A Comparison of Proteins Expressed between Human and Mouse Adipose-Derived Mesenchymal Stem Cells by a Proteome Analysis through Liquid Chromatography with Tandem Mass Spectrometry
Abstract
1. Introduction
2. Results
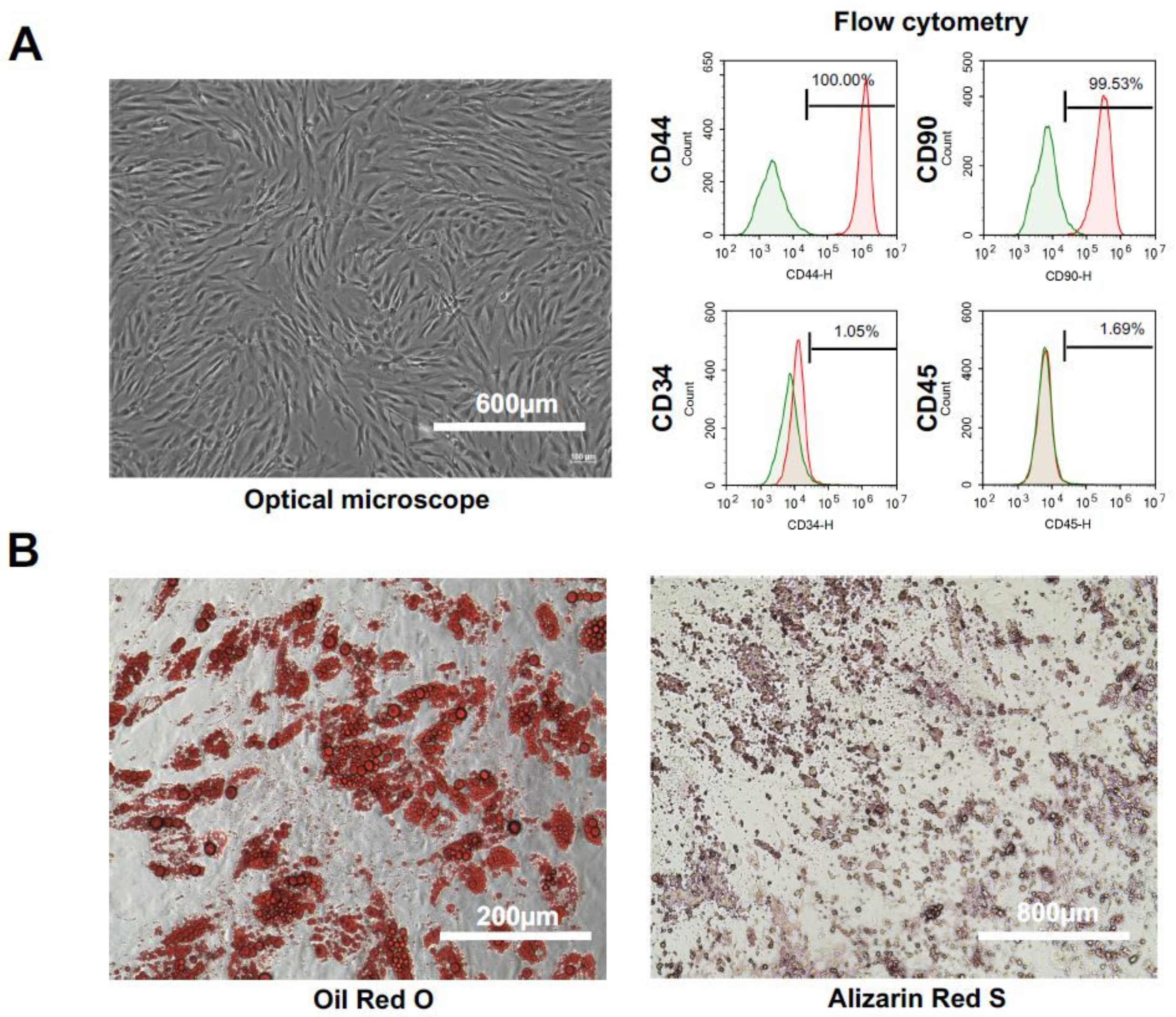
2.1. Characteristics and Cell Qualities of hADSCs
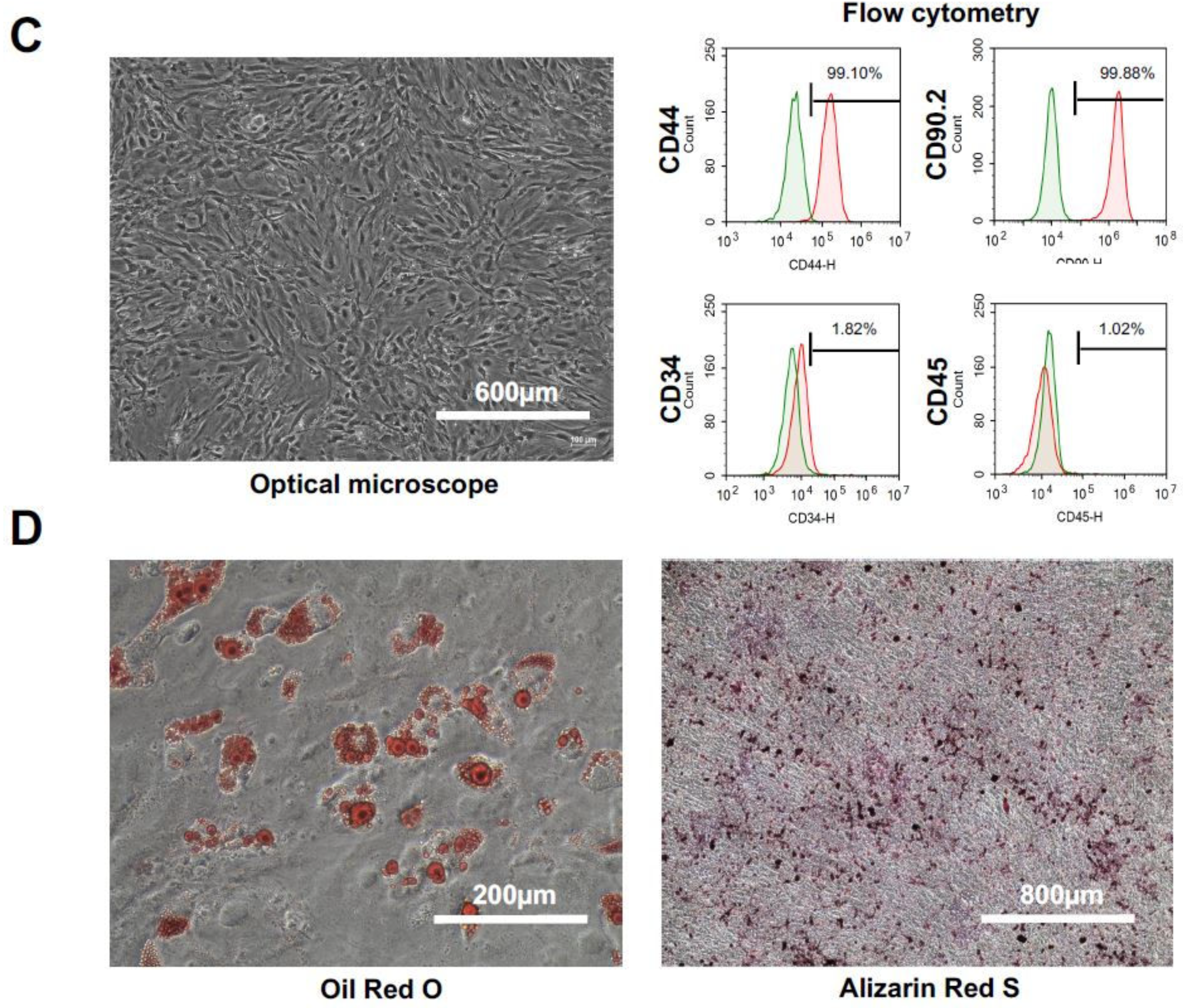
2.2. Characteristics and Cell Qualities of mADSCs
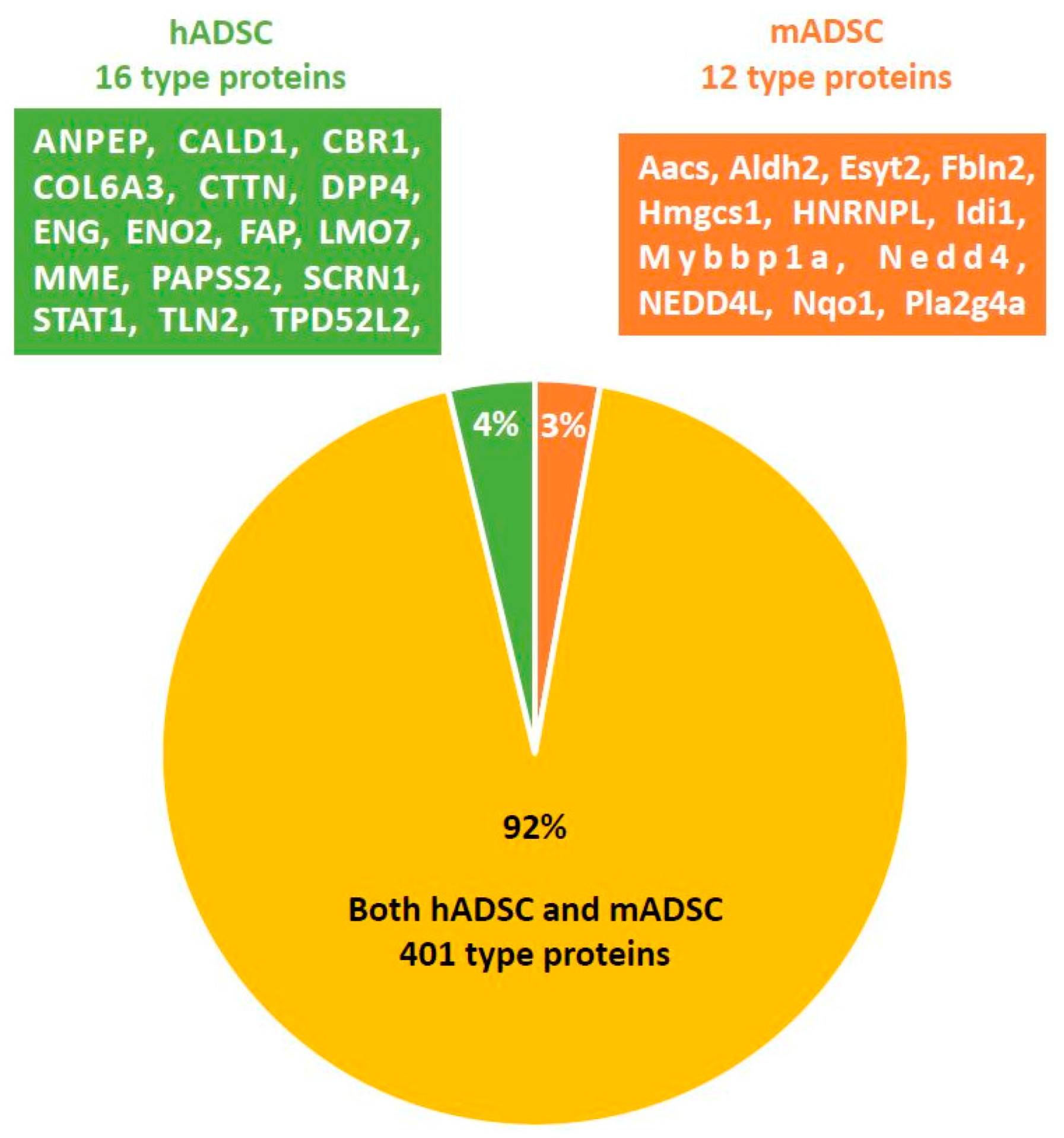
2.3. Comprehensive Protein Expression Analysis of hADSCs and mADSCs
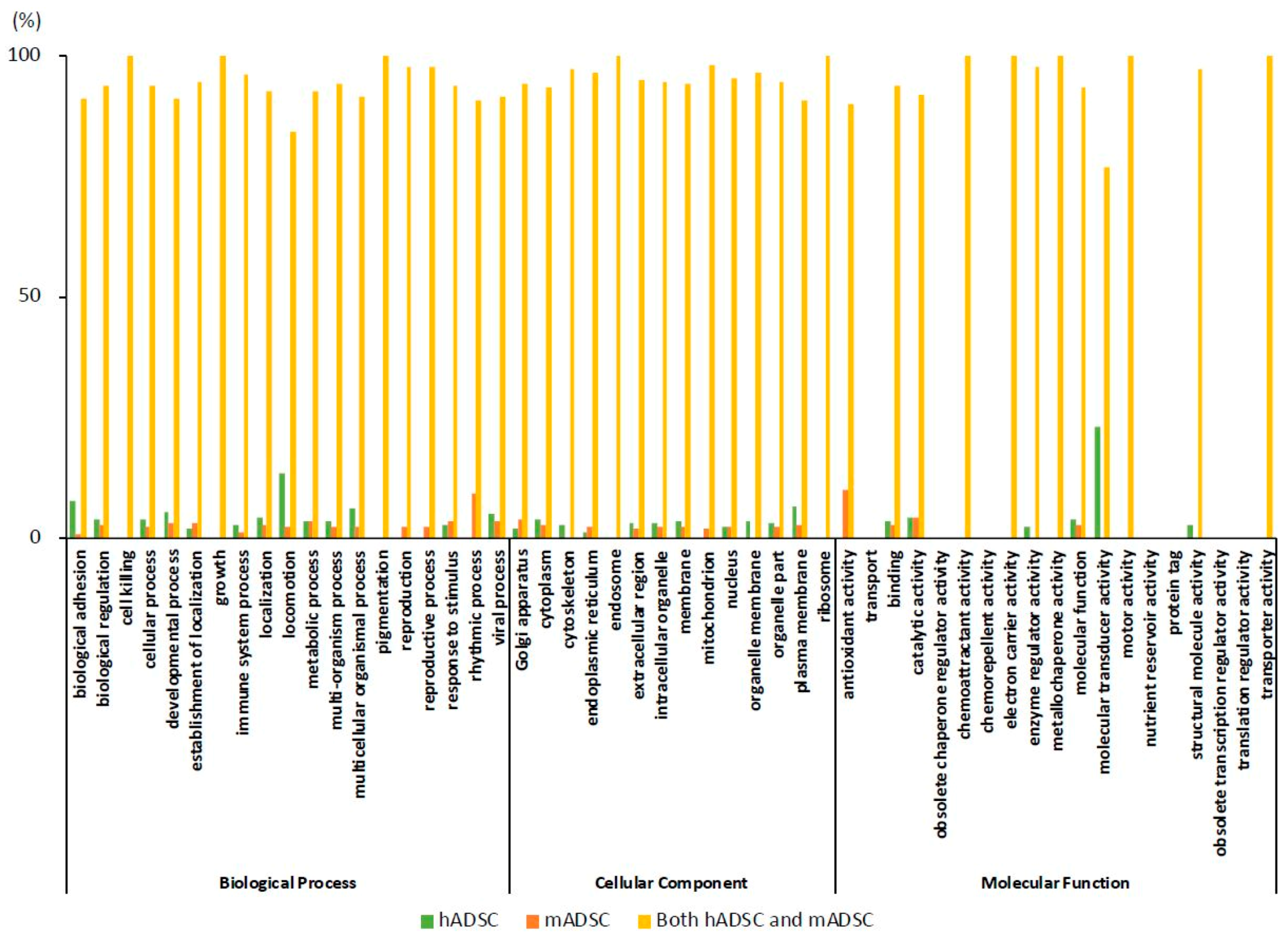
2.4. GO Classification of Proteins Expressed in hADSCs and mADSCs
2.5. Comparison of the Rates of the GO Findings
3. Discussion
4. Materials and Methods
4.1. Reagents
4.2. Animal Care
4.3. Isolation of Mouse ADSCs from Adipose Tissue
4.4. Cell Preparation and Quality Control
4.4.1. mADSCs
4.4.2. hADSCs
4.5. Cell Proliferation
4.6. Cell Differentiation
4.7. Immunofluorescence Staining
4.8. Protein Identification by Nano-LC-MS/MS
4.9. Data Analyses
4.9.1. Database Searching
4.9.2. Criteria for Protein Identification
4.9.3. The Protein GO Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Wagers, A.J.; Weissman, I.L. Plasticity of adult stem cells. Cell 2004, 116, 639–648. [Google Scholar] [CrossRef]
- Szoke, K.; Brinchmann, J.E. Concise review: Therapeutic potential of adipose tissue-derived angiogenic cells. Stem Cells Transl. Med. 2012, 1, 658–667. [Google Scholar] [CrossRef] [PubMed]
- Miyagi-Shiohira, C.; Kurima, K.; Kobayashi, N.; Saitoh, I.; Watanabe, M.; Noguchi, Y.; Matsushita, M.; Noguchi, H. Cryopreservation of Adipose-Derived Mesenchymal Stem Cells. Cell Med. 2015, 8, 3–7. [Google Scholar] [CrossRef] [PubMed]
- Lim, M.H.; Ong, W.K.; Sugii, S. The current landscape of adipose-derived stem cells in clinical applications. Expert Rev. Mol. Med. 2014, 16, e8. [Google Scholar] [CrossRef] [PubMed]
- Seki, A.; Sakai, Y.; Komura, T.; Nasti, A.; Yoshida, K.; Higashimoto, M.; Honda, M.; Usui, S.; Takamura, M.; Takamura, T.; et al. Adipose tissue-derived stem cells as a regenerative therapy for a mouse steatohepatitis-induced cirrhosis model. Hepatology 2013, 58, 1133–1142. [Google Scholar] [CrossRef] [PubMed]
- Ullah, I.; Subbarao, R.B.; Rho, G.J. Human mesenchymal stem cells—Current trends and future prospective. Biosci. Rep. 2015, 35, e00191. [Google Scholar] [CrossRef] [PubMed]
- Miyagi-Shiohira, C.; Kobayashi, N.; Saitoh, I.; Watanabe, M.; Noguchi, Y.; Matsushita, M.; Noguchi, H. Evaluation of Serum-Free, Xeno-Free Cryopreservation Solutions for Human Adipose-Derived Mesenchymal Stem Cells. Cell Med. 2017, 9, 15–20. [Google Scholar] [CrossRef] [PubMed]
- Ashburner, M.; Ball, C.A.; Blake, J.A.; Botstein, D.; Butler, H.; Cherry, J.M.; Davis, A.P.; Dolinski, K.; Dwight, S.S.; Eppig, J.T.; et al. Gene ontology: Tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000, 25, 25–29. [Google Scholar] [CrossRef] [PubMed]
- Huntley, R.P.; Sawford, T.; Martin, M.J.; O’Donovan, C. Understanding how and why the Gene Ontology and its annotations evolve: The GO within UniProt. GigaScience 2014, 3, 4. [Google Scholar] [CrossRef] [PubMed]
- Zimmerlin, L.; Donnenberg, V.S.; Rubin, J.P.; Donnenberg, A.D. Mesenchymal markers on human adipose stem/progenitor cells. Cytom. A 2013, 83, 134–140. [Google Scholar] [CrossRef] [PubMed]
- Gao, H.; Volat, F.; Sandhow, L.; Galitzky, J.; Nguyen, T.; Esteve, D.; Astrom, G.; Mejhert, N.; Ledoux, S.; Thalamas, C.; et al. CD36 Is a Marker of Human Adipocyte Progenitors with Pronounced Adipogenic and Triglyceride Accumulation Potential. Stem Cells 2017, 35, 1799–1814. [Google Scholar] [CrossRef] [PubMed]
- Suga, H.; Matsumoto, D.; Eto, H.; Inoue, K.; Aoi, N.; Kato, H.; Araki, J.; Yoshimura, K. Functional Implications of CD34 Expression in Human Adipose-Derived Stem/Progenitor Cells. Stem Cells Dev. 2009, 18, 1201–1209. [Google Scholar] [CrossRef] [PubMed]
- Ambrosi, T.H.; Scialdone, A.; Graja, A.; Gohlke, S.; Jank, A.M.; Bocian, C.; Woelk, L.; Fan, H.; Logan, D.W.; Schurmann, A.; et al. Adipocyte Accumulation in the Bone Marrow during Obesity and Aging Impairs Stem Cell-Based Hematopoietic and Bone Regeneration. Cell Stem Cell 2017, 20, 771–784. [Google Scholar] [CrossRef] [PubMed]
- Qin, Y.R.; Qin, J.L.; Zhou, C.K.; Li, J.S.; Gao, W.Q. Generation of embryonic stem cells from mouse adipose-tissue derived cells via somatic cell nuclear transfer. Cell Cycle 2015, 14, 1282–1290. [Google Scholar] [CrossRef] [PubMed]
- Ross, P.L.; Huang, Y.L.N.; Marchese, J.N.; Williamson, B.; Parker, K.; Hattan, S.; Khainovski, N.; Pillai, S.; Dey, S.; Daniels, S.; et al. Multiplexed protein quantitation in Saccharomyces cerevisiae using amine-reactive isobaric tagging reagents. Mol. Cell. Proteom. 2004, 3, 1154–1169. [Google Scholar] [CrossRef] [PubMed]
- Santos, J.; Milthorpe, B.K.; Herbert, B.R.; Padula, M.P. Proteomic Analysis of Human Adipose Derived Stem Cells during Small Molecule Chemical Stimulated Pre-neuronal Differentiation. Int. J. Stem Cells 2017, 10, 193–217. [Google Scholar] [CrossRef] [PubMed]
- Nakashima, Y.; Nahar, S.; Miyagi-Shiohira, C.; Kinjo, T.; Kobayashi, N.; Saitoh, I.; Watanabe, M.; Fujita, J.; Noguchi, H. A Liquid Chromatography with Tandem Mass Spectrometry-Based Proteomic Analysis of Cells Cultured in DMEM 10% FBS and Chemically Defined Medium Using Human Adipose-Derived Mesenchymal Stem Cells. Int. J. Mol. Sci. 2018, 19, 2042. [Google Scholar] [CrossRef] [PubMed]
- Braun, J.; Kurtz, A.; Barutcu, N.; Bodo, J.; Thiel, A.; Dong, J. Concerted Regulation of CD34 and CD105 Accompanies Mesenchymal Stromal Cell Derivation from Human Adventitial Stromal Cell. Stem Cells Dev. 2013, 22, 815–827. [Google Scholar] [CrossRef] [PubMed]
- Nakashima, Y.; Nahar, S.; Miyagi-Shiohira, C.; Kinjo, T.; Kobayashi, N.; Saitoh, I.; Watanabe, M.; Fujita, J.; Noguchi, H. A liquid chromatography with tandem mass spectrometry-based proteomic analysis of primary cultured cells and subcultured cells using mouse adipose-derived mesenchymal stem cells. Stem Cells Int. 2018. in Press. [Google Scholar]
- Niu, P.; Smagul, A.; Wang, L.; Sadvakas, A.; Sha, Y.; Perez, L.M.; Nussupbekova, A.; Amirbekov, A.; Akanov, A.A.; Galvez, B.G.; et al. Transcriptional profiling of interleukin-2-primed human adipose derived mesenchymal stem cells revealed dramatic changes in stem cells response imposed by replicative senescence. Oncotarget 2015, 6, 17938–17957. [Google Scholar] [CrossRef] [PubMed]
- Dankel, S.N.; Svard, J.; Mattha, S.; Claussnitzer, M.; Kloting, N.; Glunk, V.; Fandalyuk, Z.; Grytten, E.; Solsvik, M.H.; Nielsen, H.J.; et al. COL6A3 expression in adipocytes associates with insulin resistance and depends on PPARgamma and adipocyte size. Obesity 2014, 22, 1807–1813. [Google Scholar] [CrossRef] [PubMed]
- Oliva-Olivera, W.; Gea, A.L.; Lhamyani, S.; Coin-Araguez, L.; Torres, J.A.; Bernal-Lopez, M.R.; Garcia-Luna, P.P.; Conde, S.M.; Fernandez-Veledo, S.; El Bekay, R.; et al. Differences in the Osteogenic Differentiation Capacity of Omental Adipose-Derived Stem Cells in Obese Patients with and without Metabolic Syndrome. Endocrinology 2015, 156, 4492–4501. [Google Scholar] [CrossRef] [PubMed]
- Jaager, K.; Islam, S.; Zajac, P.; Linnarsson, S.; Neuman, T. RNA-Seq Analysis Reveals Different Dynamics of Differentiation of Human Dermis- and Adipose-Derived Stromal Stem Cells. PLoS ONE 2012, 7. [Google Scholar] [CrossRef] [PubMed]
- Chung, K.M.; Hsu, S.C.; Chu, Y.R.; Lin, M.Y.; Jiaang, W.T.; Chen, R.H.; Chen, X. Fibroblast Activation Protein (FAP) Is Essential for the Migration of Bone Marrow Mesenchymal Stem Cells through RhoA Activation. PLoS ONE 2014, 9, e88772. [Google Scholar] [CrossRef] [PubMed]
- Jang, I.K.; Jung, H.J.; Noh, O.K.; Lee, D.H.; Lee, K.C.; Park, J.E. B7H1mediated immunosuppressive properties in human mesenchymal stem cells are mediated by STAT1 and not PI3K/Akt signaling. Mol. Med. Rep. 2018, 18, 1842–1848. [Google Scholar] [PubMed]
- Lv, X.J.; Zhou, G.D.; Liu, Y.; Liu, X.; Chen, J.N.; Luo, X.S.; Cao, Y.L. In vitro proliferation and differentiation of adipose-derived stem cells isolated using anti-CD105 magnetic beads. Int. J. Mol. Med. 2012, 30, 826–834. [Google Scholar] [CrossRef] [PubMed]
- Zhong, J.X.; Kankanala, S.; Rajagopalan, S. Dipeptidyl peptidase-4 inhibition: Insights from the bench and recent clinical studies. Curr. Opin. Lipidol. 2016, 27, 484–492. [Google Scholar] [CrossRef] [PubMed]
- Heinrichs, D.; Knauel, M.; Offermanns, C.; Berres, M.L.; Nellen, A.; Leng, L.; Schmitz, P.; Bucala, R.; Trautwein, C.; Weber, C.; et al. Macrophage migration inhibitory factor (MIF) exerts antifibrotic effects in experimental liver fibrosis via CD74. Proc. Natl. Acad. Sci. USA 2011, 108, 17444–17449. [Google Scholar] [CrossRef] [PubMed]
- Nakashima, Y.; Nahar, S.; Miyagi-Shiohira, C.; Kinjo, T.; Toyoda, Z.; Kobayashi, N.; Saitoh, I.; Watanabe, M.; Fujita, J.; Noguchi, H. A Liquid Chromatography with Tandem Mass Spectrometry-Based Proteomic Analysis of the Proteins Secreted by Human Adipose-Derived Mesenchymal Stem Cells. Cell Transplant. 2018, 27, 1469–1494. [Google Scholar] [CrossRef] [PubMed]
- Miyagi-Shiohira, C.; Nakashima, Y.; Kobayashi, N.; Saitoh, I.; Watanabe, M.; Noguchi, H. Characterization of induced tissue-specific stem cells from pancreas by a synthetic self-replicative RNA. Sci. Rep. 2018, 8, 12341. [Google Scholar] [CrossRef] [PubMed]
- Nakashima, Y.; Miyagi-Shiohira, C.; Kobayashi, N.; Saitoh, I.; Watanabe, M.; Noguchi, H. A proteome analysis of pig pancreatic islets and exocrine tissue by liquid chromatography with tandem mass spectrometry. Islets 2017, 9, 159–176. [Google Scholar] [CrossRef] [PubMed]
- Shinoda, K.; Tomita, M.; Ishihama, Y. emPAI Calc-for the estimation of protein abundance from large-scale identification data by liquid chromatography-tandem mass spectrometry. Bioinformatics 2010, 26, 576–577. [Google Scholar] [CrossRef] [PubMed]
- Ishihama, Y.; Oda, Y.; Tabata, T.; Sato, T.; Nagasu, T.; Rappsilber, J.; Mann, M. Exponentially modified protein abundance index (emPAI) for estimation of absolute protein amount in proteomics by the number of sequenced peptides per protein. Mol. Cell. Proteom. 2005, 4, 1265–1272. [Google Scholar] [CrossRef] [PubMed]
- Zhu, W.H.; Smith, J.W.; Huang, C.M. Mass Spectrometry-Based Label-Free Quantitative Proteomics. J. Biomed. Biotechnol. 2010. [Google Scholar] [CrossRef] [PubMed]
- Rappsilber, J.; Ryder, U.; Lamond, A.I.; Mann, M. Large-scale proteomic analysis of the human spliceosome. Genome Res. 2002, 12, 1231–1245. [Google Scholar] [CrossRef] [PubMed]

| UniProt/SWISS- | Biological Process | Cellular Component | Molecular Function | emPAI a | |||||
|---|---|---|---|---|---|---|---|---|---|
| PROT ID | Alternate ID | Biological Adhesion | Locomotion | Rhythmic Process | Plasma Membrane | Antioxidant Activity | Molecular Transducer Activity | hMSC_P3_D | mouse_D |
| hADSC | TLN2 | cell adhesion | plasma membrane | 10.426 | 0 | ||||
| COL6A3 | cell adhesion | sarcolemma | 77.722 | 0 | |||||
| CALD1 | cell-cell adhesion | plasma membrane | 51.183 | 0 | |||||
| ANPEP | external side of plasma membrane, integral component of plasma membrane | receptor activity, virus receptor activity | 39.809 | 0 | |||||
| LMO7 | 36.966 | 0 | |||||||
| FAP | cell adhesion | endothelial cell migration | invadopodium membrane, lamellipodium membrane, plasma membrane, ruffle membrane | 35.07 | 0 | ||||
| STAT1 | cell-cell adhesion | endothelial cell migration | 28.435 | 0 | |||||
| MME | integral component of plasma membrane | 26.539 | 0 | ||||||
| CBR1 | 16.113 | 0 | |||||||
| ENG | cell adhesion | cell chemotaxis, cell migration involved in endocardial cushion formation | external side of plasma membrane, transforming growth factor beta receptor complex | transforming growth factor beta-activated receptor activity, transmembrane signaling receptor activity | 14.218 | 0 | |||
| TPD52L2 | 12.322 | 0 | |||||||
| DPP4 | cell adhesion | endothelial cell migration | apical plasma membrane, invadopodium membrane, lamellipodium membrane | virus receptor activity | 15.165 | 0 | |||
| PAPSS2 | 13.27 | 0 | |||||||
| CTHRC1 | cell migration | 12.322 | 0 | ||||||
| SCRN1 | 12.322 | 0 | |||||||
| CTTN | cell-cell adhesion, focal adhesion assembly | cell motility, substrate-dependent cell migration, cell extension | plasma membrane, voltage-gated potassium channel complex | 10.426 | 0 | ||||
| mADSC | Mybbp1a | circadian regulation of gene expression | 0 | 29.631 | |||||
| Hmgcs1 | plasma membrane | 0 | 27.514 | ||||||
| Aldh2 | 0 | 22.223 | |||||||
| Esyt2 | cell-cell adhesion | extrinsic component of cytoplasmic side of plasma membrane, integral component of plasma membrane | 0 | 15.874 | |||||
| Nedd4 | transmission of virus | plasma membrane | 0 | 33.864 | |||||
| Pla2g4a | 0 | 21.165 | |||||||
| Fbln2 | 0 | 17.99 | |||||||
| Idi1 | 0 | 16.932 | |||||||
| HNRNPL | 0 | 17.99 | |||||||
| Aacs | 0 | 14.815 | |||||||
| Nqo1 | superoxide dismutase activity | 0 | 13.757 | ||||||
| NEDD4L | plasma membrane | 0 | 14.815 | ||||||
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nahar, S.; Nakashima, Y.; Miyagi-Shiohira, C.; Kinjo, T.; Kobayashi, N.; Saitoh, I.; Watanabe, M.; Noguchi, H.; Fujita, J. A Comparison of Proteins Expressed between Human and Mouse Adipose-Derived Mesenchymal Stem Cells by a Proteome Analysis through Liquid Chromatography with Tandem Mass Spectrometry. Int. J. Mol. Sci. 2018, 19, 3497. https://doi.org/10.3390/ijms19113497
Nahar S, Nakashima Y, Miyagi-Shiohira C, Kinjo T, Kobayashi N, Saitoh I, Watanabe M, Noguchi H, Fujita J. A Comparison of Proteins Expressed between Human and Mouse Adipose-Derived Mesenchymal Stem Cells by a Proteome Analysis through Liquid Chromatography with Tandem Mass Spectrometry. International Journal of Molecular Sciences. 2018; 19(11):3497. https://doi.org/10.3390/ijms19113497
Chicago/Turabian StyleNahar, Saifun, Yoshiki Nakashima, Chika Miyagi-Shiohira, Takao Kinjo, Naoya Kobayashi, Issei Saitoh, Masami Watanabe, Hirofumi Noguchi, and Jiro Fujita. 2018. "A Comparison of Proteins Expressed between Human and Mouse Adipose-Derived Mesenchymal Stem Cells by a Proteome Analysis through Liquid Chromatography with Tandem Mass Spectrometry" International Journal of Molecular Sciences 19, no. 11: 3497. https://doi.org/10.3390/ijms19113497
APA StyleNahar, S., Nakashima, Y., Miyagi-Shiohira, C., Kinjo, T., Kobayashi, N., Saitoh, I., Watanabe, M., Noguchi, H., & Fujita, J. (2018). A Comparison of Proteins Expressed between Human and Mouse Adipose-Derived Mesenchymal Stem Cells by a Proteome Analysis through Liquid Chromatography with Tandem Mass Spectrometry. International Journal of Molecular Sciences, 19(11), 3497. https://doi.org/10.3390/ijms19113497





