-Urgel, M.; Krell, T. Assessment of the contribution of chemoreceptor-based signaling to biofilm formation. Environ. Microbiol. 2016, 18, 3355–3372. [Google Scholar] [CrossRef] [PubMed]
- Garcia, V.; Reyes-Darias, J.A.; Martin-Mora, D.; Morel, B.; Matilla, M.A.; Krell, T. Identification of a Chemoreceptor for C2 and C3 Carboxylic Acids. Appl. Environ. Microbiol. 2015, 81, 5449–5457. [Google Scholar] [CrossRef] [PubMed]
- Krell, T. Microcalorimetry: A response to challenges in modern biotechnology. Microb. Biotechnol. 2008, 1, 126–136. [Google Scholar] [CrossRef] [PubMed]
- Brewster, J.L.; McKellar, J.L.; Finn, T.J.; Newman, J.; Peat, T.S.; Gerth, M.L. Structural basis for ligand recognition by a Cache chemosensory domain that mediates carboxylate sensing in Pseudomonas syringae. Sci. Rep. 2016, 6, 35198. [Google Scholar] [CrossRef] [PubMed]
- Compton, K.K.; Hildreth, S.B.; Helm, R.F.; Scharf, B.E. Sinorhizobium meliloti chemoreceptor McpV senses short chain carboxylates via direct binding. J. Bacteriol. 2018, 200. [Google Scholar] [CrossRef] [PubMed]
- Martin-Mora, D.; Ortega, A.; Perez-Maldonado, F.J.; Krell, T.; Matilla, M.A. The activity of the C4-dicarboxylic acid chemoreceptor of Pseudomonas aeruginosa is controlled by chemoattractants and antagonists. Sci. Rep. 2018, 8, 2102. [Google Scholar] [CrossRef] [PubMed]
- Webb, B.A.; Compton, K.K.; Del Campo, J.S.M.; Taylor, D.; Sobrado, P.; Scharf, B.E. Sinorhizobium meliloti Chemotaxis to Multiple Amino Acids Is Mediated by the Chemoreceptor McpU. Mol. Plant Microbe Interact. 2017, 30, 770–777. [Google Scholar] [CrossRef] [PubMed]
- Martin-Mora, D.; Ortega, A.; Reyes-Darias, J.A.; García, V.; López-Farfán, D.; Matilla, M.A.; Krell, T. Identification of a Chemoreceptor in Pseudomonas aeruginosa that specifically mediates Chemotaxis towards alpha-Ketoglutarate. Front Microbiol. 2016, 7, 1937. [Google Scholar] [CrossRef] [PubMed]
- Upadhyay, A.A.; Fleetwood, A.D.; Adebali, O.; Finn, R.D.; Zhulin, I.B. Cache Domains That are Homologous to, but Different from PAS Domains Comprise the Largest Superfamily of Extracellular Sensors in Prokaryotes. PLoS Comput. Biol. 2016, 12, e1004862. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Hendrickson, W.A. Structural characterization of the predominant family of histidine kinase sensor domains. J. Mol. Biol. 2010, 400, 335–353. [Google Scholar] [CrossRef] [PubMed]
- Cheung, J.; Hendrickson, W.A. Crystal structures of C4-dicarboxylate ligand complexes with sensor domains of histidine kinases DcuS and DctB. J. Biol. Chem. 2008, 283, 30256–30265. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.F.; Nan, B.; Nan, J.; Ma, Q.; Panjikar, S.; Liang, Y.H.; Wang, Y.; Su, X.D. C4-dicarboxylates sensing mechanism revealed by the crystal structures of DctB sensor domain. J. Mol. Biol. 2008, 383, 49–61. [Google Scholar] [CrossRef] [PubMed]
- Matilla, M.A.; Krell, T. Chemoreceptor-based signal sensing. Curr. Opin. Biotechnol. 2017, 45, 8–14. [Google Scholar] [CrossRef] [PubMed]
- Gavira, J.A.; Ortega, A.; Martin-Mora, D.; Conejero-Muriel, M.T.; Corral-Lugo, A.; Morel, B.; Matilla, M.A.; Krell, T. Structural Basis for Polyamine Binding at the dCACHE Domain of the McpU Chemoreceptor from Pseudomonas putida. J. Mol. Biol. 2018, 430, 1950–1963. [Google Scholar] [CrossRef] [PubMed]
- Nishiyama, S.; Takahashi, Y.; Yamamoto, K.; Suzuki, D.; Itoh, Y.; Sumita, K.; Uchida, Y.; Homma, M.; Imada, K.; Kawagishi, I. Identification of a Vibrio cholerae chemoreceptor that senses taurine and amino acids as attractants. Sci. Rep. 2016, 6, 20866. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.C.; Machuca, M.A.; Beckham, S.A.; Gunzburg, M.J.; Roujeinikova, A. Structural basis for amino-acid recognition and transmembrane signalling by tandem Per-Arnt-Sim (tandem PAS) chemoreceptor sensory domains. Acta Crystallogr. D Biol. Crystallogr. 2015, 71, 2127–2136. [Google Scholar] [CrossRef] [PubMed]
- Wallace, A.C.; Laskowski, R.A.; Thornton, J.M. LIGPLOT: A program to generate schematic diagrams of protein-ligand interactions. Protein Eng. 1995, 8, 127–134. [Google Scholar] [CrossRef] [PubMed]
- Irwin, J.J.; Sterling, T.; Mysinger, M.M.; Bolstad, E.S.; Coleman, R.G. ZINC: A free tool to discover chemistry for biology. J. Chem. Inf. Model 2012, 52, 1757–1768. [Google Scholar] [CrossRef] [PubMed]
- Irwin, J.J.; Shoichet, B.K. ZINC—A free database of commercially available compounds for virtual screening. J. Chem. Inf. Model 2005, 45, 177–182. [Google Scholar] [CrossRef] [PubMed]
- Friesner, R.A.; Banks, J.L.; Murphy, R.B.; Halgren, T.A.; Klicic, J.J.; Mainz, D.T.; Repasky, M.P.; Knoll, E.H.; Shelley, M.; Perry, J.K.; et al. Glide: A new approach for rapid, accurate docking and scoring. 1. Method and assessment of docking accuracy. J. Med. Chem. 2004, 47, 1739–1749. [Google Scholar] [CrossRef] [PubMed]
- Friesner, R.A.; Murphy, R.B.; Repasky, M.P.; Frye, L.L.; Greenwood, J.R.; Halgren, T.A.; Sanschagrin, P.C.; Mainz, D.T. Extra precision glide: Docking and scoring incorporating a model of hydrophobic enclosure for protein-ligand complexes. J. Med. Chem. 2006, 49, 6177–6196. [Google Scholar] [CrossRef] [PubMed]
- Rutherford, S.T.; Bassler, B.L. Bacterial quorum sensing: Its role in virulence and possibilities for its control. Cold Spring Harb. Perspect. Med. 2012, 2. [Google Scholar] [CrossRef] [PubMed]
- Venturi, V.; Fuqua, C. Chemical signaling between plants and plant-pathogenic bacteria. Annu. Rev. Phytopathol. 2013, 51, 17–37. [Google Scholar] [CrossRef] [PubMed]
- Corral-Lugo, A.; Daddaoua, A.; Ortega, A.; Espinosa-Urgel, M.; Krell, T. Rosmarinic acid is a homoserine lactone mimic produced by plants that activates a bacterial quorum-sensing regulator. Sci. Signal. 2016, 9, ra1. [Google Scholar] [CrossRef] [PubMed]
- Annapoorani, A.; Umamageswaran, V.; Parameswari, R.; Pandian, S.K.; Ravi, A.V. Computational discovery of putative quorum sensing inhibitors against LasR and RhlR receptor proteins of Pseudomonas aeruginosa. J. Comput. Aided. Mol. Des. 2012, 26, 1067–1077. [Google Scholar] [CrossRef] [PubMed]
- Corral Lugo, A.; Daddaoua, A.; Ortega, A.; Morel, B.; Diez Pena, A.I.; Espinosa-Urgel, M.; Krell, T. Purification and characterization of Pseudomonas aeruginosa LasR expressed in acyl-homoserine lactone free Escherichia coli cultures. Protein Expr. Purif. 2017, 130, 107–114. [Google Scholar] [CrossRef] [PubMed]
- Fernandez, M.; Corral-Lugo, A.; Krell, T. The plant compound rosmarinic acid induces a broad quorum sensing response in Pseudomonas aeruginosa PAO1. Environ. Microbiol. 2018. [Google Scholar] [CrossRef] [PubMed]
- Walker, T.S.; Bais, H.P.; Deziel, E.; Schweizer, H.P.; Rahme, L.G.; Fall, R.; Vivanco, J.M. Pseudomonas aeruginosa-plant root interactions. Pathogenicity, biofilm formation, and root exudation. Plant Physiol. 2004, 134, 320–331. [Google Scholar] [CrossRef] [PubMed]
- Privett, B.R.; Pellegrini, M.; Kovacikova, G.; Taylor, R.K.; Skorupski, K.; Mierke, D.; Kull, F.J. Identification of a Small Molecule Activator for AphB, a LysR-Type Virulence Transcriptional Regulator in Vibrio cholerae. Biochemistry 2017, 56, 3840–3849. [Google Scholar] [CrossRef] [PubMed]
- Mandal, R.S.; Ta, A.; Sinha, R.; Theeya, N.; Ghosh, A.; Tasneem, M.; Bhunia, A.; Koley, H.; Das, S. Ribavirin suppresses bacterial virulence by targeting LysR-type transcriptional regulators. Sci. Rep. 2016, 6, 39454. [Google Scholar] [CrossRef] [PubMed]
- Tan, K.; Chhor, G.; Binkowski, T.A.; Jedrzejczak, R.P.; Makowska-Grzyska, M.; Joachimiak, A. Sensor domain of histidine kinase KinB of Pseudomonas: A helix-swapped dimer. J. Biol. Chem. 2014, 289, 12232–12244. [Google Scholar] [CrossRef] [PubMed]
- Dey, A.; Shree, S.; Pandey, S.K.; Tripathi, R.P.; Ramachandran, R. Crystal Structure of Mycobacterium tuberculosis H37Rv AldR (Rv2779c), a Regulator of the ald Gene: DNA binding and identification of small molecule inhibitors. J. Biol. Chem. 2016, 291, 11967–11980. [Google Scholar] [CrossRef] [PubMed]
- Pagliai, F.A.; Gardner, C.L.; Bojilova, L.; Sarnegrim, A.; Tamayo, C.; Potts, A.H.; Teplitski, M.; Folimonova, S.Y.; Gonzalez, C.F.; Lorca, G.L. The transcriptional activator LdtR from ‘Candidatus Liberibacter asiaticus’ mediates osmotic stress tolerance. PLoS Pathog. 2014, 10, e1004101. [Google Scholar] [CrossRef] [PubMed]
- Pagliai, F.A.; Gonzalez, C.F.; Lorca, G.L. Identification of a Ligand Binding Pocket in LdtR from Liberibacter asiaticus. Front Microbiol. 2015, 6, 1314. [Google Scholar] [CrossRef] [PubMed]
- Manson, M.D.; Blank, V.; Brade, G.; Higgins, C.F. Peptide chemotaxis in E. coli involves the Tap signal transducer and the dipeptide permease. Nature 1986, 321, 253–256. [Google Scholar] [CrossRef] [PubMed]
- Mowbray, S.L.; Koshland, D.E., Jr. Additive and independent responses in a single receptor: Aspartate and maltose stimuli on the tar protein. Cell 1987, 50, 171–180. [Google Scholar] [CrossRef]
- Kondoh, H.; Ball, C.B.; Adler, J. Identification of a methyl-accepting chemotaxis protein for the ribose and galactose chemoreceptors of Escherichia coli. Proc. Natl. Acad. Sci. USA 1979, 76, 260–264. [Google Scholar] [CrossRef] [PubMed]
- Hegde, M.; Englert, D.L.; Schrock, S.; Cohn, W.B.; Vogt, C.; Wood, T.K.; Manson, M.D.; Jayaraman, A. Chemotaxis to the quorum-sensing signal AI-2 requires the Tsr chemoreceptor and the periplasmic LsrB AI-2-binding protein. J. Bacteriol. 2011, 193, 768–773. [Google Scholar] [CrossRef] [PubMed]
- Anderson, J.K.; Huang, J.Y.; Wreden, C.; Sweeney, E.G.; Goers, J.; Remington, S.J.; Guillemin, K. Chemorepulsion from the Quorum Signal Autoinducer-2 Promotes Helicobacter pylori Biofilm Dispersal. MBio 2015, 6, e00379. [Google Scholar] [CrossRef] [PubMed]
- Glekas, G.D.; Mulhern, B.J.; Kroc, A.; Duelfer, K.A.; Lei, V.; Rao, C.V.; Ordal, G.W. The Bacillus subtilis chemoreceptor McpC senses multiple ligands using two discrete mechanisms. J. Biol. Chem. 2012, 287, 39412–39418. [Google Scholar] [CrossRef] [PubMed]
- Machuca, M.A.; Liu, Y.C.; Beckham, S.A.; Gunzburg, M.J.; Roujeinikova, A. The crystal structure of the tandem-PAS sensing domain of Campylobacter jejuni chemoreceptor Tlp1 suggests indirect mechanism of ligand recognition. J. Struct. Biol. 2016, 194, 205–213. [Google Scholar] [CrossRef] [PubMed]
- Ulrich, L.E.; Zhulin, I.B. Four-helix bundle: A ubiquitous sensory module in prokaryotic signal transduction. Bioinformatics 2005, 21, iii45–iii48. [Google Scholar] [CrossRef] [PubMed]
- Ortega, A.; Krell, T. The HBM domain: Introducing bimodularity to bacterial sensing. Protein Sci. 2014, 23, 332–336. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Wang, C.; Yang, G.; Sun, Z.; Guo, H.; Shao, K.; Gu, Y.; Jiang, W.; Zhang, P. Molecular mechanism of environmental d-xylose perception by a XylFII-LytS complex in bacteria. Proc. Natl. Acad. Sci. USA 2017, 114, 8235–8240. [Google Scholar] [CrossRef] [PubMed]
- Moore, J.O.; Hendrickson, W.A. An asymmetry-to-symmetry switch in signal transmission by the histidine kinase receptor for TMAO. Structure 2012, 20, 729–741. [Google Scholar] [CrossRef] [PubMed]
- Neiditch, M.B.; Federle, M.J.; Pompeani, A.J.; Kelly, R.C.; Swem, D.L.; Jeffrey, P.D.; Bassler, B.L.; Hughson, F.M. Ligand-induced asymmetry in histidine sensor kinase complex regulates quorum sensing. Cell 2006, 126, 1095–1108. [Google Scholar] [CrossRef] [PubMed]
- Rajagopala, S.V.; Sikorski, P.; Kumar, A.; Mosca, R.; Vlasblom, J.; Arnold, R.; Franca-Koh, J.; Pakala, S.B.; Phanse, S.; Ceol, A.; et al. The binary protein-protein interaction landscape of Escherichia coli. Nat. Biotechnol. 2014, 32, 285–290. [Google Scholar] [CrossRef] [PubMed]
- Van der Geer, P. Analysis of protein-protein interactions by coimmunoprecipitation. Methods Enzymol. 2014, 541, 35–47. [Google Scholar] [PubMed]
- Huttlin, E.L.; Ting, L.; Bruckner, R.J.; Gebreab, F.; Gygi, M.P.; Szpyt, J.; Tam, S.; Zarraga, G.; Colby, G.; Baltier, K.; et al. The BioPlex Network: A Systematic Exploration of the Human Interactome. Cell 2015, 162, 425–440. [Google Scholar] [CrossRef] [PubMed]
- Navare, A.T.; Chavez, J.D.; Zheng, C.; Weisbrod, C.R.; Eng, J.K.; Siehnel, R.; Singh, P.K.; Manoil, C.; Bruce, J.E. Probing the protein interaction network of Pseudomonas aeruginosa cells by chemical cross-linking mass spectrometry. Structure 2015, 23, 762–773. [Google Scholar] [CrossRef] [PubMed]
- Smith, G.P. Filamentous fusion phage: Novel expression vectors that display cloned antigens on the virion surface. Science 1985, 228, 1315–1317. [Google Scholar] [CrossRef] [PubMed]
- Boder, E.T.; Wittrup, K.D. Yeast surface display for screening combinatorial polypeptide libraries. Nat. Biotechnol. 1997, 15, 553–557. [Google Scholar] [CrossRef] [PubMed]
- Younger, D.; Berger, S.; Baker, D.; Klavins, E. High-throughput characterization of protein-protein interactions by reprogramming yeast mating. Proc. Natl. Acad. Sci. USA 2017, 114, 12166–12171. [Google Scholar] [CrossRef] [PubMed]
- Vastermark, A.; Saier, M.H., Jr. The involvement of transport proteins in transcriptional and metabolic regulation. Curr. Opin. Microbiol. 2014, 18, 8–15. [Google Scholar] [CrossRef] [PubMed]
- Neiditch, M.B.; Federle, M.J.; Miller, S.T.; Bassler, B.L.; Hughson, F.M. Regulation of LuxPQ receptor activity by the quorum-sensing signal autoinducer-2. Mol. Cell 2005, 18, 507–518. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Gardina, P.J.; Kuebler, A.S.; Kang, H.S.; Christopher, J.A.; Manson, M.D. Model of maltose-binding protein/chemoreceptor complex supports intrasubunit signaling mechanism. Proc. Natl. Acad. Sci. USA 1999, 96, 939–944. [Google Scholar] [CrossRef] [PubMed]
- Nishijyo, T.; Park, S.M.; Lu, C.D.; Itoh, Y.; Abdelal, A.T. Molecular characterization and regulation of an operon encoding a system for transport of arginine and ornithine and the ArgR regulatory protein in Pseudomonas aeruginosa. J. Bacteriol. 1998, 180, 5559–5566. [Google Scholar] [PubMed]
- Wagner, M.; Shen, L.; Albersmeier, A.; van der Kolk, N.; Kim, S.; Cha, J.; Brasen, C.; Kalinowski, J.; Siebers, B.; Albers, S.V. Sulfolobus acidocaldarius uptakes pentoses via a cut2-type ABC transporter and metabolizes them through the aldolase-independent Weimberg pathway. Appl. Environ. Microbiol. 2017. [Google Scholar] [CrossRef] [PubMed]
- Botero, L.M.; Al-Niemi, T.S.; McDermott, T.R. Characterization of two inducible phosphate transport systems in Rhizobium tropici. Appl. Environ. Microbiol. 2000, 66, 15–22. [Google Scholar] [CrossRef] [PubMed]

| Chemoreceptor Name | Bacterial Species | LBD Type | Tm without Ligand (°C) | Ligand | Tm Shift (°C) | KD (µM) | Reference |
|---|---|---|---|---|---|---|---|
| PscD | P. syringae | sCACHE | 56.1 | acetic acid | 3.3 | 31 | [63] |
| propionic acid | 2.7 | 101 | |||||
| pyruvic acid | 1.0 | 356 | |||||
| capric (decanoic) acid | 2.2 | No binding | |||||
| glycolate | 1.1 | 23 | |||||
| McpV | S. meliloti | sCACHE | 57.0 | acetate | 12.3 | 9.1 | [64] |
| propionate | 12.3 | 3.4 | |||||
| pyruvate | 11.8 | 33 | |||||
| glycolate | 9.3 | 27 | |||||
| L-lactate | 8.2 | n.d. | |||||
| acetoacetate | 6.0 | 280 | |||||
| glyoxylate | 5.8 | n.d. | |||||
| methyl-pyruvate | 5.8 | n.d. | |||||
| α-hydroxy-butyrate | 4.0 | n.d. | |||||
| α-keto-butyrate | 3.8 | n.d. | |||||
| PA2652 | P. aeruginosa | sCACHE | 45.5 | L-malic acid | 5.2 | 23 | [65] |
| citramalic acid | 2.1 | 61 | |||||
| methylsuccinic acid | n.d. | 224 | |||||
| bromosuccinic acid | 3.6 | 1240 | |||||
| citraconic acid | 2.5 | 210 | |||||
| McpH | P. putida | dCACHE | 47.2 | adenine | 3.2 | 2.4 | [51] |
| guanine | 4.2 | 4.3 | |||||
| xanthine | 3.7 | 2.7 | |||||
| hypoxanthine | n.d. | 3.6 | |||||
| purine | n.d. | 2.4 | |||||
| McpU | P. putida | dCACHE | 46.0 | putrescine | 11 | 2 | [33,60] |
| spermidine | 2 | 4.5 | |||||
| cadaverine | 10.5 | 22 | |||||
| histamine | 3.7 | 26 | |||||
| agmatine | 14 | 0.48 | |||||
| ethylenediamine | 2.3 | 39 | |||||
| PscA | P. syringae | dCACHE | 40.1 | L-Asp | 11.0 | 6.1 | [53] |
| L-Glu | 8.2 | 27 | |||||
| D-Asp | 10.0 | 2.3/19 a | |||||
| McpU | S. meliloti | dCACHE | 36.0 | arginine | 13.1 | 350 | [66] |
| phenylalanine | 12.6 | 53 | |||||
| proline | 13.2 | 42 and 104 | [38,66] | ||||
| tryptophan | 15.0 | 34 | |||||
| McpX | S. meliloti | dCACHE | 45.0 | trigoneline | 4.8 | 88 | [39] |
| choline | 9.8 | 0.14 | |||||
| glycine betaine | 9.2 | 1.3 | |||||
| betonicine | 1.2 | 2300 | |||||
| stachydrine | 6.8 | 3.8 | |||||
| proline | 4.7 | 45 | |||||
| McpK | P. aeruginosa | HBM | 38.7 | α-ketoglutarate | 5.2 | 301/81 b | [67] |
| uracil | 4.2 | No binding by ITC | |||||
| γ-aminobutyrate | 4.1 | No binding by ITC | |||||
| 5-carbamyl phosphate | 3.5 | No binding by ITC | |||||
| phenylethylamine | 3.5 | No binding by ITC | |||||
| d-glucosaminic acid | 3.5 | No binding by ITC | |||||
| McpN | P. aeruginosa | PilJ | 49 | nitrate | 3.5 | 47 | (David Martín Mora, personal communication) |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Martín-Mora, D.; Fernández, M.; Velando, F.; Ortega, Á.; Gavira, J.A.; Matilla, M.A.; Krell, T. Functional Annotation of Bacterial Signal Transduction Systems: Progress and Challenges. Int. J. Mol. Sci. 2018, 19, 3755. https://doi.org/10.3390/ijms19123755
Martín-Mora D, Fernández M, Velando F, Ortega Á, Gavira JA, Matilla MA, Krell T. Functional Annotation of Bacterial Signal Transduction Systems: Progress and Challenges. International Journal of Molecular Sciences. 2018; 19(12):3755. https://doi.org/10.3390/ijms19123755
Chicago/Turabian StyleMartín-Mora, David, Matilde Fernández, Félix Velando, Álvaro Ortega, José A. Gavira, Miguel A. Matilla, and Tino Krell. 2018. "Functional Annotation of Bacterial Signal Transduction Systems: Progress and Challenges" International Journal of Molecular Sciences 19, no. 12: 3755. https://doi.org/10.3390/ijms19123755
APA StyleMartín-Mora, D., Fernández, M., Velando, F., Ortega, Á., Gavira, J. A., Matilla, M. A., & Krell, T. (2018). Functional Annotation of Bacterial Signal Transduction Systems: Progress and Challenges. International Journal of Molecular Sciences, 19(12), 3755. https://doi.org/10.3390/ijms19123755







