Activation of CREBZF Increases Cell Apoptosis in Mouse Ovarian Granulosa Cells by Regulating the ERK1/2 and mTOR Signaling Pathways
Abstract
1. Introduction
2. Results
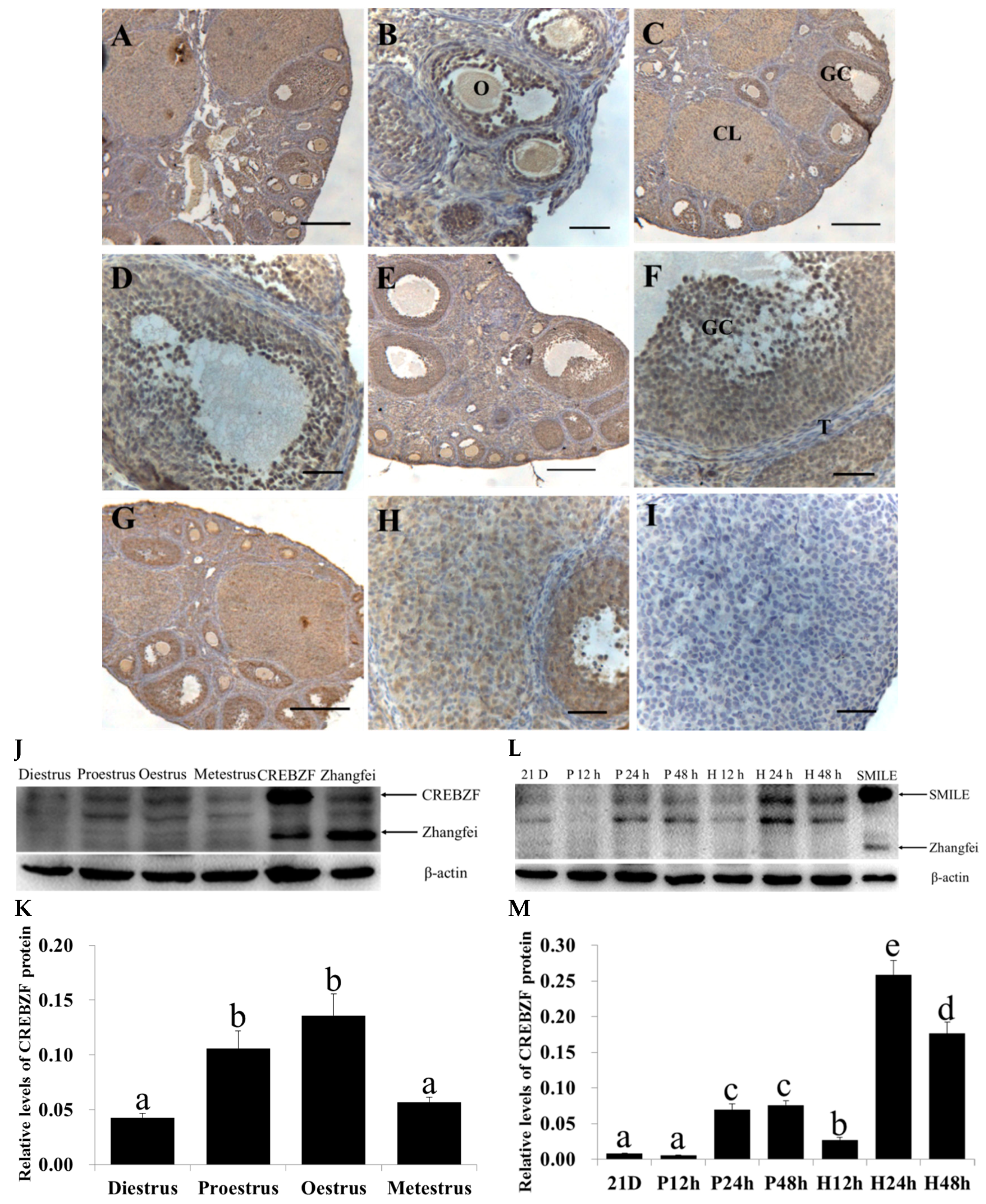
2.1. Expression of CREBZF in the Ovaries throughout the Estrous Cycle
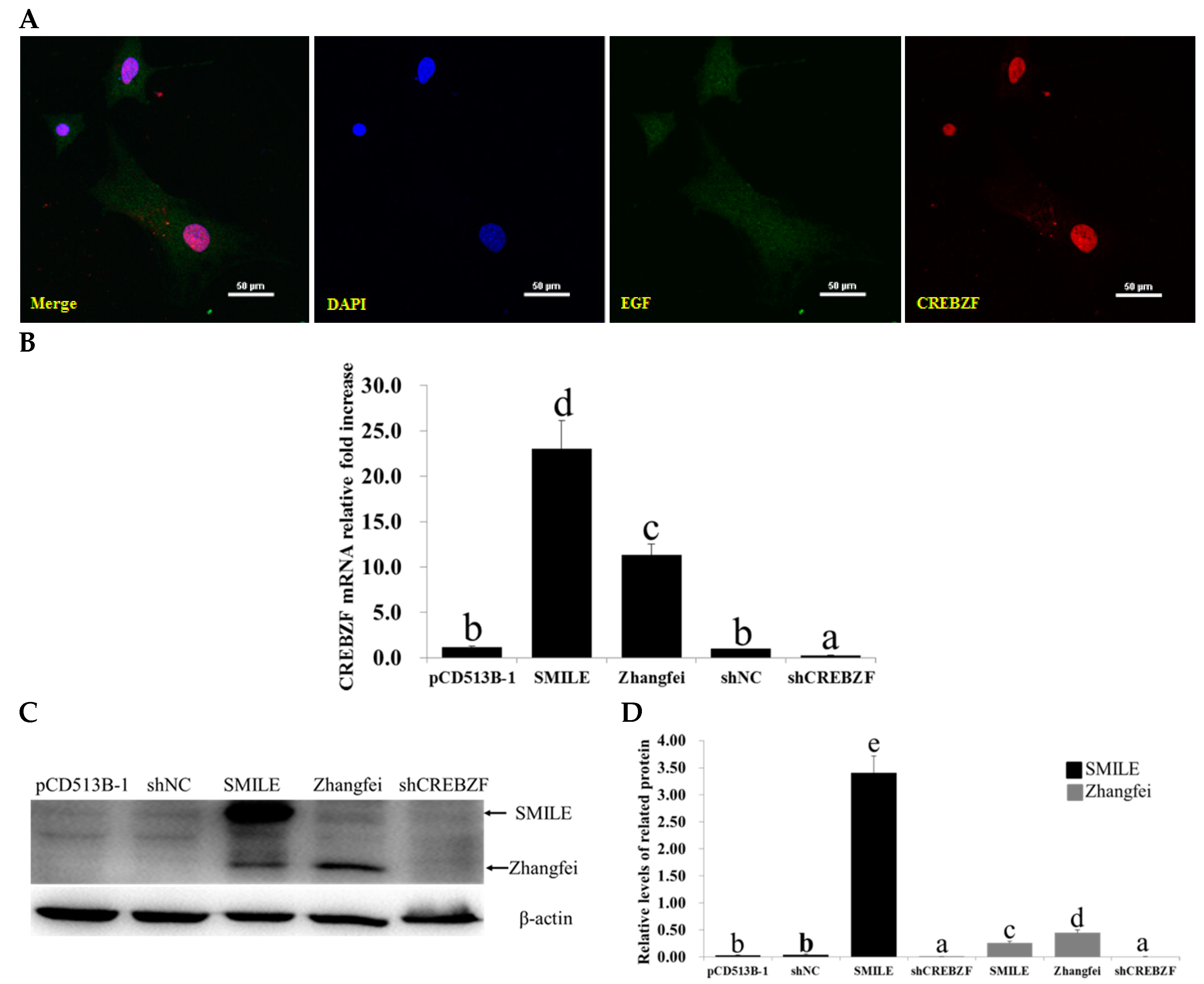
2.2. Effects of Lentivirus-Induced CREBZF Overexpression and Knockdown in Primary Granulosa Cells
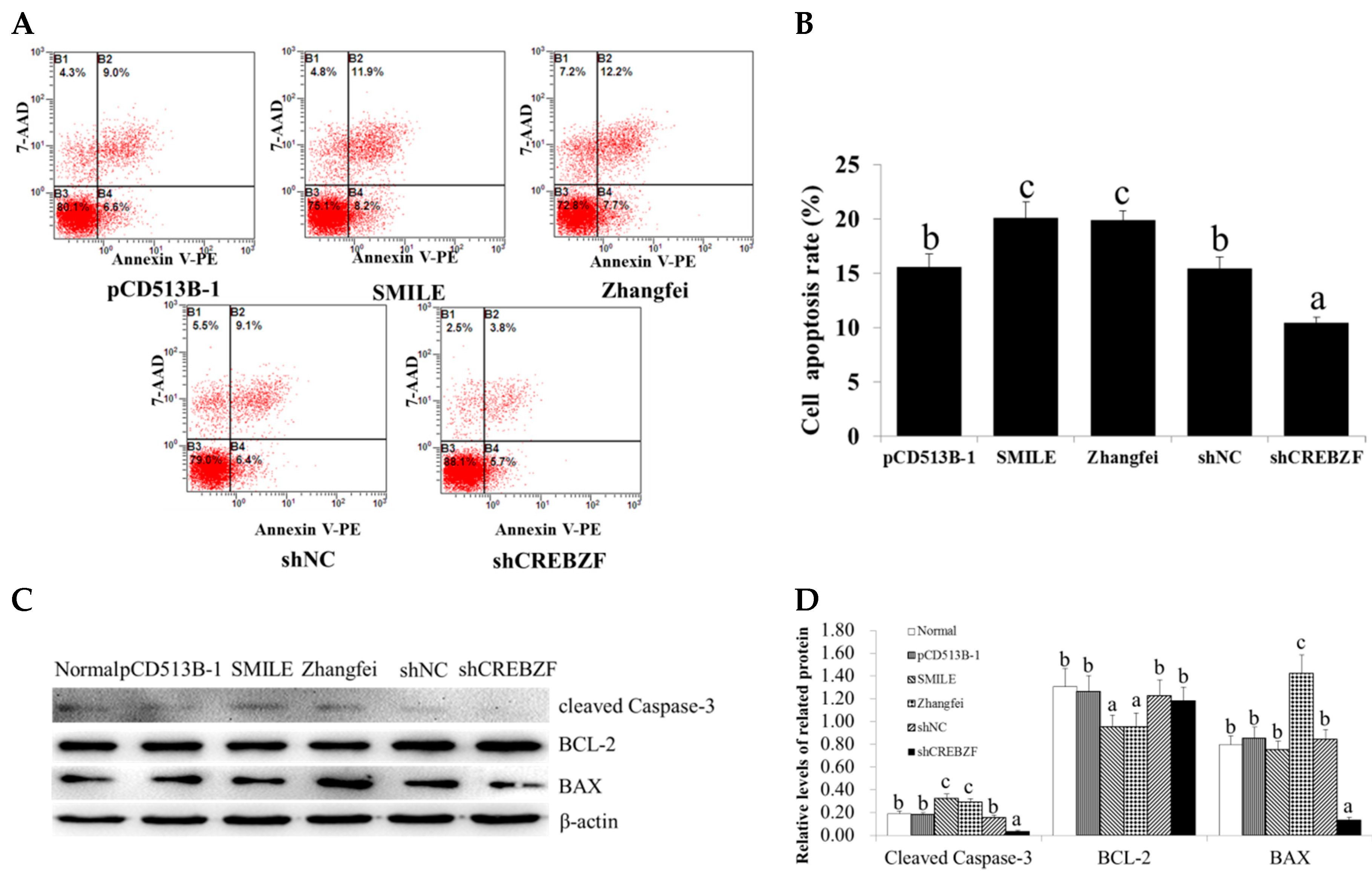
2.3. Effect of CREBZF on Ovarian Granulosa Cells Apoptosis
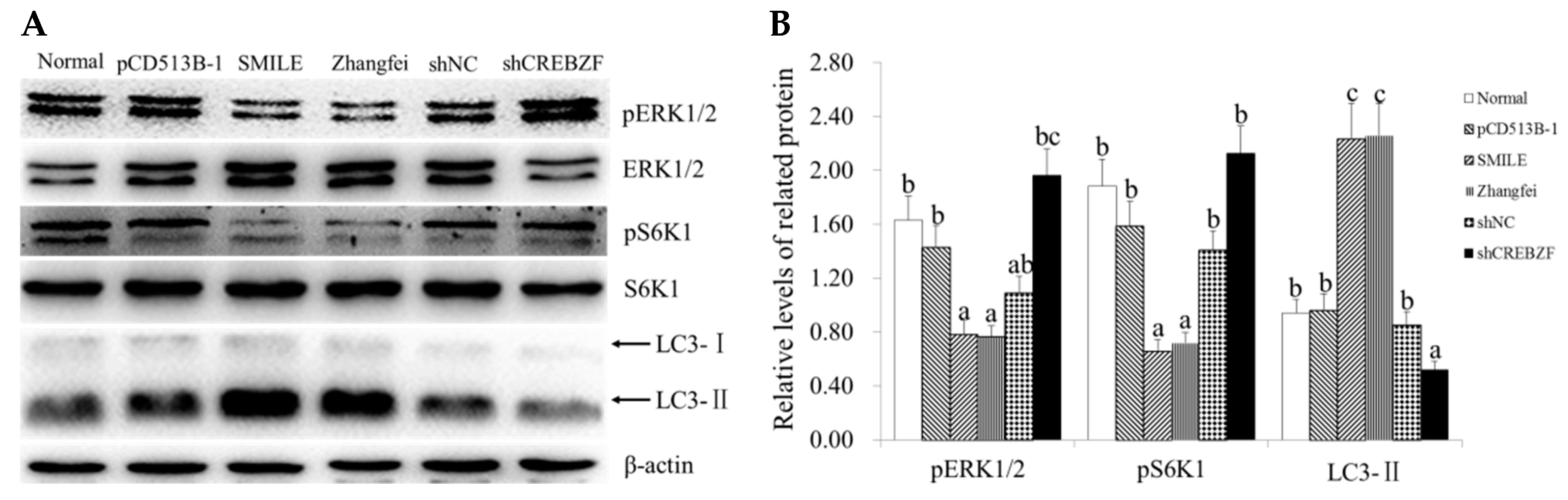
2.4. Effect of CREBZF on ERK1/2 and mTOR Signaling Pathways
3. Discussion
4. Materials and Methods
4.1. Animals and Ovaries Collection
4.2. Primary Mouse Ovarian Granulosa Cells Culture
4.3. CREBZF Overexpression or shRNAs Lentivirus Transduction
4.4. Immunohistochemistry
4.5. Immunofluorescence Staining
4.6. RNA Isolation and Quantitative Real-Time PCR Analysis
4.7. Western Blot Analysis
4.8. Apoptosis Analysis
4.9. Statistical Analyses
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| ERK1/2 | intracellular-regulated kinases 1/2 |
| S6K1 | p70 S6 kinase |
| CL | corpus luteum |
| bZIP | basic leucine zipper |
| CREB | cAMP response element-binding protein |
| ATF | activating transcription factor |
| HSV-1 | herpes simplex virus-1 |
| HCF-1 | host cell factor 1 |
| ER | endoplasmic reticulum |
| UPR | unfolded protein response |
| MAPK | mitogen-activated protein kinases |
| ATG | autophagy response genes |
| PMSG | pregnant mare serum gonadotropin |
| hCG | human chorionic gonadotropin |
| FBS | fetal bovine serum |
| PVDF | polyvinylidene fluoride |
| DAB | diaminobenzidine |
| DAPI | 4′,6-diamidino-2-phenylindole |
References
- Matsuda, F.; Inoue, N.; Manabe, N.; Ohkura, S. Follicular Growth and Atresia in Mammalian Ovaries: Regulation by Survival and Death of Granulosa Cells. J. Reprod. Dev. 2012, 58, 44–50. [Google Scholar] [CrossRef] [PubMed]
- Xuan, J.; Liu, Y.X. Follicular growth, differentiation and atresia. Chin. Sci. Bull. 2003, 48, 1786–1790. [Google Scholar]
- Nilsson, E.; Skinner, M.K. Cellular interactions that control primordial follicle development and folliculogenesis. J. Soc. Gynecol. Investig. 2001, 8, S17–S20. [Google Scholar] [CrossRef] [PubMed]
- Li, F.X.; Liu, J.; Jo, M.; Curry, T.E. A Role for Nuclear Factor Interleukin-3 (NFIL3), a Critical Transcriptional Repressor, in Down-Regulation of Periovulatory Gene Expression. Mol. Endocrinol. 2011, 25, 445–459. [Google Scholar] [CrossRef] [PubMed]
- Machtinger, R.; Laurent, L.C.; Baccarelli, A.A. Extracellular vesicles: Roles in gamete maturation, fertilization and embryo implantation. Hum. Reprod. Update 2016, 22, 182–193. [Google Scholar] [CrossRef] [PubMed]
- Hsueh, A.J.W.; Kawamura, K.; Cheng, Y.; Fauser, B.C.J.M. Intraovarian Control of Early Folliculogenesis. Endocr Rev. 2015, 36, 1–24. [Google Scholar] [CrossRef] [PubMed]
- Scaramuzzi, R.J.; Baird, D.T.; Campbell, B.K.; Driancourt, M.A.; Dupont, J.; Fortune, J.E.; Gilchrist, R.B.; Martin, G.B.; McNatty, K.P.; McNeilly, A.S.; et al. Regulation of folliculogenesis and the determination of ovulation rate in ruminants. Reprod. Fert. Dev. 2011, 23, 444–467. [Google Scholar] [CrossRef] [PubMed]
- Stocco, C.; Telleria, C.; Gibori, G. The molecular control of corpus luteum formation, function, and regression. Endocr. Rev. 2007, 28, 117–149. [Google Scholar] [CrossRef] [PubMed]
- Sterneck, E.; Tessarollo, L.; Johnson, P.F. An essential role for C/EBPbeta in female reproduction. Genes Dev. 1997, 11, 2153–2162. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.K.; Hao, X.Q.; Yang, J.; Li, J.; Zhang, M.J. CREB Activity Is Required for Luteinizing Hormone-Induced the Expression of EGF-Like Factors. Mol. Reprod. Dev. 2016, 83, 1116–1127. [Google Scholar] [CrossRef] [PubMed]
- Prasertlux, S.; Yocawibun, P.; Janpooma, S.; Klinbunga, S.; Menasveta, P.; Khamnamtong, B. Differential expression of X-box binding protein 1 during ovarian development and association between its SNP and growth-related parameters of the giant tiger shrimp Penaeus monodon. Aquaculture 2015, 448, 531–538. [Google Scholar] [CrossRef]
- Lu, R.; Misra, V. Zhangfei: A second cellular protein interacts with herpes simplex virus accessory factor HCF in a manner similar to Luman and VP16. Nucleic Acids Res. 2000, 28, 2446–2454. [Google Scholar] [CrossRef] [PubMed]
- Xie, Y.B.; Lee, O.H.; Nedumaran, B.; Seong, H.A.; Lee, K.M.; Ha, H.; Lee, I.K.; Yun, Y.; Choi, H.S. SMILE, a new orphan nuclear receptor SHP-interacting protein, regulates SHP-repressed estrogen receptor transactivation. Biochem. J. 2008, 416, 463–473. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.; Rapin, N.; Ying, Z.X.; Shklanka, E.; Bodnarchuk, T.W.; Verge, V.M.K.; Misra, V. Zhangfei/CREB-ZF—A Potential Regulator of the Unfolded Protein Response. PLoS ONE 2013, 8, e77256. [Google Scholar] [CrossRef] [PubMed]
- Misra, V.; Rapin, N.; Akhova, O.; Bainbridge, M.; Korchinski, P. Zhangfei is a potent and specific inhibitor of the host cell factor-binding transcription factor luman. J. Biol. Chem. 2005, 280, 15257–15266. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.Y.; Song, C.H.; Xie, Y.B.; Jung, C.; Choi, H.S.; Lee, K. SMILE upregulated by metformin inhibits the function of androgen receptor in prostate cancer cells. Cancer Lett. 2014, 354, 390–397. [Google Scholar] [CrossRef] [PubMed]
- Xie, Y.B.; Nedumaran, B.; Choi, H.S. Molecular characterization of SMILE as a novel corepressor of nuclear receptors. Nucleic Acids Res. 2009, 37, 4100–4115. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.M.; Seo, Y.J.; Kim, M.K.; Seo, H.A.; Jeong, J.Y.; Choi, H.S.; Lee, I.K.; Park, K.G. Mediation of glucolipotoxicity in INS-1 rat insulinoma cells by small heterodimer partner interacting leucine zipper protein (SMILE). Biochem. Biophys. Res. Commun. 2012, 419, 768–773. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.M.; Seo, W.Y.; Han, H.S.; Oh, K.J.; Lee, Y.S.; Kim, D.K.; Choi, S.; Choi, B.H.; Harris, R.A.; Lee, C.H.; et al. Insulin-Inducible SMILE Inhibits Hepatic Gluconeogenesis. Diabetes 2016, 65, 62–73. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.H.; Lee, G.T.; Kwon, S.J.; Jeong, J.; Ha, Y.S.; Kim, W.J.; Kim, I.Y. CREBZF, a novel Smad8-binding protein. Mol. Cell. Biochem. 2012, 368, 147–153. [Google Scholar] [CrossRef] [PubMed]
- Hogan, M.R.; Cockram, G.R.P.; Lu, R. Cooperative interaction of Zhangfei and ATF4 in transactivation of the cyclic AMP response element. FEBS Lett. 2006, 580, 58–62. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.; Misra, V. Zhangfei/CREBZF arrests the growth of osteosarcoma cells by displacing Mdm2 and stabilizing p53. Mol. Cancer Ther. 2013, 12, B59. [Google Scholar] [CrossRef]
- Zhang, R.; Misra, V. Effects of cyclic AMP response element binding protein-Zhangfei (CREBZF) on the unfolded protein response and cell growth are exerted through the tumor suppressor p53. Cell Cycle 2014, 13, 279–292. [Google Scholar] [CrossRef] [PubMed]
- Bodnarchuk, T.W.; Napper, S.; Rapin, N.; Misra, V. Mechanism for the induction of cell death in ONS-76 medulloblastoma cells by Zhangfei/CREB-ZF. J. Neuro-Oncol. 2012, 109, 485–501. [Google Scholar] [CrossRef] [PubMed]
- Lin, P.F.; Chen, F.L.; Wang, N.; Wang, X.G.; Li, X.; Zhou, J.H.; Jin, Y.P.; Wang, A.H. CREBZF expression and hormonal regulation in the mouse uterus. Reprod. Biol. Endocrinol. 2013, 11, 110. [Google Scholar] [CrossRef] [PubMed]
- Jang, H. Regulation of Cyclic AMP-Response Element Binding Protein Zhangfei (CREBZF) Expression by Estrogen in Mouse Uterus. Dev. Reprod. 2018, 22, 95–104. [Google Scholar] [CrossRef] [PubMed]
- Chen, F.; Lin, P.F.; Li, X.; Sun, J.; Zhang, Z.; Du, E.; Wang, A.; Jin, Y.P. Construction and expression of lentiviral vectors encoding recombinant mouse CREBZF in NIH 3T3 cells. Plasmid 2014, 76, 24–31. [Google Scholar] [CrossRef] [PubMed]
- Valderrama, X.; Rapin, N.; Verge, V.M.; Misra, V. Zhangfei induces the expression of the nerve growth factor receptor, trkA, in medulloblastoma cells and causes their differentiation or apoptosis. J. Neurooncol. 2009, 91, 7–17. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Zhang, Y.; Audas, T.E.; Martyn, A.C.; Lu, R. A CREBZF/Zhangfei isoform activates CHOP and promotes apoptosis during prolonged endoplasmic reticulum stress. Cancer Res. 2010, 70, 333. [Google Scholar] [CrossRef]
- Zhang, R.; Thamm, D.H.; Misra, V. The effect of Zhangfei/CREBZF on cell growth, differentiation, apoptosis, migration, and the unfolded protein response in several canine osteosarcoma cell lines. BMC Vet. Res. 2015, 11, 22. [Google Scholar] [CrossRef] [PubMed]
- Sirotkin, A.V.; Grossmann, R. Role of tyrosine kinase- and MAP kinase-dependent intracellular mechanisms in control of ovarian functions in the domestic fowl (Gallus domesticus) and in mediating effects of IGF-II. J. Reprod. Dev. 2003, 49, 99–106. [Google Scholar] [CrossRef] [PubMed]
- Woods, D.C.; Johnson, A.L. Modulation of mitogen activated protein kinase (MAPK)/Erk signaling up-regulates transforming growth factor beta (TGF beta) and initiates differentiation in avian granulosa cells. Biol. Reprod. 2005, 23–25, 246–247. [Google Scholar]
- Ye, D.F.; Li, M.F.; Zhang, Y.H.; Wang, X.H.; Liu, H.; Wu, W.T.; Ma, W.Y.; Quan, K.W.; Ng, E.H.Y.; Wu, X.K.; et al. Cryptotanshinone Regulates Androgen Synthesis through the ERK/c-Fos/CYP17 Pathway in Porcine Granulosa Cells. Evid. Based Complement. Altern. Med. 2017, 2017, 5985703. [Google Scholar] [CrossRef] [PubMed]
- Donaubauer, E.M.; Hunzicker-Dunn, M.E. Extracellular Signal-regulated Kinase (ERK)-dependent Phosphorylation of Y-Box-binding Protein 1 (YB-1) Enhances Gene Expression in Granulosa Cells in Response to Follicle-stimulating Hormone (FSH). J. Biol. Chem. 2016, 291, 12145–12160. [Google Scholar] [CrossRef] [PubMed]
- Kayampilly, P.P.; Menon, K.M.J. AMPK Activation by Dihydrotestosterone Reduces FSH-Stimulated Cell Proliferation in Rat Granulosa Cells by Inhibiting ERK Signaling Pathway. Endocrinology 2012, 153, 2831–2838. [Google Scholar] [CrossRef] [PubMed]
- Kayampilly, P.P.; Menon, K.M.J. Evidence That Activation of AMPK by DHT Inhibits Granulosa Cell Proliferation by Reducing ERK Phosphorylation. Biol. Reprod. 2011, 85, 644. [Google Scholar] [CrossRef]
- Ryan, K.E.; Glister, C.; Martin, F.; Knight, P.G.; Evans, A.C.O. Effect of FSH and IGF on Akt and Erk pathways, estradiol, and inhibin-A secretion, and survival/proliferation of bovine granulosa cells in vitro. Biol. Reprod. 2003, 68, 286. [Google Scholar]
- Makarevich, A.V.; Sirotkin, A.V.; Chrenek, P.; Bulla, J. Effect of epidermal growth factor (EGF) on steroid and cyclic nucleotide secretion, proliferation and ERK-related MAP-kinase in cultured rabbit granulosa cells. Exp. Clin. Endocr. Diabet. 2002, 110, 124–129. [Google Scholar] [CrossRef] [PubMed]
- Peter, A.T.; Dhanasekaran, N. Apoptosis of granulosa cells: A review on the role of MAPK-signalling modules. Reprod. Domest. Anim. 2003, 38, 209–213. [Google Scholar] [CrossRef] [PubMed]
- Han, Y.; Luo, H.N.; Wang, H.; Cai, J.; Zhang, Y.S. SIRT1 induces resistance to apoptosis in human granulosa cells by activating the ERK pathway and inhibiting NF-kappa B signaling with anti-inflammatory functions. Apoptosis 2017, 22, 1260–1272. [Google Scholar] [CrossRef] [PubMed]
- Gebauer, G.; Peter, A.T.; Onesime, D.; Dhanasekaran, N. Apoptosis of ovarian granulosa cells: Correlation with the reduced activity of ERK-signaling module. J. Cell. Biochem. 1999, 75, 547–554. [Google Scholar] [CrossRef]
- Rak-Mardyla, A.; Gregoraszczuk, E.L. Erk 1/2 and Pi-3 Kinase Pathways as a Potential Mechanism of Ghrelin Action on Cell Proliferation and Apoptosis in the Porcine Ovarian Follicular Cells. J. Physiol. Pharmacol. 2010, 61, 451–458. [Google Scholar] [PubMed]
- Rak, A.; Drwal, E.; Wrobel, A.; Gregoraszczuk, E.L. Resistin is a survival factor for porcine ovarian follicular cells. Reproduction 2015, 150, 343–355. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.M.; Li, L.; Xu, J.J.; Wang, N.; Liu, W.J.; Lin, X.H.; Fu, Y.C.; Luo, L.L. Rapamycin preserves the follicle pool reserve and prolongs the ovarian lifespan of female rats via modulating mTOR activation and sirtuin expression. Gene 2013, 523, 82–87. [Google Scholar] [CrossRef] [PubMed]
- Sirotkin, A.V.; Alexa, R.; Dekanova, P.; Kadasi, A.; Stochmalova, A.; Grossmann, R.; Alwasel, S.H.; Harrath, A.H. The mTOR System Can Affect Basic Ovarian Cell Functions and Mediate the Effect of Ovarian Hormonal Regulators. Int. J. Pharmacol. 2015, 11, 570–578. [Google Scholar] [CrossRef]
- Song, W.J.; Shi, X.B.; Zhang, J.; Chen, L.; Fu, S.X.; Ding, Y.L. Akt-mTOR Signaling Mediates Abnormalities in the Proliferation and Apoptosis of Ovarian Granulosa Cells in Patients with Polycystic Ovary Syndrome. Gynecol. Obstet. Investig. 2018, 83, 124–132. [Google Scholar] [CrossRef] [PubMed]
- Sirotkin, A.V. The Role and Application of Sirtuins and mTOR Signaling in the Control of Ovarian Functions. Cells 2016, 5, 42. [Google Scholar] [CrossRef] [PubMed]
- Makker, A.; Goel, M.M.; Mahdi, A.A. PI3K/PTEN/Akt and TSC/mTOR signaling pathways, ovarian dysfunction, and infertility: An update. J. Mol. Endocrinol. 2014, 53, R103–R118. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.; Yaba, A.; Kasiman, C.; Thomson, T.; Johnson, J. mTOR Controls Ovarian Follicle Growth by Regulating Granulosa Cell Proliferation. PLoS ONE 2011, 6, e21415. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.C.; Guan, K.L. mTOR: A pharmacologic target for autophagy regulation. J. Clin. Investig. 2015, 125, 25–32. [Google Scholar] [CrossRef] [PubMed]
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, F.; Wen, X.; Lin, P.; Chen, H.; Wang, A.; Jin, Y. Activation of CREBZF Increases Cell Apoptosis in Mouse Ovarian Granulosa Cells by Regulating the ERK1/2 and mTOR Signaling Pathways. Int. J. Mol. Sci. 2018, 19, 3517. https://doi.org/10.3390/ijms19113517
Chen F, Wen X, Lin P, Chen H, Wang A, Jin Y. Activation of CREBZF Increases Cell Apoptosis in Mouse Ovarian Granulosa Cells by Regulating the ERK1/2 and mTOR Signaling Pathways. International Journal of Molecular Sciences. 2018; 19(11):3517. https://doi.org/10.3390/ijms19113517
Chicago/Turabian StyleChen, Fenglei, Xin Wen, Pengfei Lin, Huatao Chen, Aihua Wang, and Yaping Jin. 2018. "Activation of CREBZF Increases Cell Apoptosis in Mouse Ovarian Granulosa Cells by Regulating the ERK1/2 and mTOR Signaling Pathways" International Journal of Molecular Sciences 19, no. 11: 3517. https://doi.org/10.3390/ijms19113517
APA StyleChen, F., Wen, X., Lin, P., Chen, H., Wang, A., & Jin, Y. (2018). Activation of CREBZF Increases Cell Apoptosis in Mouse Ovarian Granulosa Cells by Regulating the ERK1/2 and mTOR Signaling Pathways. International Journal of Molecular Sciences, 19(11), 3517. https://doi.org/10.3390/ijms19113517




