Genome-Wide Transcriptome Analysis Reveals Conserved and Distinct Molecular Mechanisms of Al Resistance in Buckwheat (Fagopyrum esculentum Moench) Leaves
Abstract
:1. Introduction
2. Results
2.1. Buckwheat Leaves Rapidly Accumulate Al
2.2. De Novo Assembly of the Transcripts and Annotation
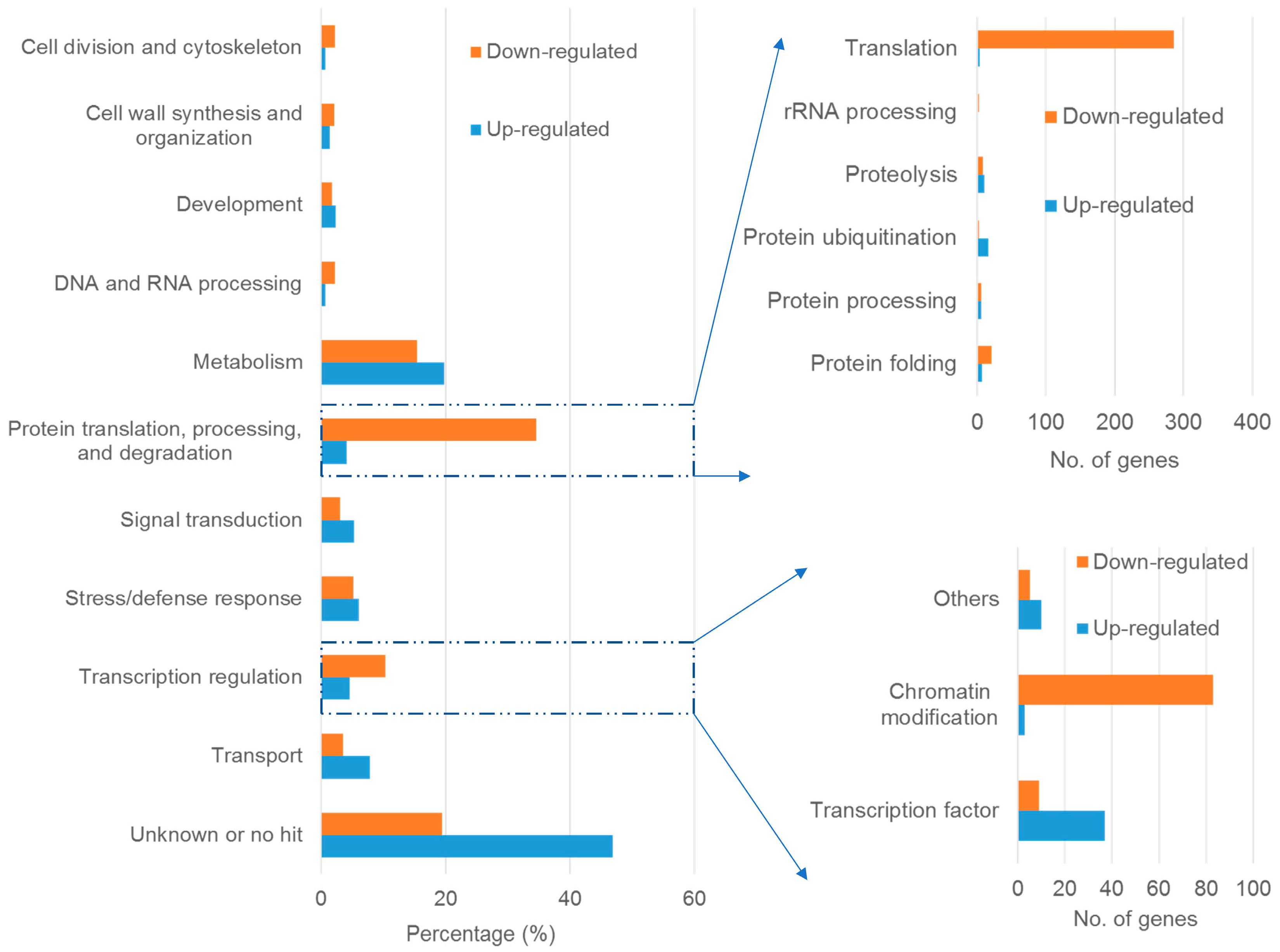
2.3. Global Effect of Al Stress on Gene Expression
2.4. Genes Involved in Protein Translation, Processing and Degradation
2.5. Genes Involved in Transcription Regulation
2.6. Genes Encoding Transporters
2.7. Genes Associated with Development
2.8. Coregulated Genes between Roots and Leaves under Al Stress
2.9. Validation of Gene Expression by qRT-PCR Analysis
3. Discussion
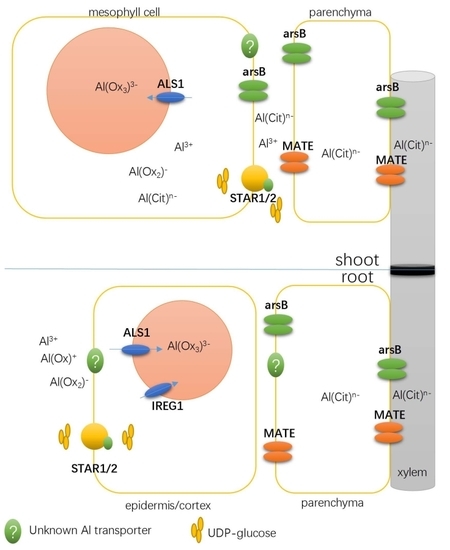
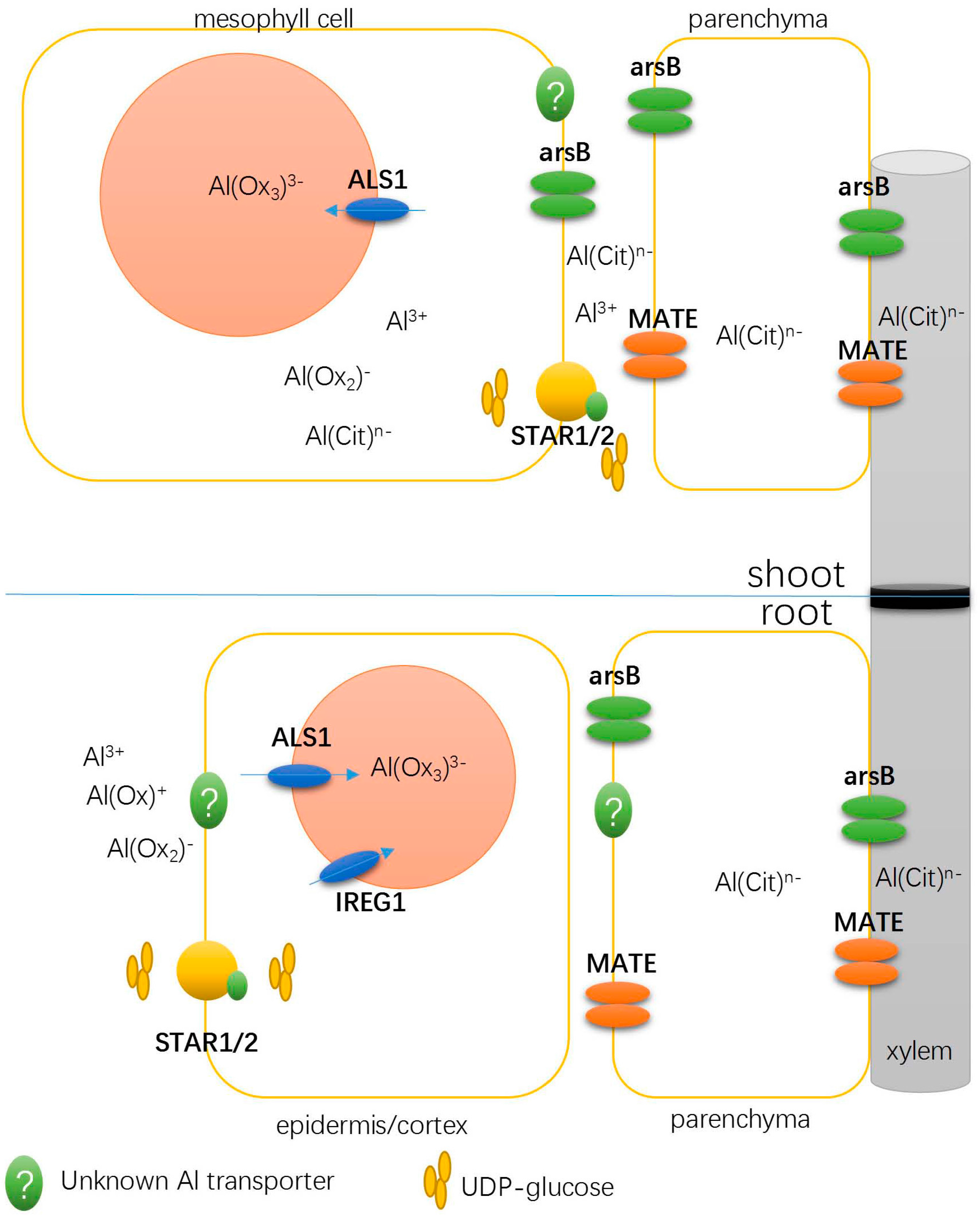
3.1. Detoxifying Apoplastic Al with a Bacterial-Type ABC Transporter
3.2. Transporters Possibly Involved in Xylem Al Unloading and Sequestration
3.3. Flowering Regulation in Response to Al Stress
3.4. Chromatin Regulation in Response to Al Stress
4. Materials and Methods
4.1. Plant Materials and Growth Conditions
4.2. Determination of Al Accumulation
4.3. RNA Isolation and Solexa Sequencing
4.4. Sequence Assembly and Annotation
4.5. Differential Gene Expression Analysis and Gene Ontology Biological Processes Analysis
4.6. Quantitative RT-PCR Analysis
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Von Uexküll, H.R.; Mutert, E. Global extent, development and economic impact of acid soils. In Plant-Soil Interactions at Low pH: Principles and Management; Date, R.A., Grundon, N.J., Rayment, G.E., Probert, M.E., Eds.; Springer: Dordrecht, The Netherlands, 1995; Volume 64, pp. 5–19. [Google Scholar]
- Kochian, L.V. Cellular mechanisms of aluminum toxicity and resistance in plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1995, 46, 237–260. [Google Scholar] [CrossRef]
- Ma, J.F.; Furukawa, J. Recent progress in the research of external Al detoxification in higher plants: A minireview. J. Inorg. Biochem. 2003, 97, 46–51. [Google Scholar] [CrossRef]
- Kochian, L.V.; Piñeros, M.A.; Hoekenga, O.A. The physiology, genetics and molecular biology of plant aluminum resistance and toxicity. Plant Soil 2005, 274, 175–195. [Google Scholar] [CrossRef]
- Liu, J.; Piñeros, M.A.; Kochian, L.V. The role of aluminum sensing and signaling in plant aluminum resistance. J. Integr. Plant Biol. 2014, 56, 221–230. [Google Scholar] [CrossRef] [PubMed]
- Ryan, P.R.; Delhaize, E.; Jones, D.L. Function and mechanism of organic anion exudation from plant roots. Ann. Rev. Plant Mol. Biol. 2001, 52, 527–560. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, T.; Osaki, M. Mechanisms of adaptation to high aluminum condition in native plant species growing in acid soils: A review. Commun. Soil Sci. Plan. 2002, 33, 1247–1260. [Google Scholar] [CrossRef]
- Kochian, L.V.; Hoekenga, O.A.; Piñeros, M.A. How do crop plants tolerate acid soils? Mechanisms of aluminum tolerance and phosphorous efficiency. Annu. Rev. Plant Biol. 2004, 55, 459–493. [Google Scholar] [CrossRef] [PubMed]
- Matsumoto, H.; Hirasawa, E.; Morimura, S.; Takahashi, E. Localization of aluminium in tea leaves. Plant Cell Physiol. 1976, 17, 627–631. [Google Scholar]
- Watanabe, T.; Osaki, M.; Tadano, T. Aluminum-induced growth stimulation in relation to calcium, magnesium, and silicate nutrition in Melastoma malabathricum L. Soil Sci. Plant Nutr. 1997, 43, 827–837. [Google Scholar] [CrossRef]
- Ma, J.F.; Hiradate, S.; Nomoto, K.; Iwashita, T.; Matsumoto, H. Internal detoxification mechanism of Al in hydrangea (Identification of Al form in the leaves). Plant Physiol. 1997, 113, 1033–1039. [Google Scholar] [CrossRef] [PubMed]
- Zheng, S.J.; Ma, J.F.; Matsumoto, H. High aluminum resistance in buckwheat. I. Al-induced specific secretion of oxalic acid from root tips. Plant Physiol. 1998, 117, 745–751. [Google Scholar] [CrossRef]
- Zheng, S.J.; Yang, J.L.; He, Y.F.; Yu, X.H.; Zhang, L.; You, J.F.; Shen, R.F.; Matsumoto, H. Immobilization of aluminum with phosphorus in roots is associated with high aluminum resistance in buckwheat. Plant Physiol. 2005, 138, 297–303. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.L.; Zheng, S.J.; He, Y.F.; You, J.F.; Zhang, L.; Yu, X.H. Comparative studies on the effect of a protein-synthesis inhibitor on aluminium-induced secretion of organic acids from Fagopyrum esculentum Moench and Cassia tora L. roots. Plant Cell Environ. 2006, 29, 240–246. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.F.; Zheng, S.J.; Matsumoto, H.; Hiradate, S. Detoxifying aluminum with buckwheat. Nature 1997, 390, 569–570. [Google Scholar] [CrossRef]
- Shen, R.; Iwashita, T.; Ma, J.F. Form of Al changes with Al concentration in leaves of buckwheat. J. Exp. Bot. 2004, 55, 131–136. [Google Scholar] [CrossRef] [PubMed]
- Yokosho, K.; Yamaji, N.; Ma, J.F. Global transcriptome analysis of Al-induced genes in an Al-accumulating species, common buckwheat (Fagopyrum esculentum Moench). Plant Cell Physiol. 2014, 55, 2077–2091. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.F.; Yamaji, N.; Mitani, N.; Yano, M.; Nagamura, Y.; Ma, J.F. A bacterial-type ABC transporter is involved in aluminum tolerance in rice. Plant Cell 2009, 21, 655–667. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.L.; Zhang, L.; Li, Y.Y.; You, J.F.; Wu, P.; Zheng, S.J. Citrate transporters play a critical role in aluminium-stimulated citrate efflux in rice bean (Vigna umbellata) roots. Ann. Bot. 2006, 97, 579–584. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.M.; Fan, W.; Jin, J.F.; Lou, H.Q.; Chen, W.W.; Yang, J.L.; Zheng, S.J. Transcriptome analysis of Al-induced genes in buckwheat (Fagopyrum esculentum Moench) root apex: New insight into Al toxicity and resistance mechanisms in an Al accumulating species. Front. Plant Sci. 2017, 8, 1141. [Google Scholar] [CrossRef] [PubMed]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q.; et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011, 29, 644–652. [Google Scholar] [CrossRef] [PubMed]
- Mortazavi, A.; Williams, B.A.; McCue, K.; Schaeffer, L.; Wold, B. Mapping and quantifying mammalian transcriptomes by RNA-seq. Nat. Methods 2008, 5, 621–628. [Google Scholar] [CrossRef] [PubMed]
- Fan, W.; Lou, H.Q.; Gong, Y.L.; Liu, M.Y.; Wang, Z.Q.; Yang, J.L.; Zheng, S.J. Identification of early Al-responsive genes in rice bean (Vigna umbellata) roots provides new clues to molecular mechanisms of Al toxicity and tolerance. Plant Cell Environ. 2014, 37, 1586–1597. [Google Scholar] [CrossRef] [PubMed]
- Mitsuda, N.; Ohme-Takagi, M. Functional analysis of transcription factors in Arabidopsis. Plant Cell Physiol. 2009, 50, 1232–1248. [Google Scholar] [CrossRef] [PubMed]
- Asensi-Fabado, M.A.; Amtmann, A.; Perrella, G. Plant responses to abiotic stress: The chromatin context of transcriptional regulation. Biochemi. Biophys. Acta 2017, 1860, 106–122. [Google Scholar] [CrossRef] [PubMed]
- Larsen, P.B.; Cancel, J.; Rounds, M.; Ochoa, V. Arabidopsis ALS1 encodes a root tip and stele localized half type ABC transporter required for root growth in an aluminum toxic environment. Planta 2007, 225, 1447. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.F.; Yamaji, N.; Chen, Z.C.; Ma, J.F. A tonoplast-localized half-size ABC transporter is required for internal detoxification of Al in rice. Plant J. 2011, 69, 857–867. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Magalhaes, J.V.; Shaff, J.; Kochian, L.V. Aluminum-activated citrate and malate transporters from the MATE and ALMT families function independently to confer Arabidopsis aluminum tolerance. Plant J. 2009, 57, 389–399. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.F.; Ryan, P.R.; Delhaize, E. Aluminium tolerance in plants and the complexing role of organic acids. Trends Plant Sci. 2001, 6, 273–278. [Google Scholar] [CrossRef]
- Lou, H.Q.; Fan, W.; Xu, J.M.; Gong, Y.L.; Jin, J.F.; Chen, W.W.; Liu, L.Y.; Hai, M.R.; Yang, J.L.; Zheng, S.J. An oxalyl-CoA synthetase is involved in oxalate degradation and aluminum tolerance. Plant Physiol. 2016, 172, 1679–1690. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.W.; Fan, W.; Lou, H.Q.; Yang, J.L.; Zheng, S.J. Regulating cytoplasmic oxalate homeostasis by acyl activating enzyme 3 is critical for plant Al tolerance. Plant Signal. Behav. 2017, 12, e1276688. [Google Scholar] [CrossRef] [PubMed]
- Zhu, H.; Wang, H.; Zhu, Y.; Zou, J.; Zhao, F.J.; Huang, C.F. Genome-wide transcriptomic and phylogenetic analyses reveal distinct aluminum-tolerance mechanisms in the aluminum-accumulating species buckwheat (Fagopyrum tatarucum). BMC Plant Biol. 2015, 15, 16. [Google Scholar] [CrossRef] [PubMed]
- Dong, J.; Piñeros, M.A.; Li, X.; Yang, H.; Liu, Y.; Muphy, A.S.; Kochian, L.V.; Liu, D. An Arabidopsis ABC transporter mediates phosphate deficiency-induced remodeling of root architecture by modulating iron homeostasis in roots. Mol. Plant 2017, 10, 244–259. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.F.; Yamaji, N.; Ma, J.F. Knockout of a bacterial-type ATP-binding cassette transporter gene, AtSTAR1, results in increased aluminum sensitivity in Arabidopsis. Plant Physiol. 2010, 153, 1669–1677. [Google Scholar] [CrossRef] [PubMed]
- Klug, B.; Horst, W.J. Oxalate exudation into the root-tip water free space confers protection from aluminum toxicity and allows aluminum accumulation in the symplast in buckwheat (Fagopyrum esculentum). New Phytol. 2010, 187, 380–391. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.F.; Hiradate, S.; Matsumoto, H. High aluminum resistance in buckwheat II. Oxalic acid detoxifies aluminum internally. Plant Physiol 1998, 117, 753–759. [Google Scholar] [CrossRef]
- Shen, R.; Ma, J.F.; Kyo, M.; Iwashita, T. Compartmentation of aluminium in leaves of an Al-accumulator, Fagopyrum esculentum Moench. Planta 2002, 215, 394–398. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.F.; Hiradate, S. Form of aluminium for uptake and translocation in buckwheat (Fagopyrum esculentum Moench). Planta 2000, 211, 355–360. [Google Scholar] [CrossRef] [PubMed]
- Negishi, T.; Oshima, K.; Hattori, M.; Yoshida, K. Plasma membrane-localized Al-transporter from blue hydrangea sepals is a member of the anion permease family. Genes Cells 2013, 18, 341–352. [Google Scholar] [CrossRef] [PubMed]
- Lei, G.J.; Yokosho, K.; Yamaji, N.; Fujii-Kashino, M.; Ma, J.F. Functional characterization of two half-size ABC transporter genes in aluminum-accumulating buckwheat. New Phytol. 2017, 215, 1080–1089. [Google Scholar] [CrossRef] [PubMed]
- Yokosho, K.; Yamaji, N.; Mitani-Ueno, N.; Shen, R.F.; Ma, J.F. An aluminum-inducible IREG gene is required for internal detoxification of aluminum in buckwheat. Plant Cell Physiol. 2016, 57, 976–985. [Google Scholar] [CrossRef] [PubMed]
- Negishi, T.; Oshima, K.; Hattori, M.; Kana, M.; Mano, S.; Nishimura, M.; Yoshida, K. Tonoplast- and plasma membrane-localized aquaporin-family transporters in blue hydrangea sepals of aluminum hyperaccumulating plant. PLoS ONE 2012, 7, e43189. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Gingrich, D.K.; Deng, Y.; Hong, Z. A nucleostemin-like GTPase required for normal apical and floral meristem development in Arabidopsis. Mol. Biol. Cell. 2012, 23, 1446–1456. [Google Scholar] [CrossRef] [PubMed]
- Schuster, C.; Gaillochet, C.; Lohmann, J. U. Arabidopsis HECATE genes function in phytohormone control during gynoecium development. Development 2015, 142, 3343–3350. [Google Scholar] [CrossRef] [PubMed]
- Sawa, M.; Nusinow, D.A.; Kay, S.A.; Imaizumi, T. FKF1 and GIGANTEA complex formation is required for day-length measurement in Arabidopsis. Science 2007, 318, 261–264. [Google Scholar] [CrossRef] [PubMed]
- Corrales, A.R.; Carrillo, L.; Lasierra, P.; Nebauer, S.G.; Dominguez-Figueroa, J.; Renau-Morata, B.; Pollmann, S.; Granell, A.; Molina, R.V.; Vicente-Carbajosa, J.; et al. Multifaceted role of cycling DOF factors 3 (CDF3) in the regulation of flowering time and abiotic stress responses in Arabidopsis. Plant Cell Environ. 2017, 40, 748–764. [Google Scholar] [CrossRef] [PubMed]
- Kania, T.; Russenberger, D.; Peng, S.; Apel, K.; Melzer, S. FPF1 promotes flowering in Arabidopsis. Plant Cell 1997, 9, 1327–1338. [Google Scholar] [CrossRef] [PubMed]
- Doyle, M.R.; Davis, S.J.; Bastow, R.M.; McWatters, H.G.; Kozma-Bognár, L.; Nagy, F.; Millar, A.J.; Amasino, R.M. The ELF4 gene controls circadian rhythms and flowering time in Arabidopsis thaliana. Nature 2002, 419, 74–77. [Google Scholar] [CrossRef] [PubMed]
- Sanchez-Bermejo, E.; Balasubramanian, S. Natural variation involving deletion alleles of FRIGIDA modulate temperature-sensitive flowering responses in Arabidopsis thaliana. Plant Cell Environ. 2016, 39, 1353–1365. [Google Scholar] [CrossRef] [PubMed]
- Castro Marín, I.; Loef, I.; Bartetzko, L.; Searle, I.; Coupland, G.; Stitt, M.; Osuna, D. Nitrate regulates floral induction in Arabidopsis, acting independently of light, gibberellin and autonomous pathways. Planta 2010, 233, 539–552. [Google Scholar] [CrossRef] [PubMed]
- Liu, T.; Li, Y.; Ren, J.; Qian, Y.; Yang, X.; Duan, W.; Hou, X. Nitrate or NaCl regulates floral induction in Arabidopsis thaliana. Biologia 2013, 68, 215–222. [Google Scholar] [CrossRef]
- Franks, S.J.; Sim, S.; Weis, A.E. Rapid evolution of flowering time by an annual plant in response to a climate fluctuation. Proc. Natl. Acad. Sci. USA 2007, 104, 1278–1282. [Google Scholar] [CrossRef] [PubMed]
- Franks, S.J. Plasticity and evolution in drought avoidance and escape in the annual plant Brassica rapa. New Phytol. 2011, 190, 249–257. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.M.; To, T.K.; Ishida, J.; Morosawa, T.; Kawashima, M.; Matsui, A.; Toyoda, T.; Kimura, H.; Shinozaki, K.; Seki, M. Alterations of lysine modifications on the histone H3 N-tail under drought stress conditions in Arabidopsis thaliana. Plant Cell Physiol. 2008, 49, 1580–1588. [Google Scholar] [CrossRef] [PubMed]
- Kim, J. M.; To, T.K.; Ishida, J.; Matsui, A.; Kimura, H.; Seki, M. Transition of chromatin status during the process of recovery from drought stress in Arabidopsis thaliana. Plant Cell Physiol. 2012, 53, 847–856. [Google Scholar] [CrossRef] [PubMed]
- Zong, W.; Zhong, X.; You, J.; Xiong, L. Genome-wide profiling of histone H3K4-tri-methylation and gene expression in rice under drought stress. Plant Mol. Biol. 2013, 81, 175–188. [Google Scholar] [CrossRef] [PubMed]
- Van Dijk, K.; Ding, Y.; Malkaram, S.; Riethoven, J.J.; Liu, R.; Yang, J.; Laczko, P.; Chen, H.; Xia, Y.; Ladunga, I.; et al. Dynamic changes in genome-wide histone H3 lysine 4 methylation patterns in response to dehydration stress in Arabidopsis thaliana. BMC Plant Biol. 2010, 10, 238. [Google Scholar] [CrossRef] [PubMed]
| Assembly Summary | −Al | +Al |
|---|---|---|
| Total number of reads | 89,531,252 | 59,073,878 |
| Total base pairs (bp) | 7,162,500,160 | 4,725,910,240 |
| Average read length (bp) | 80 | 80 |
| Total number of transcripts | 50,782 | |
| Mean length of transcripts | 716 | |
| Total number of unigenes | 33,931 | |
| Mean length of unigenes | 677 | |
| Sequence with E-value | 26,300 | |
| Gene ID | Fold Change | TAIR ID | Description | Category 1 | Biological Processes 2 |
|---|---|---|---|---|---|
| Upregulated | |||||
| comp24276_c0_seq2 | 1.391 | AT1G46768 | Related to AP2 1 | AP2-EREBP | Ethylene-activated signaling pathway, response to cold, response to water deprivation |
| comp28160_c0_seq1 | 1.029 | AT4G36900 | Related to AP2 10 | AP2-EREBP | Ethylene-activated signaling pathway |
| comp27009_c0_seq1 | 1.045 | ||||
| comp4672_c0_seq1 | 1.329 | AT1G76110 | HMG (high mobility group) box protein with ARID/BRIGHT DNA-binding domain | ARID | Karyogamy, polar nucleus fusion |
| comp13872_c0_seq1 | 1.257 | ||||
| comp28434_c0_seq1 | 1.029 | AT5G49700 | AT-HOOK motif nuclear localized protein 17 | AT-hook | Unknown |
| comp20160_c0_seq1 | 1.651 | AT5G43700 | IAA4 | AUX/IAA | Response to auxin |
| comp32831_c0_seq1 | 1.326 | ||||
| comp27076_c0_seq2 | 1.084 | AT1G09530 | Phytochrome interacting factor 3 | bHLH | De-etiolation, gibberellic acid-mediated signaling pathway, positive regulation of anthocyanin metabolic process, red or far-red light signaling pathway |
| comp17528_c0_seq2 | 1.750 | AT3G19860 | BHLH121 | bHLH | Unknown |
| comp2972_c0_seq1 | 1.898 | ||||
| comp28503_c0_seq1 | 1.511 | AT5G61270 | Basic helix-loop-helix (bHLH) phytochrome interacting factor | bHLH | De-etiolation, red, far-red light phototransduction |
| comp16875_c0_seq1 | 1.294 | AT5G67060 | Encodes a bHLH transcription factor | bHLH | Carpel formation, gynoecium development, regulation of auxin polar transport, transmitting tissue development |
| comp24549_c0_seq1 | 1.005 | AT3G17609 | HY5-homolog | bZIP | Red, far-red light phototransduction, response to UV-B, response to karrikin |
| comp21671_c0_seq1 | 1.015 | ||||
| comp31175_c0_seq1 | 1.074 | AT5G28770 | Basic leucine zipper 63 | bZIP | Response to abscisic acid, response to glucose |
| comp30394_c0_seq1 | 1.113 | ||||
| comp18077_c0_seq1 | 1.106 | AT1G68520 | B-Box domain protein 14 | C2C2-CO-LIKE | Unknown |
| comp31981_c0_seq1 | 1.432 | AT5G24930 | Constans-like 4 | C2C2-CO-LIKE | Regulation of flower development |
| comp31991_c0_seq2 | 1.037 | AT5G57660 | Constans-like 5 | C2C2-CO-LIKE | Regulation of flower development |
| comp31991_c0_seq1 | 1.291 | ||||
| comp5736_c0_seq1 | 1.243 | AT3G21270 | DOF zinc finger protein 2 | C2C2-Dof | Unknown |
| comp13725_c0_seq1 | 1.216 | ||||
| comp60617_c0_seq1 | 1.144 | AT3G47500 | Cycling DOF factor 3 | C2C2-Dof | Flowering |
| comp27353_c0_seq1 | 1.010 | AT2G05160 | CCCH-type zinc finger family protein with RNA-binding domain | C3H | Unknown |
| comp30562_c0_seq1 | 1.351 | AT2G20570 | Golden 2-like 1 | G2-like | Chloroplast organization, negative regulation of flower development, negative regulation of leaf senescence, regulation of chlorophyll biosynthetic process |
| comp32323_c0_seq1 | 1.064 | AT3G04450 | Homeodomain-like superfamily protein | G2-like | Unknown |
| comp25041_c0_seq1 | 1.177 | AT2G46680 | Homeobox 7 | HB | Response to abscisic acid, response to water deprivation |
| comp28163_c0_seq1 | 1.160 | ||||
| comp32393_c0_seq2 | 1.239 | AT5G04760 | Duplicated homeodomain-like superfamily protein | MYB | Unknown |
| comp110794_c0_seq1 | 1.074 | AT2G46830 | Circadian clock-associated 1 | MYB-related (flower) | Flowering |
| comp80633_c0_seq1 | 1.173 | ||||
| comp30095_c0_seq1 | 1.245 | AT1G01720 | ATAF1 | NAC | Negative regulation of abscisic acid-activated signaling pathway |
| comp47425_c0_seq1 | 2.096 | AT3G15500 | ANAC055 | NAC | Jasmonic acid-mediated signaling pathway, response to water deprivation |
| comp26124_c0_seq1 | 2.605 | AT3G15510 | ANAC056 | NAC | Jasmonic acid-mediated signaling pathway, response to water deprivation |
| comp27368_c0_seq1 | 1.295 | AT1G32700 | PLATZ transcription factor family protein | PLATZ | Unknown |
| comp28592_c0_seq2 | 1.021 | AT4G17900 | PLATZ transcription factor family protein | PLATZ | Unknown |
| comp83456_c0_seq1 | 1.048 | AT4G02020 | SET domain-containing protein 10 | SET | Endosperm development |
| comp94475_c0_seq1 | 1.981 | AT5G13080 | WRKY75 | WRKY | Atrichoblast differentiation, lateral root development, induced by Pi starvation |
| comp87094_c0_seq1 | 2.190 | ||||
| Downregulated | |||||
| comp23084_c0_seq1 | −1.004 | AT4G34590 | Basic leucine-zipper 11 | bZIP | Response to sucrose |
| comp7947_c0_seq1 | −1.088 | AT1G75710 | C2H2-like zinc finger protein | C2H2 | Unknown |
| comp18294_c0_seq1 | −1.002 | AT5G28640 | GRF1-interacting factor 1 | GIF | Adaxial/abaxial pattern specification, leaf development |
| comp30187_c0_seq2 | −1.260 | AT4G24540 | Agamous-like 24 | MADS | Flowering |
| comp30099_c0_seq1 | −1.224 | ||||
| comp30187_c0_seq1 | −1.219 | ||||
| comp17187_c0_seq1 | −1.042 | AT1G73230 | Nascent polypeptide-associated complex NAC | TF_GTF | Response to salt stress |
| comp27260_c0_seq1 | −1.051 | ||||
| comp27190_c0_seq1 | −1.004 | ||||
| Gene ID | Fold Change | TAIR ID | Description | Transport Substrate 1 |
|---|---|---|---|---|
| Upregulated | ||||
| comp31346_c0_seq4 | 1.044 | AT1G02260 | Divalent ion symporter | Al |
| comp29520_c0_seq1 | 1.250 | AT5G39040 | Aluminum-sensitive 1 | Al |
| comp3719_c0_seq1 | 1.065 | |||
| comp32696_c0_seq2 | 1.060 | |||
| comp18915_c0_seq1 | 1.333 | AT2G41190 | Transmembrane amino acid transporter family protein | Amino acid |
| comp14037_c0_seq1 | 1.054 | |||
| comp18529_c0_seq1 | 1.266 | AT4G01100 | Adenine nucleotide transporter 1 | AMP; ADP; ATP |
| comp3758_c0_seq1 | 1.016 | AT5G17400 | Endoplasmic reticulum-adenine nucleotide transporter 1 | AMP; ADP; ATP |
| comp29175_c0_seq1 | 1.263 | AT5G15410 | Cyclic nucleotide gated channel 2 | Ca2+ |
| comp29835_c0_seq1 | 1.124 | |||
| comp15571_c0_seq1 | 1.857 | AT2G23790 | Protein of unknown function (DUF607) | Ca2+ |
| comp9425_c0_seq1 | 1.860 | |||
| comp68193_c0_seq1 | 1.851 | |||
| comp31329_c0_seq1 | 1.403 | AT5G61520 | Major facilitator superfamily protein | Carbohydrate |
| comp27968_c0_seq1 | 1.499 | |||
| comp29178_c0_seq1 | 1.403 | AT5G18840 | Major facilitator superfamily protein | Carbohydrate |
| comp7291_c0_seq1 | 1.167 | |||
| comp23114_c0_seq1 | 1.510 | |||
| comp98352_c0_seq1 | 1.678 | AT1G51340 | MATE efflux family protein | Citrate |
| comp11267_c0_seq1 | 2.039 | |||
| comp96590_c0_seq1 | 1.764 | |||
| comp88250_c0_seq1 | 1.705 | |||
| comp81877_c0_seq1 | 1.967 | AT3G21090 | ABCG15 | Cutin |
| comp56152_c0_seq1 | 2.033 | |||
| comp96281_c0_seq1 | 2.336 | |||
| comp22596_c0_seq2 | 1.350 | AT5G16150 | Putative plastidic glucose transporter | Glucose |
| comp22596_c0_seq1 | 1.314 | |||
| comp25276_c0_seq1 | 1.121 | |||
| comp49196_c0_seq1 | 2.598 | AT3G47960 | NRT1/ PTR family 2.10 | Glucosinolate |
| comp26699_c0_seq1 | 2.651 | |||
| comp28002_c0_seq1 | 2.654 | |||
| comp91186_c0_seq1 | 3.385 | |||
| comp24147_c0_seq1 | 1.022 | AT4G24120 | Yellow stripe-like 1 | Iron-nicotianamine; oligopeptide |
| comp106142_c0_seq1 | 1.425 | AT1G31120 | K+ uptake permease 10 | K+ |
| comp30691_c0_seq1 | 1.012 | AT2G35060 | K+ uptake permease 11 | K+ |
| comp12992_c0_seq1 | 2.012 | AT3G02850 | Stellar K+ outward rectifier | K+ |
| comp16659_c0_seq1 | 1.954 | AT5G37500 | Gated outwardly-rectifying K+ channel | K+ |
| comp84025_c0_seq1 | 2.025 | K+ | ||
| comp3908_c0_seq1 | 1.328 | AT5G46240 | Potassium channel protein (KAT1) | K+ |
| comp69280_c0_seq1 | 1.332 | AT3G18830 | Polyol/monosaccharide transporter 5 | Linear polyols; myo-inositol; monosaccharides |
| comp7769_c0_seq1 | 1.787 | AT4G21120 | Cationic amino acid transporter 1 | Lys, Arg and Glu |
| comp10794_c0_seq1 | 1.790 | |||
| comp13115_c0_seq1 | 1.429 | |||
| comp8282_c0_seq1 | 1.398 | |||
| comp25372_c0_seq1 | 1.911 | |||
| comp32005_c0_seq2 | 1.502 | AT1G25480 | Aluminum-activated malate transporter family protein | Malate |
| comp33002_c0_seq1 | 1.168 | AT1G77210 | Sugar transport protein 14 | Monosaccharide |
| comp5166_c0_seq1 | 1.145 | AT3G54140 | Peptide transporter 1 | Peptide |
| comp15597_c0_seq1 | 1.121 | AT2G38940 | Phosphate transporter 1;4 | Pi |
| comp14809_c0_seq1 | 1.128 | |||
| comp32347_c0_seq1 | 1.111 | AT3G26570 | Phosphate transporter 2;1 | Pi |
| comp22508_c0_seq1 | 1.293 | AT5G43350 | Phosphate transporter 1 | Pi |
| comp26614_c0_seq1 | 1.078 | AT5G54800 | Glucose 6-phosphate/phosphate translocator 1 | Pi; PEP, triose phosphate; glucose 6-phosphate |
| comp30882_c0_seq12 | 1.154 | AT4G18210 | Purine permease 10 | Purine; purine derivatives |
| comp30860_c0_seq2 | 1.241 | |||
| comp48026_c0_seq1 | 1.237 | AT1G71880 | Sucrose-proton symporter 1 | Sucrose |
| comp18524_c0_seq1 | 1.234 | AT5G19600 | Sulfate transporter 3;5 | Sulfate |
| comp25104_c0_seq5 | 1.224 | AT3G23560 | Aberrant lateral root formation 5 | Toxins |
| comp25104_c0_seq4 | 1.197 | |||
| comp25244_c0_seq5 | 1.140 | |||
| comp25244_c0_seq2 | 1.123 | |||
| comp26932_c0_seq1 | 2.705 | AT4G30420 | Usually multiple acids move in and out transporter 34 | Unknown |
| comp69634_c0_seq1 | 1.345 | AT4G36670 | Polyol/monosaccharide transporter 6 | Glucose; hexose |
| comp29397_c0_seq1 | 1.032 | AT5G64410 | Oligopeptide transporter 4 | Oligopeptide |
| comp25749_c0_seq1 | 3.018 | AT1G67940 | AtSTAR1 | UDP-glucose |
| comp26709_c0_seq1 | 2.987 | |||
| comp30641_c0_seq1 | 2.916 | AT2G37330 | Aluminum sensitive 3 | UDP-glucose |
| comp30389_c0_seq1 | 2.852 | |||
| comp8633_c0_seq1 | 1.022 | AT5G15640 | Mitochondrial substrate carrier family protein | Unknown |
| comp81941_c0_seq1 | 1.012 | |||
| comp21728_c0_seq1 | 1.064 | AT1G74780 | Nodulin-like/major facilitator superfamily protein | Unknown |
| comp17931_c0_seq1 | 1.231 | AT2G39210 | Major facilitator superfamily protein | Unknown |
| comp5953_c0_seq1 | 1.419 | |||
| comp12616_c0_seq1 | 1.249 | |||
| comp29405_c0_seq1 | 1.204 | AT4G23400 | PIP1;5 | Water |
| comp50926_c0_seq1 | 1.895 | AT1G51500 | ABCG12 | Wax |
| comp6165_c0_seq1 | 1.950 | |||
| comp92864_c0_seq1 | 1.903 | |||
| Downregulated | ||||
| comp29871_c0_seq3 | −1.038 | AT4G28390 | ADP/ATP carrier 3 | ADP; ATP |
| comp30078_c0_seq2 | −1.144 | |||
| comp30078_c0_seq1 | −1.137 | |||
| comp29871_c0_seq1 | −1.003 | |||
| comp31976_c0_seq1 | −1.058 | AT2G36910 | ABCB1/P-glycoprotein 1 | Auxin |
| comp22423_c0_seq1 | −1.066 | AT1G75500 | Usually multiple acids move in and out transporter 5 | Auxin |
| comp28292_c0_seq1 | −1.120 | AT1G53210 | Na+/Ca2+ exchanger | Ca2+ |
| comp33125_c0_seq2 | −2.085 | AT3G51860 | Cation exchanger 3 | Ca2+ and H+ |
| comp23194_c0_seq1 | −1.036 | AT4G32390 | Nucleotide–sugar transporter family protein | Carbohydrate |
| comp25558_c0_seq1 | −1.269 | AT5G19760 | Encodes a novel mitochondrial carrier | Dicarboxylates; tricarboxylates |
| comp27413_c0_seq1 | −1.285 | AT4G27720 | Major facilitator superfamily protein | Mo6+ |
| comp23196_c0_seq1 | −1.057 | AT1G64650 | Major facilitator superfamily protein | Mo6+ |
| comp27697_c0_seq1 | −1.264 | AT2G26690 | NRT1/PTR family 6.2 | Oligopeptide |
| comp32440_c0_seq1 | −1.426 | AT5G33320 | Phosphate/phosphoenolpyruvate translocator | PEP |
| comp20485_c0_seq1 | −1.269 | AT5G14040 | Mitochondrial phosphate transporter 3 | Pi |
| comp25757_c0_seq1 | −1.264 | AT3G01550 | Phosphoenolpyruvate/phosphate translocator 2 | Pi and PEP |
| comp17525_c0_seq1 | −1.012 | AT5G62890 | Xanthine/uracil permease family protein | Unknown |
| comp23073_c0_seq1 | −2.726 | AT4G35100 | PIP2;7 | Water |
| comp27116_c0_seq1 | −1.065 | AT3G16240 | TIP2;1 | Water |
| comp24264_c0_seq1 | −1.383 | AT2G36830 | TIP1;1 | Water |
| comp28684_c0_seq1 | −1.186 | AT3G04090 | SIP 1A | Water |
| comp29502_c0_seq1 | −1.146 | |||
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, W.W.; Xu, J.M.; Jin, J.F.; Lou, H.Q.; Fan, W.; Yang, J.L. Genome-Wide Transcriptome Analysis Reveals Conserved and Distinct Molecular Mechanisms of Al Resistance in Buckwheat (Fagopyrum esculentum Moench) Leaves. Int. J. Mol. Sci. 2017, 18, 1859. https://doi.org/10.3390/ijms18091859
Chen WW, Xu JM, Jin JF, Lou HQ, Fan W, Yang JL. Genome-Wide Transcriptome Analysis Reveals Conserved and Distinct Molecular Mechanisms of Al Resistance in Buckwheat (Fagopyrum esculentum Moench) Leaves. International Journal of Molecular Sciences. 2017; 18(9):1859. https://doi.org/10.3390/ijms18091859
Chicago/Turabian StyleChen, Wei Wei, Jia Meng Xu, Jian Feng Jin, He Qiang Lou, Wei Fan, and Jian Li Yang. 2017. "Genome-Wide Transcriptome Analysis Reveals Conserved and Distinct Molecular Mechanisms of Al Resistance in Buckwheat (Fagopyrum esculentum Moench) Leaves" International Journal of Molecular Sciences 18, no. 9: 1859. https://doi.org/10.3390/ijms18091859
APA StyleChen, W. W., Xu, J. M., Jin, J. F., Lou, H. Q., Fan, W., & Yang, J. L. (2017). Genome-Wide Transcriptome Analysis Reveals Conserved and Distinct Molecular Mechanisms of Al Resistance in Buckwheat (Fagopyrum esculentum Moench) Leaves. International Journal of Molecular Sciences, 18(9), 1859. https://doi.org/10.3390/ijms18091859