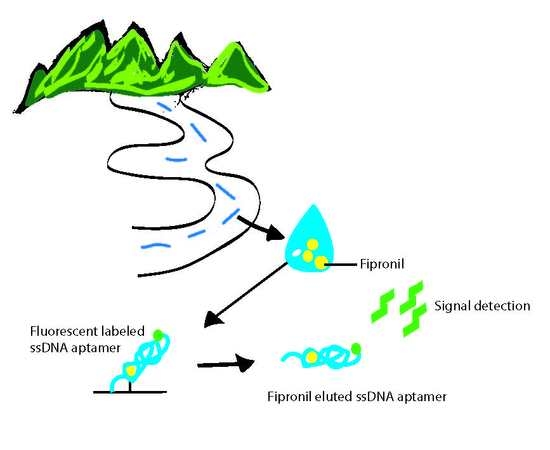
In Vitro Selection of a Single-Stranded DNA Molecular Recognition Element against the Pesticide Fipronil and Sensitive Detection in River Water
Abstract
1. Introduction
2. Results and Discussion
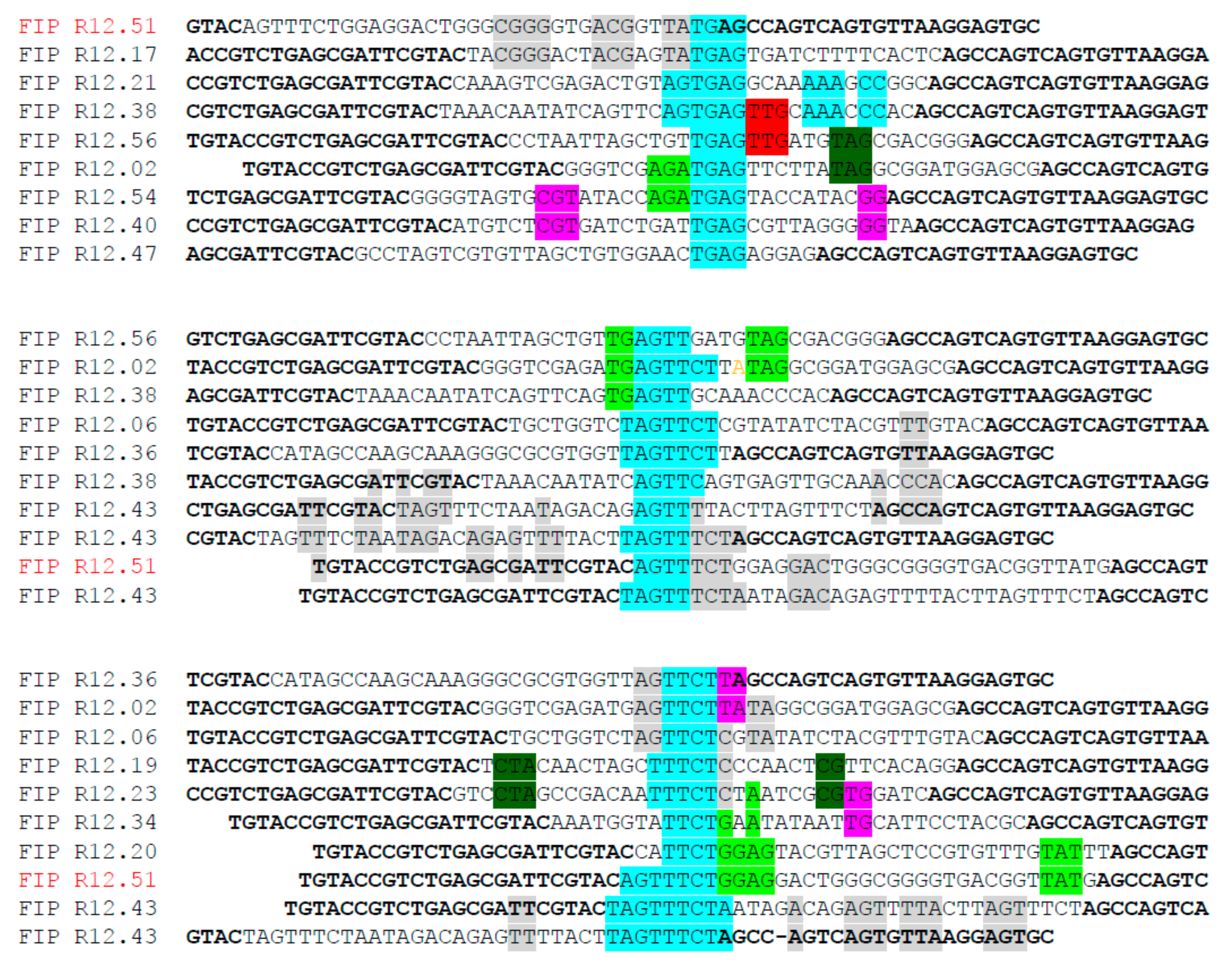
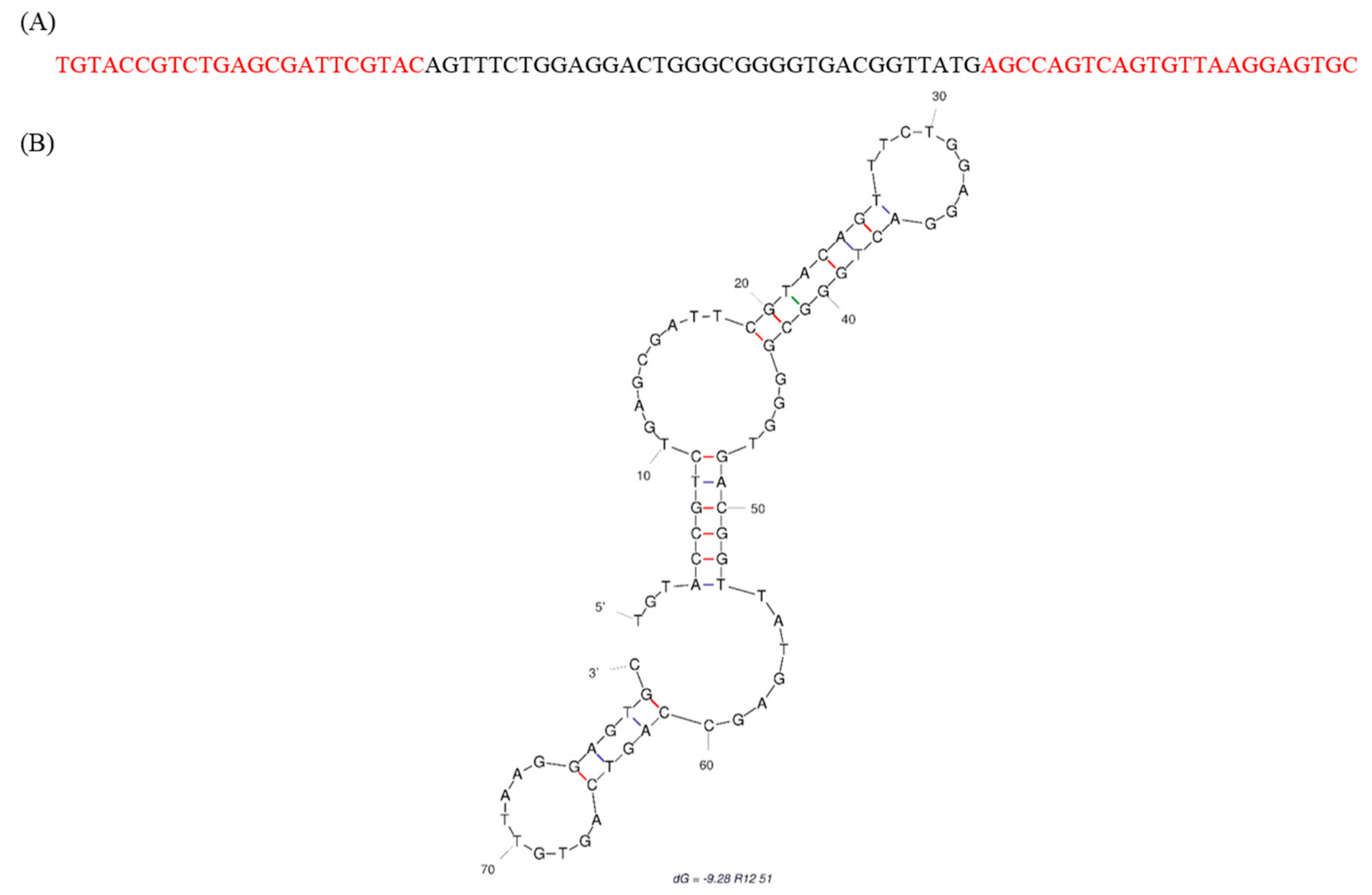
2.1. Identification of Fipronil Specific ssDNA MRE
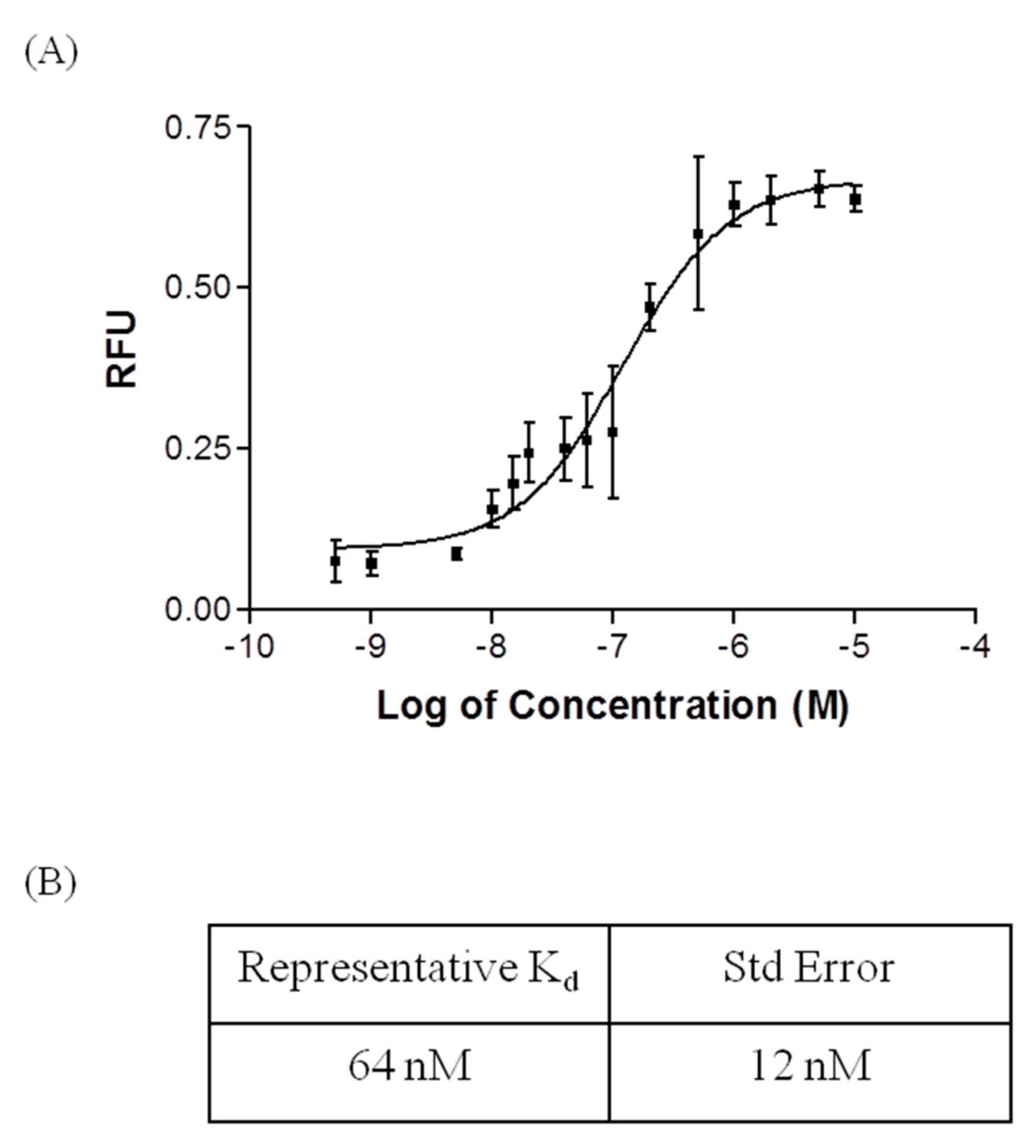
2.2. Affinity and Specifcity of Fipronil-Specific ssDNA MRE in Selection Buffer and Buffered River Water
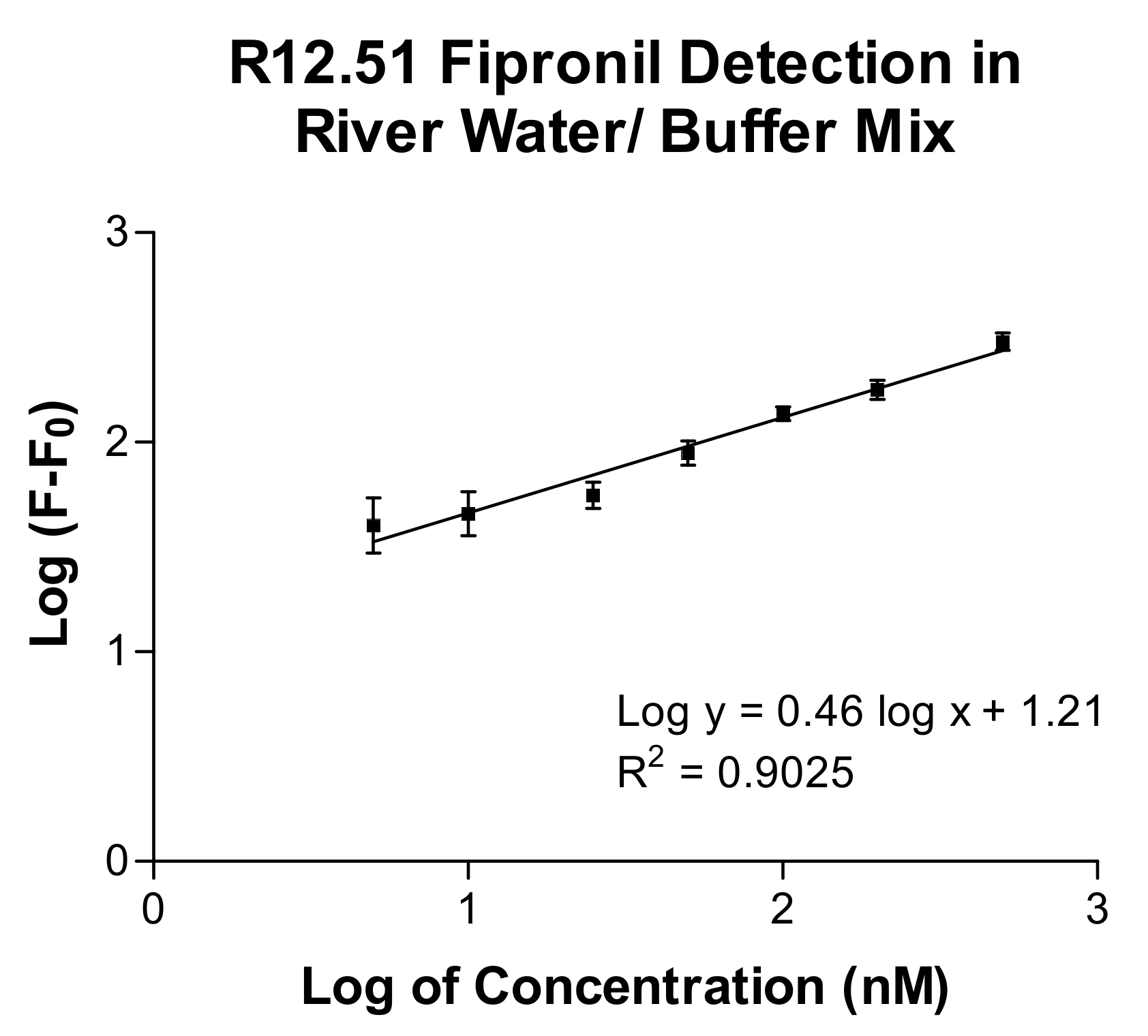
2.3. Biosensing Application of Fipronil-Specific MRE in River Water
3. Materials and Methods
3.1. In Vitro Selection for Fipronil-Specific MREs
3.2. Cloning and Sequencing of the Fipronil-Binding MREs
3.3. Fipronil-Binding MREs Binding Assays in Selection Buffer
3.4. Fipronil Cross-Binding Assay in River Water
3.5. Fipronil Detection Assay in River Water
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| MRE | Molecular Recognition Element |
| BSA | Bovine Serum Albumin |
| IS | Immobilizing Substrate |
| IT | Immobilized Target |
| LOD | Limit of Detection |
References
- U.S. Environmental Protection Agency, Office of Prevention. New-Pesticide Fact. Sheet—Fipronil; Pesticides and Toxic Substances, Office of Pesticide Programs, U.S. Government Printing Office: Washington, DC, USA, 1996; pp. 1–10.
- Caboni, P.; Sammelson, R.E.; Casida, J.E. Phenylpyrazole insecticide photochemistry, metabolism, and gabaergic action: Ethiprole compared with fipronil. J. Agric. Food Chem. 2003, 51, 7055–7061. [Google Scholar] [CrossRef] [PubMed]
- Stone, W.W.; Gilliom, R.J.; Ryberg, K.R. Pesticides in U.S. Streams and rivers: Occurrence and trends during 1992–2011. Environ. Sci. Technol. 2014, 48, 11025–11030. [Google Scholar] [CrossRef] [PubMed]
- Moran, K.D. Urban Use of the Insecticide Fipronil—Water Quality Implications; TDC Environmental: San Mateo, CA, USA, 2007; pp. 1–14. [Google Scholar]
- Simon-Delso, N.; Amaral-Rogers, V.; Belzunces, L.P.; Bonmatin, J.M.; Chagnon, M.; Downs, C.; Furlan, L.; Gibbons, D.W.; Giorio, C.; Girolami, V.; et al. Systemic insecticides (neonicotinoids and fipronil): Trends, uses, mode of action and metabolites. Environ. Sci. Pollut. Res. Int. 2015, 22, 5–34. [Google Scholar] [CrossRef] [PubMed]
- Aquatic Life Benchmarks and Ecological Risk Assessments for Registered Pesticides. Available online: https://www.epa.gov/pesticide-science-and-assessing-pesticide-risks/aquatic-life-benchmarks-and-ecological-risk#benchmarks (accessed on 30 October 2017).
- Chau, N.D.; Sebesvari, Z.; Amelung, W.; Renaud, F.G. Pesticide pollution of multiple drinking water sources in the mekong delta, vietnam: Evidence from two provinces. Environ. Sci. Pollut. Res. Int. 2015, 22, 9042–9058. [Google Scholar] [CrossRef] [PubMed]
- Weston, D.P.; Lydy, M.J. Toxicity of the insecticide fipronil and its degradates to benthic macroinvertebrates of urban streams. Environ. Sci. Technol. 2014, 48, 1290–1297. [Google Scholar] [CrossRef] [PubMed]
- Bonmatin, J.M.; Giorio, C.; Girolami, V.; Goulson, D.; Kreutzweiser, D.P.; Krupke, C.; Liess, M.; Long, E.; Marzaro, M.; Mitchell, E.A.; et al. Environmental fate and exposure; neonicotinoids and fipronil. Environ. Sci. Pollut. Res. Int. 2015, 22, 35–67. [Google Scholar] [CrossRef] [PubMed]
- U.S. Environmental Protection Agency, Healthy Effects Division. Fipronil: Third Reevaluation—Report of the Hazard Identification Assessment Review Committee; U.S. Government Printing Office: Washington, DC, USA, 2000; pp. 1–24.
- Li, P.; Akk, G. The insecticide fipronil and its metabolite fipronil sulphone inhibit the rat α1β2γ2l GABAA receptor. Br. J. Pharmacol. 2008, 155, 783–794. [Google Scholar] [CrossRef] [PubMed]
- Van der Sluijs, J.P.; Amaral-Rogers, V.; Belzunces, L.P.; van Bijleveld Lexmond, M.F.; Bonmatin, J.M.; Chagnon, M.; Downs, C.A.; Furlan, L.; Gibbons, D.W.; Giorio, C.; et al. Conclusions of the worldwide integrated assessment on the risks of neonicotinoids and fipronil to biodiversity and ecosystem functioning. Environ. Sci. Pollut. Res. Int. 2015, 22, 148–154. [Google Scholar] [CrossRef] [PubMed]
- Bichon, E.; Richard, C.A.; Le Bizec, B. Development and validation of a method for fipronil residue determination in ovine plasma using 96-well plate solid-phase extraction and gas chromatography-tandem mass spectrometry. J. Chromatogr. A 2008, 1201, 91–99. [Google Scholar] [CrossRef] [PubMed]
- Duhan, A.; Kumari, B.; Duhan, S. Determination of residues of fipronil and its metabolites in cauliflower by using gas chromatography-tandem mass spectrometry. Bull. Environ. Contam. Toxicol. 2015, 94, 260–266. [Google Scholar] [CrossRef] [PubMed]
- Jimenez, J.J.; Bernal, J.L.; del Nozal, M.J.; Martin, M.T.; Mayo, R. Sample preparation methods to analyze fipronil in honey by gas chromatography with electron-capture and mass spectrometric detection. J. Chromatogr. A 2008, 1187, 40–45. [Google Scholar] [CrossRef] [PubMed]
- Lacroix, M.Z.; Puel, S.; Toutain, P.L.; Viguie, C. Quantification of fipronil and its metabolite fipronil sulfone in rat plasma over a wide range of concentrations by LC/UV/MS. J. Chromatogr. B Anal. Technol. Biomed. Life Sci. 2010, 878, 1934–1938. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Yan, C.; Dong, J.; Yu, X.; Xu, D. Poly- and monoclonal antibody-based elisas for fipronil. J. Agric. Food Chem. 2007, 55, 226–230. [Google Scholar] [CrossRef] [PubMed]
- Mohamed, F.; Senarathna, L.; Percy, A.; Abeyewardene, M.; Eaglesham, G.; Cheng, R.; Azher, S.; Hittarage, A.; Dissanayake, W.; Sheriff, M.H.; et al. Acute human self-poisoning with the N-phenylpyrazole insecticide fipronil—A GABAA-gated chloride channel blocker. J. Toxicol. Clin. Toxicol. 2004, 42, 955–963. [Google Scholar] [CrossRef] [PubMed]
- Vasylieva, N.; Ahn, K.C.; Barnych, B.; Gee, S.J.; Hammock, B.D. Development of an immunoassay for the detection of the phenylpyrazole insecticide fipronil. Environ. Sci. Technol. 2015, 49, 10038–10047. [Google Scholar] [CrossRef] [PubMed]
- Bordeaux, J.; Welsh, A.; Agarwal, S.; Killiam, E.; Baquero, M.; Hanna, J.; Anagnostou, V.; Rimm, D. Antibody validation. Biotechniques 2010, 48, 197–209. [Google Scholar] [CrossRef] [PubMed]
- Mairal, T.; Ozalp, V.C.; Lozano Sanchez, P.; Mir, M.; Katakis, I.; O’Sullivan, C.K. Aptamers: Molecular tools for analytical applications. Anal. Bioanal. Chem. 2008, 390, 989–1007. [Google Scholar] [CrossRef] [PubMed]
- Tuerk, C.; Gold, L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science 1990, 249, 505–510. [Google Scholar] [CrossRef] [PubMed]
- Hong, K.L.; Yancey, K.; Battistella, L.; Williams, R.M.; Hickey, K.M.; Bostick, C.D.; Gannett, P.M.; Sooter, L.J. Selection of single-stranded DNA molecular recognition elements against exotoxin a using a novel decoy-selex method and sensitive detection of exotoxin a in human serum. BioMed Res. Int. 2015, 2015, 417641. [Google Scholar] [CrossRef] [PubMed]
- Williams, R.M.; Kulick, A.R.; Yedlapalli, S.; Battistella, L.; Hajiran, C.J.; Sooter, L.J. In vitro selection of a single-stranded DNA molecular recognition element specific for bromacil. J. Nucleic Acids 2014, 2014, 102968. [Google Scholar] [CrossRef] [PubMed]
- Williams, R.M.; Crihfield, C.L.; Gattu, S.; Holland, L.A.; Sooter, L.J. In vitro selection of a single-stranded DNA molecular recognition element against atrazine. Int. J. Mol. Sci. 2014, 15, 14332–14347. [Google Scholar] [CrossRef] [PubMed]
- Williams, R.M.; Maher, E.; Sooter, L.J. In vitro selection of a single-stranded DNA molecular recognition element for the pesticide malathion. Comb. Chem. High Throughput Screen. 2014, 17, 694–702. [Google Scholar] [CrossRef] [PubMed]
- Zuker, M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003, 31, 3406–3415. [Google Scholar] [CrossRef] [PubMed]
- Connell, G.J.; Illangesekare, M.; Yarus, M. Three small ribooligonucleotides with specific arginine sites. Biochemistry 1993, 32, 5497–5502. [Google Scholar] [CrossRef] [PubMed]
- Ellington, A.D.; Khrapov, M.; Shaw, C.A. The scene of a frozen accident. RNA 2000, 6, 485–498. [Google Scholar] [CrossRef] [PubMed]
- Hall, B.; Hesselberth, J.R.; Ellington, A.D. Computational selection of nucleic acid biosensors via a slip structure model. Biosens. Bioelectron. 2007, 22, 1939–1947. [Google Scholar] [CrossRef] [PubMed]
- Lozupone, C.; Changayil, S.; Majerfeld, I.; Yarus, M. Selection of the simplest RNA that binds isoleucine. RNA 2003, 9, 1315–1322. [Google Scholar] [CrossRef] [PubMed]
- Majerfeld, I.; Yarus, M. Isoleucine: RNA sites with associated coding sequences. RNA 1998, 4, 471–478. [Google Scholar] [PubMed]
- Alsager, O.A.; Kumar, S.; Willmott, G.R.; McNatty, K.P.; Hodgkiss, J.M. Small molecule detection in solution via the size contraction response of aptamer functionalized nanoparticles. Biosens. Bioelectron. 2014, 57, 262–268. [Google Scholar] [CrossRef] [PubMed]
- Mehta, J.; Rouah-Martin, E.; van Dorst, B.; Maes, B.; Herrebout, W.; Scippo, M.L.; Dardenne, F.; Blust, R.; Robbens, J. Selection and characterization of PCB-binding DNA aptamers. Anal. Chem. 2012, 84, 1669–1676. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, V.T.; Kwon, Y.S.; Kim, J.H.; Gu, M.B. Multiple go-selex for efficient screening of flexible aptamers. Chem. Commun. 2014, 50, 10513–10516. [Google Scholar] [CrossRef] [PubMed]
- Hong, K.L.; Battistella, L.; Salva, A.D.; Williams, R.M.; Sooter, L.J. In vitro selection of single-stranded DNA molecular recognition elements against S. aureus α toxin and sensitive detection in human serum. Int. J. Mol. Sci. 2015, 16, 2794–2809. [Google Scholar] [CrossRef] [PubMed]
- Hong, K.L.; Maher, E.; Williams, R.M.; Sooter, L.J. In vitro selection of a single-stranded DNA molecular recognition element against clostridium difficile toxin B and sensitive detection in human fecal matter. J. Nucleic Acids 2015, 2015, 808495. [Google Scholar] [CrossRef] [PubMed]
- Department of Environmental Protection, Commonwealth of Pennsylvania. 2012–2013 Susquehanna River Sampling and Assessment Report; Commonwealth of Pennsylvania: Harrisburg, PA, USA, 2014; pp. 1–147.
- Miyakawa, S.; Nomura, Y.; Sakamoto, T.; Yamaguchi, Y.; Kato, K.; Yamazaki, S.; Nakamura, Y. Structural and molecular basis for hyperspecificity of RNA aptamer to human immunoglobulin G. RNA 2008, 14, 1154–1163. [Google Scholar] [CrossRef] [PubMed]
- Tingle, C.C.; Rother, J.A.; Dewhurst, C.F.; Lauer, S.; King, W.J. Fipronil: Environmental fate, ecotoxicology, and human health concerns. Rev. Environ. Contam. Toxicol. 2003, 176, 1–66. [Google Scholar] [PubMed]
- Armbruster, D.A.; Pry, T. Limit of blank, limit of detection and limit of quantitation. Clin. Biochem. Rev. 2008, 29, S49–S52. [Google Scholar] [PubMed]
- Miyake, S.; Uchigashima, M.; Kadowaki, A. Kit for Measurement of Termite Insecticide Active Ingredient by Immunoassay Method. U.S. Patent 8,323,904 B2, 4 December 2012. [Google Scholar]
- Fan, L.; Zhao, G.; Shi, H.; Liu, M.; Li, Z. A highly selective electrochemical impedance spectroscopy-based aptasensor for sensitive detection of acetamiprid. Biosens. Bioelectron. 2013, 43, 12–18. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Chen, W.; Ma, W.; Liu, L.; Ma, W.; Zhao, Y.; Zhu, Y.; Xu, L.; Kuang, H.; Xu, C. Fluorescent strip sensor for rapid determination of toxins. Chem. Commun. 2011, 47, 1574–1576. [Google Scholar] [CrossRef] [PubMed]
- Barthelmebs, L.; Jonca, J.; Hayat, A.; Prieto-Simon, B.; Marty, J.-L. Enzyme-linked aptamer assays (ELAAs), based on a competition format for a rapid and sensitive detection of ochratoxin a in wine. Food Control 2011, 22, 737–743. [Google Scholar] [CrossRef]
- Madianos, L.; Tsekenis, G.; Skotadis, E.; Patsiouras, L.; Tsoukalas, D. A highly sensitive impedimetric aptasensor for the selective detection of acetamiprid and atrazine based on microwires formed by platinum nanoparticles. Biosens. Bioelectron. 2018, 101, 268–274. [Google Scholar] [CrossRef] [PubMed]
- Bujacz, A. Structures of bovine, equine and leporine serum albumin. Acta Crystallogr. Sect. D Biol. Crystallogr. 2012, 68, 1278–1289. [Google Scholar] [CrossRef] [PubMed]
- De-Los-Santos-Álvarez, N.; Lobo-Castañón, M.J.; Miranda-Ordieres, A.J.; Tuñón-Blanco, P. Modified-RNA aptamer-based sensor for competitive impedimetric assay of neomycin B. J. Am. Chem. Soc. 2007, 129, 3808–3809. [Google Scholar] [CrossRef] [PubMed]
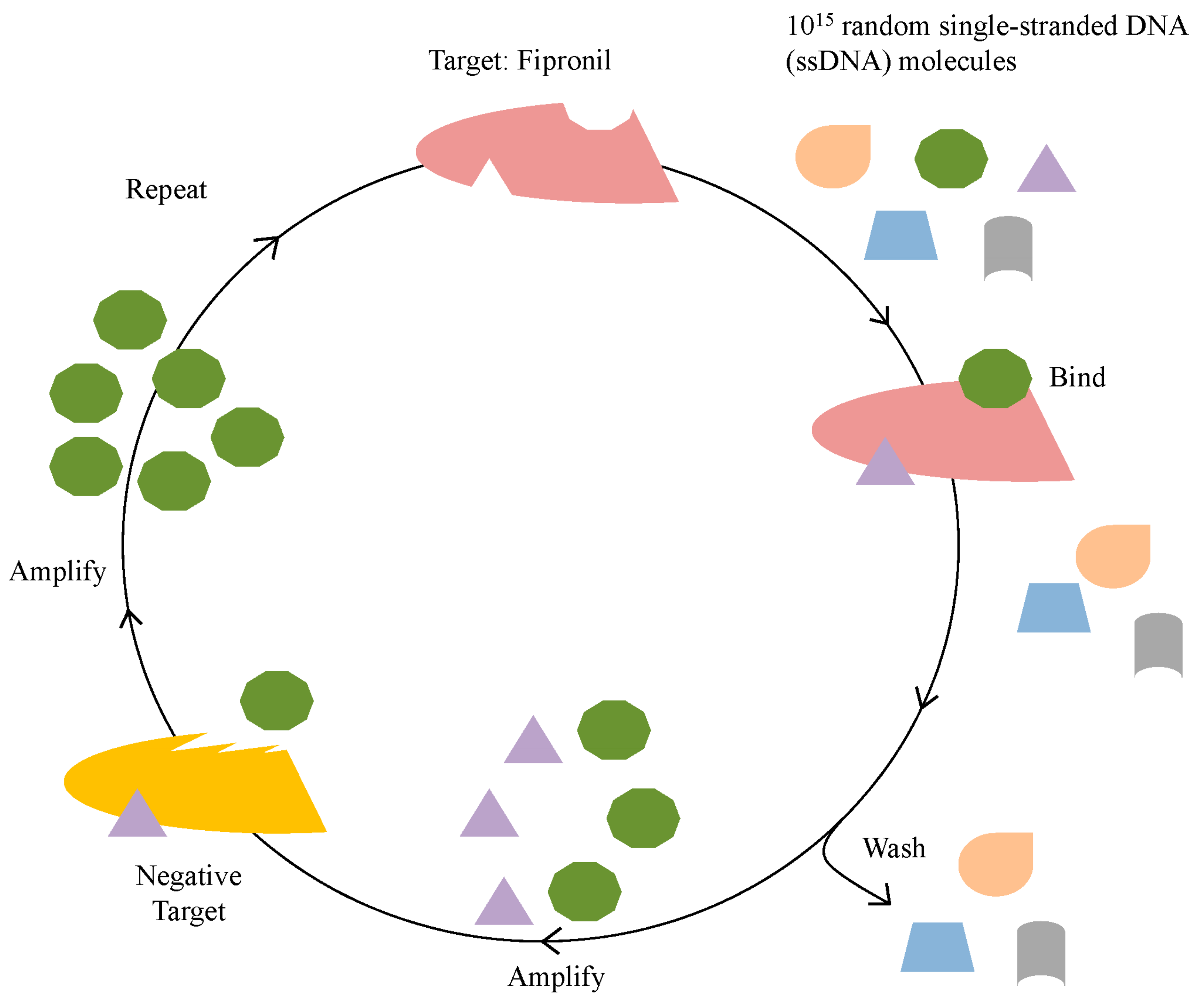
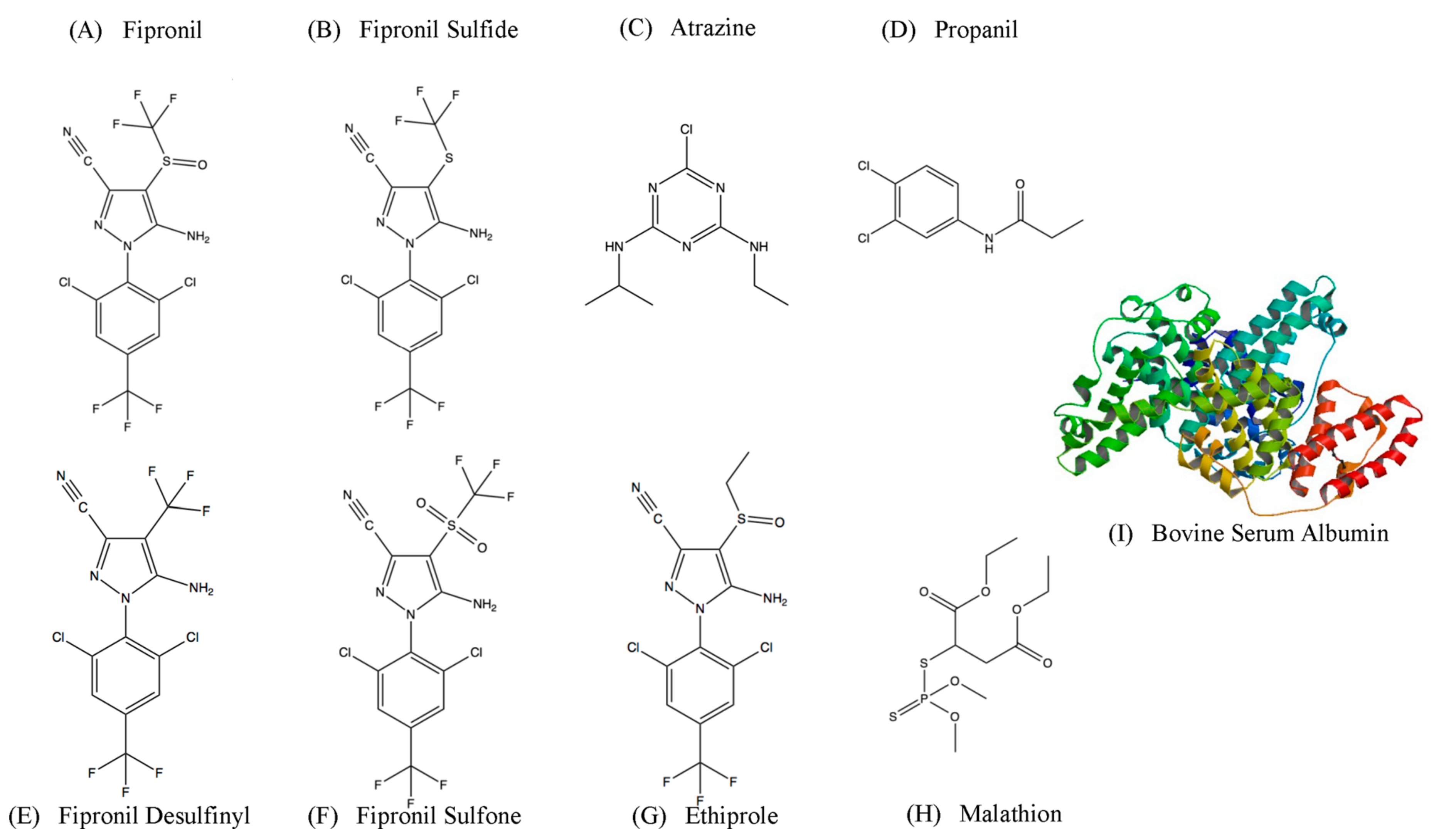
| Round | Positive Selection | Time | Negative Selection | Time |
|---|---|---|---|---|
| 1 | Immobilized Target (IT) | 48 h | Immobilizing substrate (IS) | 24 h |
| 2 | IT | 22 h | Immobilized Negative Target (INT) Fipronil Sulfide | 24 h |
| 3 | IT | 16 h | INT Fipronil Desulfinyl | 24 h |
| 4 | IT | 11 h | INT Fipronil Sulfone | 24 h |
| 5 | IT | 6.5 h | INT Ethiprole | 24 h |
| 6 | IT | 3 h | IT/ 1 mM of BSA Free Elution (FE) | 3 h/ 24 h |
| 7 | IT/ Competitive Elution with 500 µM free Fipronil | 3 h/ 1 h | IT/ 1 µM of Atrazine FE/1 µM of Propanil FE | 3 h/ 24 h/ 24 h |
| 8 | IT/ Competitive Elution with 500 µM free Fipronil | 1 h/ 30 min | IT/ 1 µM of Fipronil Sulfone FE/ 1 µM of Fipronil Desulfinyl FE | 3 h/ 24 h/ 24 h |
| 9 | IT/ Competitive Elution with 100 µM free Fipronil | 15 min/ 5 min | IT/ 1 µM of Fipronil Sulfide FE | 15 min/ 15 min |
| 10 | IT/ Competitive Elution with 10 µM free Fipronil | 1 min/ 1 min | IT/ 1 µM of Ethiprole Free Elution | 1 min/ 1 min |
| 11 | IT/ Competitive Elution with 1 µM free Fipronil | Immediate/ Immediate | IT/ 1 mM of BSA FE/ 1 µM of Malathion FE | 1 min/ 6 h/ 30 min |
| 12 | IT/ Competitive Elution with 100 nM free Fipronil | Immediate/ Immediate | - |
| Target | Normalized Average Fluorescence | Standard Deviation | p-Value | Selective Ratio |
|---|---|---|---|---|
| Fipronil | 2.413 | 0.194 | - | - |
| Fipronil Sulfone | 2.248 | 0.388 | 0.279 | 1.07 |
| Fipronil Sulfide | 2.726 | 0.460 | 0.169 | 0.89 |
| Fipronil Desulfinyl | 2.088 | 0.623 | 0.219 | 1.16 |
| Ethiprole | 2.185 | 0.217 | 0.124 | 1.10 |
| Atrazine | 1.963 | 0.227 | 0.030 | 1.23 |
| Propanil | 1.587 | 0.243 | 0.005 | 1.52 |
| Malathion | 2.048 | 0.184 | 0.039 | 1.18 |
| BSA | 8.687 | 0.360 | 5.99 × 10−6 | 0.28 |
| Selection Buffer (No Targets) | 0.247 | 0.214 | 1.02 × 10−4 | 9.74 |
| Target | Normalized Average Fluorescence | Standard Deviation | p-Value | Selective Ratio |
|---|---|---|---|---|
| Fipronil | 3.863 | 0.090 | - | - |
| Fipronil Sulfone | 2.863 | 0.180 | 4.98 × 10−4 | 1.34 |
| Fipronil Sulfide | 4.333 | 0.944 | 2.19 × 10−1 | 0.89 |
| Fipronil Desulfinyl | 2.902 | 0.245 | 15.5 × 10−3 | 1.33 |
| Ethiprole | 3.157 | 0.148 | 1.06 × 10−3 | 1.22 |
| Atrazine | 0.804 | 0.302 | 3.66 × 10−5 | 4.80 |
| Propanil | 0.764 | 0.509 | 2.44 × 10−4 | 5.05 |
| Malathion | 0.725 | 0.355 | 5.98 × 10−5 | 5.32 |
| BSA | 20.41 | 1.118 | 6.95 × 10−6 | 0.19 |
| River water/Buffer Mix (No Targets) | −0.549 | 1.779 | 6.37 × 10−3 | −7.035 |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hong, K.L.; Sooter, L.J. In Vitro Selection of a Single-Stranded DNA Molecular Recognition Element against the Pesticide Fipronil and Sensitive Detection in River Water. Int. J. Mol. Sci. 2018, 19, 85. https://doi.org/10.3390/ijms19010085
Hong KL, Sooter LJ. In Vitro Selection of a Single-Stranded DNA Molecular Recognition Element against the Pesticide Fipronil and Sensitive Detection in River Water. International Journal of Molecular Sciences. 2018; 19(1):85. https://doi.org/10.3390/ijms19010085
Chicago/Turabian StyleHong, Ka L., and Letha J. Sooter. 2018. "In Vitro Selection of a Single-Stranded DNA Molecular Recognition Element against the Pesticide Fipronil and Sensitive Detection in River Water" International Journal of Molecular Sciences 19, no. 1: 85. https://doi.org/10.3390/ijms19010085
APA StyleHong, K. L., & Sooter, L. J. (2018). In Vitro Selection of a Single-Stranded DNA Molecular Recognition Element against the Pesticide Fipronil and Sensitive Detection in River Water. International Journal of Molecular Sciences, 19(1), 85. https://doi.org/10.3390/ijms19010085